Abstract
NADH:quinone oxidoreductase (complex I) plays a pivotal role in cellular energy production. It employs a series of redox cofactors to couple electron transfer to the generation of a proton-motive force across the inner mitochondrial or bacterial cytoplasmic membrane. Complex I contains a noncovalently bound flavin mononucleotide at the active site for NADH oxidation and eight or nine iron–sulfur clusters to transfer electrons between the flavin and a quinone-binding site. Understanding the mechanism of complex I requires the properties of these clusters to be defined, both individually and as an ensemble. Most functional information on the clusters has been gained from EPR spectroscopy, but some clusters are not observed by EPR and attributing the observed signals to the structurally defined clusters is difficult. The current consensus picture relies on correlating the spectra from overexpressed subunits (containing one to four clusters) with those from intact complexes I. Here, we analyze spectra from the overexpressed NuoG subunit from Escherichia coli complex I and compare them with spectra from the intact enzyme. Consequently, we propose that EPR signals N4 and N5 have been misassigned: signal N4 is from NuoI (not NuoG) and signal N5 is from the conserved cysteine-ligated [4Fe-4S] cluster in NuoG (not from the cluster with a histidine ligand). The consequences of reassigning the EPR signals and their associated functional information on the free energy profile for electron transfer through complex I are discussed.
Keywords: complex I, mitochondria, electron transfer
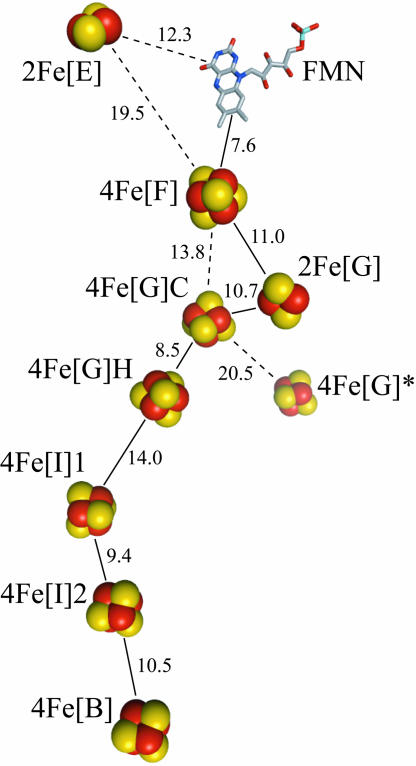
NADH:quinone oxidoreductase (complex I) is a complicated, membrane-bound, proton-pumping redox enzyme, through which electrons enter the respiratory chain (1–4). Recently, structural analysis of the hydrophilic domain of complex I from Thermus thermophilus showed that it contains nine iron–sulfur (FeS) clusters, seven of which transfer electrons between the spatially separated sites of NADH oxidation and quinone reduction (5, 6). Fig. 1 depicts the arrangement of the cofactors in complex I and defines the nomenclature used here. Sequence analysis has confirmed that eight FeS clusters are conserved in all known complexes I, including those from mammalian mitochondria, but 4Fe[G]*, which is not part of the link between the two active sites, is found in only a few species including T. thermophilus and Escherichia coli (1, 3, 7). 2Fe[E] is located on the opposite side of the flavin mononucleotide (FMN) from the cluster chain, and whether it has any functional role is not yet known.
Fig. 1.
Arrangement of the FMN and the FeS clusters in T. thermophilus complex I (5, 6). The clusters are named according to their nuclearity (2Fe or 4Fe), their subunit location (using the nomenclature for E. coli complex I, Table 1), and, when necessary, as ligated by four Cys (C) or three Cys and one His (H). The 4Fe[G]* cluster is not conserved in all species. Important edge-to-edge distances are indicated (Å), and the most likely electron transfer pathway is denoted by solid lines.
Before the structure became available, knowledge about the FeS clusters in complex I was derived from sequence homology and FeS-binding motifs (1, 3), overexpressing and studying cofactor-containing subunits (8–12), and EPR spectroscopy (7). However, not all of the clusters have been detected by EPR. Consequently, the reduction potentials and properties of these clusters are unknown, and studies that rely on the properties of the cluster ensemble are compromised. Assigning the observed EPR signals to the structurally defined clusters presents further challenges.
Most EPR studies have focused on complex I from bovine mitochondria: five reduced FeS clusters are typically observed, and their signals are referred to as N1b, N2, N3, N4, and N5 (7). Studies of E. coli complex I reveal N1a also, but N5 is not observed (13). Table 1 presents the current consensus assignment of the EPR signals to the structurally defined FeS clusters. Signals N3 and N2 are attributed to 4Fe[F] and 4Fe[B], respectively, because interactions with the flavin semiquinone and ubisemiquinones have been observed (14, 15). Signal N1a is from 2Fe[E], because it is exhibited by the overexpressed subunits from several species and by bovine subcomplex Fp (the 24- and 51-kDa subunits) (16, 17). Consequently, signal N1b, from a different [2Fe-2S] cluster, is from 2Fe[G], consistent with results from overexpressed NuoG homologues (9, 12, 18, 19). According to current consensus, signals N4, N5, and N7 are from the three [4Fe-4S] clusters in NuoG, whereas the two clusters in NuoI have not been observed by EPR spectroscopy on any intact enzyme. They are commonly referred to as “N6a and N6b” on the basis of an EPR signal from the “connecting fragment” of E. coli complex I (20).
Table 1.
The iron–sulfur clusters in complex I and the assignment of their EPR signals
| Cluster | Subunit E. coli | Subunit Bos taurus | EPR signal (consensus) | EPR signal (revised) |
|---|---|---|---|---|
| 2Fe[E] | NuoE | 24 kDa | N1a | N1a |
| 4Fe[F] | NuoF | 51 kDa | N3 | N3† |
| 2Fe[G] | NuoG | 75 kDa | N1b | N1b |
| 4Fe[G]C | NuoG | 75 kDa | N4 | N5 |
| 4Fe[G]H | NuoG | 75 kDa | N5 | — |
| 4Fe[G]* | NuoG | — | N7 | N7 |
| 4Fe[I]1 | NuoI | TYKY | N6a or N6b | } N4 |
| 4Fe[I]2 | NuoI | TYKY | N6a or N6b | |
| 4Fe[B] | NuoB | PSST | N2 | N2† |
—, not present or observed.
†No data pertaining to the assignment of signals N2 or N3 are presented here.
Here, we compare EPR spectra from the overexpressed E. coli NuoG subunit (EcNuoG) and intact E. coli complex I, and we reexamine the results of previous studies. Consequently, we propose an alternative assignment for signals N4 and N5 (and thus of all data pertaining to them). We assign signal N4 to the NuoI subunit and assign signal N5 to the 4Fe[G]C cluster. Our conclusions have direct implications for understanding electron transfer in complex I, for the correct interpretation of extant data, and for future investigations of the catalytic mechanism.
Results and Discussion
Overexpression of the E. coli NuoG Subunit (EcNuoG).
To optimize the preparation of a cofactor-containing NuoG subunit from complex I, we overexpressed a number of different constructs in E. coli, applying the general strategy described in refs. 9 and 19. The full-length homologues from E. coli and Paracoccus denitrificans [plasmid from T. Yano (University of Pennsylvania, Philadelphia, PA); ref. 9] both gave promising results, but truncated forms containing only the FeS domain (P. denitrificans residues 1–219 and 1–237, or T. thermophilus 1–371, designed using sequence analysis; ref. 18) were much less successful. For each construct, we varied the coexpression system (see Materials and Methods) and the growth conditions (temperature, aeration, induction point and concentration, supplementation with iron salts or cysteine) to maximize soluble protein expression and iron incorporation. Addition of a six-histidine tag to the N terminus of the E. coli homologue facilitated its rapid purification and was not detrimental to holoprotein expression or stability. In the structure of the hydrophilic domain of T. thermophilus complex I, the N terminus of the NuoG homologue (Nqo3) is solvent-exposed and free of interactions with other subunits (6). Subsequently, the His-tagged E. coli homologue (denoted EcNuoG) was chosen for further study because it expressed to comparatively high levels as a holoprotein, because it could be isolated rapidly under anaerobic conditions, and, importantly, because intact complex I from E. coli was available for direct comparison. EPR samples were prepared immediately upon elution of EcNuoG from the affinity column. Although SDS/PAGE analysis revealed several impurities, more stringent purification resulted only in uniform changes in spectrum intensity. The spectra presented are from a preparation that is 40–45% EcNuoG protein, with a molar iron:protein ratio of ≈6:1. Oxidized and dithionite-reduced UV-visible spectra were consistent with the presence of [2Fe-2S] and [4Fe-4S] clusters, with characteristic features at ≈320, 420, 460, and 550 nm in the oxidized state (21).
EPR spectra of EcNuoG.
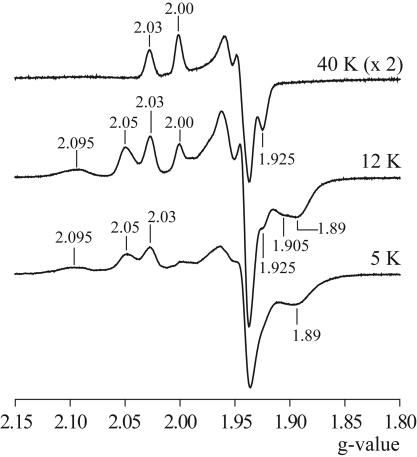
Fig. 2 shows EPR spectra from EcNuoG reduced by dithionite at low microwave power (0.1 mW) to minimize saturation distortion. At 40 K, an axial [2Fe-2S] cluster spectrum is evident, with g‖⊥ = 2.030, 1.939. At 12 and 5 K, two rhombic [4Fe-4S] spectra are apparent also, with gzyx = 2.048, ≈1.94, 1.916 and gzyx = 2.074, ≈1.95, 1.885. Fig. 2B shows that the spectrum at 12 K is reproduced well by simulation using these parameters. Double integration procedures aimed at spin quantitation suggest that each cluster is present at approximately the same concentration. However, ratios were subject to up to 50% variation, resulting from different modifications of the [2Fe-2S] spectral line shape, background correction, and uncertainty in attributing the gy region.
Fig. 2.
EPR spectra of EcNuoG. EcNuoG (≈20 mg·ml−1) was reduced anaerobically with 1 mM dithionite and frozen immediately. (A) The spectrum at 40 K shows predominantly the spectrum of one [2Fe-2S] cluster; the spectra at 12 and 5 K show the spectra of two [4Fe-4S] clusters also. g values for the major features are marked. Conditions are as follows: microwave power, 0.1 mW; conversion time, 81.92 ms; time constant, 20.48 ms; modulation amplitude, 10 G; microwave frequency, ≈9.38 MHz. (B) A model of the 12-K spectrum using the following parameters: gzyx = 2.074, 1.950, 1.887, Lzyx = 40, 25, 40 G (Gaussian); gzyx = 2.048, 1.942, 1.914, Lzyx = 13, 15, 25 G (Gaussian); gzyx = 2.030, 1.939, 1.937, Lzyx = 15, 20, 20 G (Lorentzian). The gz and gx pairs from the two [4Fe-4S] clusters are indicated.
The EPR spectra reported here agree closely with those described previously from overexpressed NuoG homologues from P. denitrificans, T. thermophilus, and E. coli (denoted PdNqo3, TtNqo3-maltose binding protein (MBP), and EcNuoG-MBP, respectively). In 1995, PdNqo3 was found to contain one [2Fe-2S] (g‖⊥ = 2.026, 1.934) and one [4Fe-4S] cluster (gzyx = 2.063, 1.928, 1.892), matching closely to our results (P. denitrificans does not contain 4Fe[G]*) (9). Subsequently, the FeS content of PdNqo3 was improved, and it was suggested that the [4Fe-4S] signal actually comprises two overlapping signals with very similar g values but with different relaxation properties (19) (see below). In addition, a putative g ≈ 5 signal was proposed to originate from a higher spin state cluster; no such signal could be detected here. In 2002, EPR spectra from a MBP-fused construct of TtNqo3 (TtNqo3-MBP) were reported and were consistent with one [2Fe-2S] cluster (g‖⊥ = 2.028, 1.940) and two [4Fe-4S] clusters (features at g = 2.072, 2.046, 1.940, and 1.87) (18). However, it was necessary to reconstitute the clusters chemically, and the individual [4Fe-4S] signals could not be distinguished. In 2005, the E. coli homologue was expressed as an MBP fusion (EcNuoG-MBP), and mutations were created in each FeS binding motif. The clusters required chemical reconstitution, and no [2Fe-2S] signals were observed, even in the wild-type protein. However, two [4Fe-4S] signals were described, with gzyx = 2.06, 1.94, 1.89 and gzyx = 2.05, 1.94, 1.91, very similar to those described here (12).
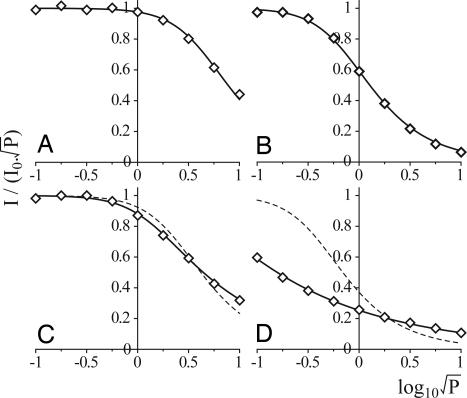
Eq. 1 was used to explore the dependence of signal intensity on microwave power (22), to further characterize the individual signals, and to help to distinguish signals that may overlap.
I is the spectrum intensity, P is the microwave power, P½ is the microwave power at half-saturation, b is 1 for an inhomogeneously broadened line, and I0 is the limiting normalized intensity (I/√P as P → 0). The signal intensity at g = 2.03 fits closely to Eq. 1 at each temperature: P½ = 20 mW at 40 K (Fig. 3A), 0.2 mW at 12 K, and ≈0.02 mW at 5 K (I0 was not attained at 5 K, so the fit was by extrapolation). The signal intensities at g = 2.048 and 1.916 (12 K only) also fit closely to Eq. 1: P½ = 0.54 mW at 12 K (Fig. 3B) and 0.008 mW at 5 K (I0 not attained). The fact that both g values produce the same P½ confirms that they originate from the same cluster. Similarly, the signal intensities at g = 2.074 and 1.885 display the same dependence on microwave power at both 12 and 5 K, but they do not fit well to Eq. 1 with b = 1 (Fig. 3 C and D). It is possible to fit these data by adding two contributions with different relaxation properties (as proposed in ref. 19) or by decreasing b below 1 (Fig. 3). We favor the latter, because our spectra display no further evidence at any temperature or microwave power to suggest that more than one signal contributes to these features. Although there is no physical justification for b being below 1, similar behavior has been observed previously in, for example, the tyrosine radical YD· in photosystem II (23) and the flavin radical in NO synthase (24). Low apparent b values are produced from powder spectra when a dipolar interaction with a neighboring paramagnetic center mediates spin-lattice relaxation: a distribution of relaxation rates is observed, because the orientation of the interspin vector varies with respect to the static magnetic field (23, 25, 26). As four clusters are ligated in close proximity by NuoG, dipolar interactions are a very plausible explanation for these data.
Fig. 3.
Dependence of EcNuoG signal intensity on microwave power. (A) g = 2.03 at 40 K (modeled with Eq. 1, P½ = 20 mW, b = 1). (B) g = 2.05 at 12 K (P½ = 0.54 mW, b = 1). (C) g = 2.074 at 12 K [P½ = 5.8 mW, b = 1 (dashed) or P½ = 1.7 mW, b = 0.55 (solid line)]. (D) g = 2.074 at 5 K [P½ = 0.16 mW, b = 1 (dashed) or P½ = 0.6 μW, b = 0.37 (solid line)].
Assignment of the EPR Spectra from EcNuoG.
The structure of the hydrophilic domain of T. thermophilus complex I (6) shows that EcNuoG coordinates four FeS clusters, but only three spectra are observed here. The current consensus (Table 1) is that the four clusters in NuoG (or its homologues) give rise to EPR spectra N1b, N4, N5 and N7 (4, 6, 7, 19). However, Table 2 presents the g values of N1b, N4, and N5 from a range of species to aid in assigning the spectra from EcNuoG.
Table 2.
g values of signals N1b, N4, and N5 reported in the literature
| EPR signal and species | gz | gy | gx | Refs. |
|---|---|---|---|---|
| N1b | ||||
| E. coli | 2.03 | 1.94 | 1.94 | 13 |
| P. denitrificans | 2.02–2.04 | 1.94 | 1.92–1.94 | 19, 27 |
| N. crassa | 2.02 | 1.93–1.94 | 1.93 | 20, 28 |
| Y. lipolytica | 2.02 | 1.95 | 1.93 | 29 |
| Solanum tuberosum | 2.02 | 1.94 | 1.94 | 30 |
| B. taurus | 2.02 | 1.94 | 1.92 | 7 |
| N4 | ||||
| E. coli | 2.09 | 1.93 | 1.89 | 13 |
| P. denitrificans | 2.09–2.10 | 1.94 | 1.88 | 19, 27 |
| N. crassa | 2.10 | 1.92 | 1.88 | 20, 28 |
| Y. lipolytica | 2.10–2.11 | 1.93 | 1.89 | 29, 31 |
| S. tuberosum | 2.11 | 1.93 | 1.88 | 30 |
| B. taurus | 2.10 | 1.93–1.94 | 1.88–1.89 | 7 |
| N5 | ||||
| Y. lipolytica | 2.06 | 1.93 | 1.89 | 29, 31 |
| B. taurus | 2.07 | 1.93 | 1.90 | 7 |
The 2Fe[G] cluster produces signal N1b (g‖⊥ = 2.030, 1.939).
The observed g values of the [2Fe-2S] cluster match well to all values reported for signal N1b (Table 2) and to those of one of the two [2Fe-2S] clusters observed in E. coli complex I at 40 K (Figs. 4 and 5). Thus, (in accordance with current consensus) we attribute EPR signal N1b to 2Fe[G] (and, by elimination, signal N1a in complex I to 2Fe[E]).
Fig. 4.
EPR spectra of isolated E. coli complex I. Complex I was dialyzed anaerobically against 20 mM Tris·HCl pH 7.5, 0.1 mM NADH for 1 h at 0°C, then reduced further by the addition of 1 mM dithionite and frozen immediately. (Top) Spectrum at 40 K comprising two [2Fe-2S] clusters. (Middle) Spectrum at 12 K comprising, in addition, at least two [4Fe-4S] clusters. (Bottom) Spectrum at 5 K comprising at least two [4Fe-4S] clusters and one [2Fe-2S] cluster. Conditions were as follows: microwave power, 0.1 mW; conversion time, 81.92 ms; time constant, 20.48 ms; modulation amplitude 10 G; microwave frequency, ≈9.38 MHz.
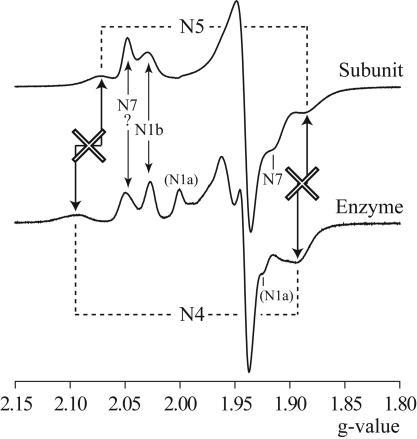
Fig. 5.
Comparison of EPR spectra from EcNuoG and E. coli complex I at 12 K. The two spectra are from Figs. 2 and 4. Arrows indicate correspondence between signals from the two spectra; crosses indicate that the signals do not match.
The 4Fe[G]* cluster produces signal N7 (gzyx = 2.048, ≈1.94, 1.916).
Signal N7 has not been observed in the EPR spectrum from any intact complex I. We attribute the spectrum with gzyx = 2.048, ≈1.94, 1.916 to 4Fe[G]* because the g values match closely to those from the overexpressed 4Fe[G]*-only domain of T. thermophilus Nqo3 (g‖⊥ = 2.045, ≈1.94) (18) but are clearly distinct from those of N4 and N5 (Table 2). Consequently, in complex I, gz (or g‖) from N7 would overlap with g‖ from N2 and gz from N3 (13): although both EcNuoG and E. coli complex I exhibit a signal at g = 2.05 (Fig. 5), the two signals do not necessarily have the same origin. If N7 is absent from the spectrum of complex I, it is most likely that 4Fe[G]* is oxidized because it has a low reduction potential, because electron transfer, even over 20.5 Å (Fig. 1), is fast on our experimental timescale (32).
Either the 4Fe[G]C cluster or the 4Fe[G]H cluster produces signal N5 (gzyx = 2.074, ≈1.95, 1.885); signal N4 is absent.
Comparison of the g values of the second [4Fe-4S] spectrum from EcNuoG with the values in Table 2 suggests strongly that it is N5, not N4. Fig. 5 compares directly the spectra of EcNuoG and E. coli complex I (in which N4 is clearly visible), displaying the obvious mismatch, particularly in gz. Consequently, we propose that the signal exhibited by EcNuoG is N5 and that N4 does not arise from NuoG.
Our interpretation clearly conflicts with those of previous studies of overexpressed NuoG homologues. Initially, the single [4Fe-4S] cluster detected in PdNqo3 (gzyx = 2.063, 1.928, 1.892) was attributed to complex I signal N4 (9), because the g values were more consistent with N4 than N2 or N3 (N5 was not then recognized as a constituent of the bacterial enzyme). EPR signals observed subsequently were essentially identical and, following the initial study, were matched to N4 also (12, 18, 19). Similarly, although N4 was identified, EPR spectra of the NADH dehydrogenase fragment of E. coli complex I (NuoE, F, and G) (33, 34) reveal no intensity in the region of the true N4 gz signal (2.09–2.11, Table 2). Further evidence to support the assignment of N4 to NuoG is scant and comprises only the results of mutating the cluster ligands. As described below, reevaluation of these results actually supports the proposal that only N5 (not N4) should be attributed to NuoG. First, however, our revised interpretation raises two further questions:
How accurately should the g values from an overexpressed subunit match those from intact complex I?
The g values of 2Fe[E] in the overexpressed E. coli subunit are identical to those from intact complex I [gzyx = 2.00, 1.95, 1.92 (13, 16)]. Cluster assembly of 4Fe[F] has been achieved only in NuoEF subcomplexes to low levels, but the g values [E. coli gzyx = 2.05, 1.95, 1.90 (35) and P. denitrificans gzyx = 2.04, 1.94, 1.87 (10)] are close to those from intact E. coli complex I [gzyx = 2.045, 1.94, 1.88 (13)]. The g values from the bovine Fp subcomplex (gzyx = 2.05, 1.95, 1.86) also correlate closely to those from the intact enzyme (gzyx = 2.04, ≈1.93, 1.86) (7, 17). 2Fe[G] in EcNuoG exhibits g values that match very closely to those from intact complex I, although its signal is broader (see above and Fig. 5). Therefore, g values typically match closely, although small variations may originate from structural perturbations in the isolated subunits: although the g values of the [4Fe-4S] cluster in EcNuoG correlate very well to N5, not to N4, a significant perturbation of the cluster environment in the isolated subunit, causing severe modification of EPR signal N4, cannot be dismissed unambiguously.
Why is N5 exhibited by EcNuoG but not by E. coli complex I?
It is most likely that the apparent reduction potential of the cluster is lower in complex I than in EcNuoG: adjacent subunits may affect the local structure and electrostatic environment, dynamics, and solvent accessibility. Note that the EPR spectra of different complexes I vary considerably. When bovine complex I is reduced by NADH, five reduced clusters are observed as N1b, N2, N3, N4 and N5 (7); when E. coli complex I is reduced by NADH, a different set of five signals are observed (N1a, N1b, N2, N3, and N4) (13). 2Fe[E] (N1a) has a significantly higher potential in E. coli than in bovine complex I (16, 36). Thus, different patterns of reduction most likely reflect variations in intrinsic cluster potentials, modulated by the ensemble, as the individual clusters respond to the redox states of nearby clusters (37). Alternatively, it is possible that additional intercluster interactions increase further the relaxation rate of N5 in E. coli complex I, such that the signal, which is already fast-relaxing in mitochondrial complex I (7, 38), becomes too broad to be distinguished.
Assigning EPR Signals to the FeS Clusters in NuoG and NuoI.
In PdNqo3, mutation of the His ligand of 4Fe[G]H to Cys produced a protein containing the [2Fe-2S] cluster and a [4Fe-4S] cluster with g values matching those of N5 (gzyx = 2.070, 1.93, 1.89) (19). Mutating the same ligand in EcNuoG-MBP provided spectra indistinguishable from those of the wild type also. However, mutating the ligands of 4Fe[G]C provided spectra that lacked the gzyx = 2.06, 1.94, 1.89 signal of N5 (but retained the gzyx = 2.05, 1.94, 1.91 signal of N7) (12). Neither N4 or N5 was affected when the His ligand of 4Fe[G]H was mutated to Ala in Yarrowia lipolytica complex I (31). These results suggest strongly that N5 originates from 4Fe[G]C (rather than 4Fe[G]H), and they confirm that a signal from 4Fe[G]H is not observed. Note that 4Fe[G]C is located between three (or four) other clusters (Fig. 1), and interactions with these clusters may contribute to the fast relaxation of N5.
[4Fe-4S] clusters with a His ligand are unusual, but several examples have been identified and characterized. In nitrate reductase, cluster FS0 has an S = 3/2 ground state (39). Thus, it exhibits no g ≈ 2 signal and was identified first by structural analysis (40). The EPR signal from the His-ligated cluster in NiFe hydrogenase (41) is complicated by interactions with a [3Fe-4S]0 cluster (42). In Fe-only hydrogenase, the relationship between the EPR signals and the structurally defined clusters is unclear, precluding definition of the signal from the His-ligated cluster (43, 44). However, in 4-hydroxybutyryl-CoA dehydratase, the His-ligated [4Fe-4S] cluster (45) exhibits a typical [4Fe-4S] spectrum (46). Although no corresponding signal could be detected here, further analyses of complex I from a variety of species are required before the possibility that 4Fe[G]H exists in the S = 3/2 ground state can be ruled out completely. Alternatively, a g ≈ 2 signal may not have been observed from 4Fe[G]H because it has an unusually low reduction potential or fast relaxation rate.
Our observations suggest strongly that EPR signal N4 does not originate in NuoG. Thus, by elimination, it originates from NuoI (see Table 1). Several previous observations support our conclusion. A signal commensurate with N4 (with features at g = 2.09 and 1.88) was exhibited by the connecting fragment from E. coli complex I (NuoB, CD, and I) (20). Chloramphenicol-poisoned Neurospora crassa formed a “small complex I,” which lacked N2 and in which N4 was altered [N1(b) and N3 were unchanged] (28). Recently, a mutation in NuoCD in E. coli complex I affected both N2 and the signal at g = 2.087, corresponding to N4 (47). The structure of the hydrophilic domain of T. thermophilus complex I (6) indicates that the mutation (E. coli R274A) is at the interface between NuoCD, NuoI, and NuoB, close to 4Fe[I]2 and 4Fe[B]. Unfortunately, overexpression of Nqo9 from P. denitrificans (the NuoI homologue) required chemical reconstitution of the clusters and provided only a weak and poorly defined spectrum (g values ≈ 2.08, 2.05, 1.92, and 1.89), which does not strongly support or dispute our argument (11). Finally, we are unable to attribute N4 specifically to 4Fe[I]1 or 4Fe[I]2 or to rule out the possibility that both clusters contribute. Point mutants of the Cys residues in E. coli NuoI have been created, but only one variant (C102A, where C102 is a ligand of 4Fe[I]2) was amenable to study by EPR, and the N4 signal remained clearly visible (48).
Implications for Electron Transfer in Complex I.
Revising the relationship between the structurally defined FeS clusters and their EPR signals (Table 1) dictates that the “energy profile” for electron transfer along the cluster chain be revised accordingly, with clear implications for our understanding of electron flux. Redox titrations of mitochondrial complex I have shown that signal N2 titrates at approximately −0.1 V and that signals N3, N1b, N4, and N5 titrate at approximately −0.25 V (7); in E. coli complex I signals, N1a, N1b, N2, N3, and N4 all titrate between −0.2 and −0.3 V (13). Combining this data with Table 1 and Fig. 1 now shows that, starting from the FMN, the first three clusters, 4Fe[F], 2Fe[G], and 4Fe[G]C, present an essentially flat energy profile. The potential of the fourth cluster, 4Fe[G]H, is not known. It may be similar to those of the preceding clusters, or it may be much lower: single out-of-line potentials are common features of electron-transfer chains and present little kinetic hindrance (32). The fifth and sixth clusters (4Fe[I]1 and 4Fe[I]2) contribute signal N4. There are two possibilities: one cluster is reduced and observed as N4 and the other has a much lower potential, or both have similar intrinsic potentials but they reduce as a pair (the first electron is distributed between the two; the second electron is not added until a much lower potential) (49). In the former case, it is not possible currently to determine which cluster is which; however, it is unlikely that both 4Fe[G]H and 4Fe[I]1 have out-of-line potentials, because this would slow electron transfer significantly. The seventh cluster in the chain has the highest potential, so it is reduced exergonically before the electrons are transferred to quinone in a reaction with an unknown mechanism. Thus, in opposition to current consensus of opinion (4), it is not acceptable to simply assume that all of the clusters between FMN and N2 are isopotential or to construct mechanistic interpretations accordingly. Further investigations of the properties of the EPR silent clusters, using alternative strategies, are clearly required, to complete the energy profile for intramolecular electron transfer through complex I and to allow comprehensive and consistent interpretations of data from both past and future studies of this important component of the enzyme's catalytic mechanism.
Materials and Methods
Preparation of EcNuoG.
The gene encoding NuoG from E. coli (50) was cloned into the expression plasmid pRUN, a derivative of pRK172 (51). DNA was amplified by PCR from E. coli DNA by using expand DNA polymerase (Roche Diagnostics, Mannheim, Germany) and the following primers: forward, TAGggatccATGCATCATCACCATCATCACGCTACAATTCATGT A G A C G GCAAAG; reverse, CGAgaattcTTATTGTTGTGCCTCCTTGAGATC (the codons for the initiator Met, the six His of the N-terminal tag, and the stop codon are in italics). After digestion with the restriction enzymes BamHI and EcoRI (sites in lowercase; New England Biolabs, Hitchin, U.K.), the insert was ligated into the expression vector and checked by direct sequencing of both strands.
Electrocompetent E. coli cells, strain C41 (DE3) (52), were transformed with the EcNuoG plasmid and either pACYC184 (New England Biolabs) containing the thioredoxin gene from E. coli (TRX) (53) or a plasmid carrying the “iron–sulfur gene cluster” (ISC) (a gift from Y. Takahashi, Osaka University, Osaka, Japan) (54). Single colonies were grown overnight at 37°C on TYE-agar plates containing 100 μg·ml−1 ampicillin and either 35 μg·ml−1 chloramphenicol (TRX) or 15 μg·ml−1 tetracycline (ISC) (Melford Laboratories, Ipswich, U.K.). Both secondary plasmids significantly increased holoprotein expression. Then an isolated colony was used to inoculate 5 ml of 2× TY medium containing the appropriate antibiotics, and the cells were grown aerobically to stationary phase at 37°C. One-milliliter aliquots were used to inoculate 0.5-liter volumes of 2× TY supplemented with 100 μg·ml−1 ferric ammonium citrate and the appropriate antibiotics. A range of growth conditions were explored; the best results were obtained when the cells were grown aerobically at 37°C to OD600 ≈1, then the temperature decreased to 25°C upon induction (0.4 mM isopropyl β-d-thiogalactopyranoside; Melford Laboratories). After overnight growth, the cells were harvested by centrifugation, resuspended in 32 mM Tris·HCl (pH 7.8) containing 1 mM phenylmethanesulfonyl chloride, 2 mM DTT, 250 mM nondetergent sulfobetaine-201 (NDSB-201; Fluka, Buchs, Switzerland) (55), and a protease inhibitor cocktail (Roche Diagnostics), and stored at −20°C.
Cells were thawed on ice and lysed at 30,000 psi by using a cell disrupter (Constant Systems, Northants, U.K.). Insoluble debris was removed by centrifugation (48,000 × g for 15 min.), then the supernatant was collected, filtered (0.2 μm, Minisart filter; Sartorius, Goettingen, Germany), and transferred quickly into an anaerobic glovebox (O2 < 2 ppm; Belle Technology, Dorset, U.K.). Using gravity flow, the supernatant was loaded onto a 10 ml Ni-NTA superflow column (Qiagen, Hilden, Germany), washed with 30 ml of buffer A [32 mM Tris·HCl (pH 7.8) containing 250 mM NDSB-201], 30 ml of buffer A containing 15 mM imidazole, and then eluted in buffer A containing 250 mM imidazole. The protein was diluted 10-fold in buffer A, then concentrated using a YM30 membrane (Millipore, Billerica, MA). Approximately 3 mg were obtained from 1 liter of cell culture.
Protein purity was assessed by SDS/PAGE analysis. Nonheme iron contents were determined colorimetrically by using 4,7-diphenyl-1,10-phenanthroline (Sigma–Aldrich, Dorset, U.K.) (56), and protein concentrations were determined by the Pierce (Rockford, IL) bicinchoninic acid assay. UV-visible spectra were recorded using a UV-1601 Shimadzu (Kyoto, Japan) spectrometer, with the samples sealed anaerobically in quartz SUPRASIL precision cuvettes (Hellma, Mullheim, Germany).
Preparation of Complex I from E. coli.
Complex I was purified from E. coli strain BL21 as described in ref. 57, except that a DEAE-Sepharose fast flow column was used for the second ion-exchange chromatography step, and the gel filtration column was a Superose 6 10/300 GL column (GE Healthcare Biosciences, Uppsala, Sweden).
EPR Spectroscopy.
EPR samples of EcNuoG (20 mg·ml−1 protein in buffer A) were prepared anaerobically by the addition of 1 mM sodium dithionite (Sigma–Aldrich) and frozen immediately. Complex I was concentrated to ≈8 mg·ml−1 and dialyzed anaerobically at 4°C against 20 mM Tris·HCl (pH 7.5) and 0.1 mM NADH for 1 h (to remove trace O2 and NAD+), before being reduced further by the addition of 1 mM dithionite and frozen immediately. EPR spectra were recorded on a Bruker (Karlsruhe, Germany) EMX X-band spectrometer by using an ER 4119HS high-sensitivity cavity maintained at low temperature by an Oxford Instruments (Abingdon, U.K.) ESR900 continuous-flow liquid helium cryostat; sample temperature was measured with a calibrated Cernox resistor (Lake Shore Cryotronics, Westerville, OH).
Acknowledgments
This research was funded by the Medical Research Council and the European Union Mitocombat Program.
Abbreviations
- FMN
flavin mononucleotide
- MBP
maltose-binding protein.
Footnotes
The authors declare no conflict of interest.
References
- 1.Walker JE. Q Rev Biophys. 1992;25:253–324. doi: 10.1017/s003358350000425x. [DOI] [PubMed] [Google Scholar]
- 2.Yagi T, Matsuno-Yagi A. Biochemistry. 2003;42:2266–2274. doi: 10.1021/bi027158b. [DOI] [PubMed] [Google Scholar]
- 3.Hirst J, Carroll J, Fearnley IM, Shannon RJ, Walker JE. Biochim Biophys Acta. 2003;1604:135–150. doi: 10.1016/s0005-2728(03)00059-8. [DOI] [PubMed] [Google Scholar]
- 4.Sazanov LA. Biochemistry. 2007;46:2275–2288. doi: 10.1021/bi602508x. [DOI] [PubMed] [Google Scholar]
- 5.Hinchliffe P, Sazanov LA. Science. 2005;309:771–774. doi: 10.1126/science.1113988. [DOI] [PubMed] [Google Scholar]
- 6.Sazanov LA, Hinchliffe P. Science. 2006;311:1430–1436. doi: 10.1126/science.1123809. [DOI] [PubMed] [Google Scholar]
- 7.Ohnishi T. Biochim Biophys Acta. 1998;1364:186–206. doi: 10.1016/s0005-2728(98)00027-9. [DOI] [PubMed] [Google Scholar]
- 8.Yano T, Sled VD, Ohnishi T, Yagi T. Biochemistry. 1994;33:494–499. doi: 10.1021/bi00168a014. [DOI] [PubMed] [Google Scholar]
- 9.Yano T, Yagi T, Sled VD, Ohnishi T. J Biol Chem. 1995;270:18264–18270. doi: 10.1074/jbc.270.31.18264. [DOI] [PubMed] [Google Scholar]
- 10.Yano T, Sled VD, Ohnishi T, Yagi T. J Biol Chem. 1996;271:5907–5913. doi: 10.1074/jbc.271.10.5907. [DOI] [PubMed] [Google Scholar]
- 11.Yano T, Magnitsky S, Sled VD, Ohnishi T, Yagi T. J Biol Chem. 1999;274:28598–28605. doi: 10.1074/jbc.274.40.28598. [DOI] [PubMed] [Google Scholar]
- 12.Nakamaru-Ogiso E, Yano T, Yagi T, Ohnishi T. J Biol Chem. 2005;280:301–307. doi: 10.1074/jbc.M410377200. [DOI] [PubMed] [Google Scholar]
- 13.Leif H, Sled VD, Ohnishi T, Weiss H, Friedrich T. Eur J Biochem. 1995;230:538–548. doi: 10.1111/j.1432-1033.1995.tb20594.x. [DOI] [PubMed] [Google Scholar]
- 14.Sled VD, Rudnitzky NI, Hatefi Y, Ohnishi T. Biochemistry. 1994;33:10069–10075. doi: 10.1021/bi00199a034. [DOI] [PubMed] [Google Scholar]
- 15.Yano T, Dunham WR, Ohnishi T. Biochemistry. 2005;44:1744–1754. doi: 10.1021/bi048132i. [DOI] [PubMed] [Google Scholar]
- 16.Zu Y, di Bernardo S, Yagi T, Hirst J. Biochemistry. 2002;41:10056–10069. doi: 10.1021/bi026026f. [DOI] [PubMed] [Google Scholar]
- 17.Barker CD, Reda T, Hirst J. Biochemistry. 2007;46:3454–3464. doi: 10.1021/bi061988y. [DOI] [PubMed] [Google Scholar]
- 18.Nakamaru-Ogiso E, Yano T, Ohnishi T, Yagi T. J Biol Chem. 2002;277:1680–1688. doi: 10.1074/jbc.M108796200. [DOI] [PubMed] [Google Scholar]
- 19.Yano T, Sklar J, Nakamaru-Ogiso E, Takahashi Y, Yagi T, Ohnishi T. J Biol Chem. 2003;278:15514–15522. doi: 10.1074/jbc.M212275200. [DOI] [PubMed] [Google Scholar]
- 20.Rasmussen T, Scheide D, Brors B, Kintscher L, Weiss H, Friedrich T. Biochemistry. 2001;40:6124–6131. doi: 10.1021/bi0026977. [DOI] [PubMed] [Google Scholar]
- 21.Orme-Johnson WH, Orme-Johnson NR. In: Iron-Sulfur Proteins. Spiro TG, editor. New York: Wiley Interscience; 1982. pp. 67–96. [Google Scholar]
- 22.Beinert H, Orme-Johnson WH. In: Magentic Resonance in Biological Systems. Ehrenberg A, Malmström BG, Vänngård T, editors. Oxford: Pergamon; 1967. pp. 221–247. [Google Scholar]
- 23.Hirsh DJ, Beck WF, Innes JB, Brudvig GW. Biochemistry. 1992;31:532–541. doi: 10.1021/bi00117a033. [DOI] [PubMed] [Google Scholar]
- 24.Galli C, MacArthur R, Abu-Soud HM, Clark P, Stuehr DJ, Brudvig GW. Biochemistry. 1996;35:2804–2810. doi: 10.1021/bi9520444. [DOI] [PubMed] [Google Scholar]
- 25.Galli C, Innes JB, Hirsh DJ, Brudvig GW. J Magn Reson B. 1996;110:284–287. doi: 10.1006/jmrb.1996.0044. [DOI] [PubMed] [Google Scholar]
- 26.Lakshmi KV, Brudvig GW. In: Biological Magnetic Resonance. Berliner LJ, Eaton SS, Eaton GR, editors. New York: Plenum; 2000. pp. 513–567. [Google Scholar]
- 27.Meinhardt SW, Kula T, Yagi T, Lillich T, Ohnishi T. J Biol Chem. 1987;262:9147–9153. [PubMed] [Google Scholar]
- 28.Wang DC, Meinhardt SW, Sackmann U, Weiss H, Ohnishi T. Eur J Biochem. 1991;197:257–264. doi: 10.1111/j.1432-1033.1991.tb15906.x. [DOI] [PubMed] [Google Scholar]
- 29.Kerscher S, Kashani-Poor N, Zwicker K, Zickermann V, Brandt U. J Bioenerg Biomembr. 2001;33:187–196. doi: 10.1023/a:1010726818165. [DOI] [PubMed] [Google Scholar]
- 30.Lin T-I, Sled VD, Ohnishi T, Brennicke A, Grohmann L. Eur J Biochem. 1995;230:1032–1036. doi: 10.1111/j.1432-1033.1995.tb20652.x. [DOI] [PubMed] [Google Scholar]
- 31.Waletko A, Zwicker K, Abdrakhmanova A, Zickermann V, Brandt U, Kerscher S. J Biol Chem. 2005;280:5622–5625. doi: 10.1074/jbc.M411488200. [DOI] [PubMed] [Google Scholar]
- 32.Page CC, Moser CC, Chen X, Dutton PL. Nature. 1999;402:47–52. doi: 10.1038/46972. [DOI] [PubMed] [Google Scholar]
- 33.Bungert S, Krafft B, Schlesinger R, Friedrich T. FEBS Lett. 1999;460:207–211. doi: 10.1016/s0014-5793(99)01341-1. [DOI] [PubMed] [Google Scholar]
- 34.Braun M, Bungert S, Friedrich T. Biochemistry. 1998;37:1861–1867. doi: 10.1021/bi971176p. [DOI] [PubMed] [Google Scholar]
- 35.Velaquez I, Nakamaru-Ogiso E, Yano T, Ohnishi T, Yagi T. FEBS Lett. 2005;579:3164–3168. doi: 10.1016/j.febslet.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 36.Uhlmann M, Friedrich T. Biochemistry. 2005;44:1653–1658. doi: 10.1021/bi048136n. [DOI] [PubMed] [Google Scholar]
- 37.Kurnikov IV, Ratner MA, Pacheco AA. Biochemistry. 2005;44:1856–1863. doi: 10.1021/bi048060v. [DOI] [PubMed] [Google Scholar]
- 38.Ohnishi T. In: Membrane Proteins in Energy Transduction. Capaldi RA, editor. New York: Dekker; 1979. pp. 1–87. [Google Scholar]
- 39.Rothery RA, Bertero MG, Cammack R, Palak M, Blasco F, Strynadka NCJ, Weiner JH. Biochemistry. 2004;43:5324–5333. doi: 10.1021/bi049938l. [DOI] [PubMed] [Google Scholar]
- 40.Bertero MG, Rothery RA, Palak M, Hou C, Lim D, Blasco F, Weiner JH, Strynadka NCJ. Nat Struct Biol. 2003;10:681–687. doi: 10.1038/nsb969. [DOI] [PubMed] [Google Scholar]
- 41.Volbeda A, Charon M-H, Piras C, Hatchikian EC, Frey M, Fontecilla-Camps JC. Nature. 1995;373:580–587. doi: 10.1038/373580a0. [DOI] [PubMed] [Google Scholar]
- 42.Teixeira M, Moura I, Xavier AV, Moura JJG, LeGall J, DerVartanian DV, Peck HD, Huynh BH. J Biol Chem. 1989;264:16435–16450. [PubMed] [Google Scholar]
- 43.Peters JW, Lanzilotta WN, Lemon BJ, Seefeldt LC. Science. 1998;282:1853–1858. doi: 10.1126/science.282.5395.1853. [DOI] [PubMed] [Google Scholar]
- 44.Bennett B, Lemon BJ, Peters JW. Biochemistry. 2000;39:7455–7460. doi: 10.1021/bi992583z. [DOI] [PubMed] [Google Scholar]
- 45.Martins BM, Dobbek H, Çinkaya I, Buckel W, Messerschmidt A. Proc Natl Acad Sci USA. 2004;101:15645–15649. doi: 10.1073/pnas.0403952101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Müh U, Çinkaya I, Albracht SPJ, Buckel W. Biochemistry. 1996;35:11710–11718. doi: 10.1021/bi9601363. [DOI] [PubMed] [Google Scholar]
- 47.Belevich G, Euro L, Wikström M, Verkhovskaya M. Biochemistry. 2007;46:526–533. doi: 10.1021/bi062062t. [DOI] [PubMed] [Google Scholar]
- 48.Flemming D, Schlitt A, Spehr V, Bischof T, Friedrich T. J Biol Chem. 2003;278:47602–47609. doi: 10.1074/jbc.M308967200. [DOI] [PubMed] [Google Scholar]
- 49.Mathews R, Charlton S, Sands RH, Palmer G. J Biol Chem. 1974;249:4326–4328. [PubMed] [Google Scholar]
- 50.Weidner U, Geier S, Ptock A, Friedrich T, Leif H, Weiss H. J Mol Biol. 1993;233:109–122. doi: 10.1006/jmbi.1993.1488. [DOI] [PubMed] [Google Scholar]
- 51.Silvester JA, Kane Dickson V, Runswick MJ, Leslie AGW, Walker JE. Acta Crystallogr F. 2006;62:530–533. doi: 10.1107/S1744309106015338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Miroux B, Walker JE. J Mol Biol. 1996;260:289–298. doi: 10.1006/jmbi.1996.0399. [DOI] [PubMed] [Google Scholar]
- 53.Leggate EJ. Cambridge, UK: Cambridge University; 2003. PhD thesis. [Google Scholar]
- 54.Nakamura M, Saeki K, Takahashi Y. J Biochem. 1999;126:10–18. doi: 10.1093/oxfordjournals.jbchem.a022409. [DOI] [PubMed] [Google Scholar]
- 55.Vuillard L, Braun-Breton C, Rabilloud T. Biochem J. 1995;305:337–343. doi: 10.1042/bj3050337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Doeg KA, Ziegler DM. Arch Biochem Biophys. 1962;97:37–40. doi: 10.1016/0003-9861(62)90041-3. [DOI] [PubMed] [Google Scholar]
- 57.Sazanov LA, Carroll J, Holt P, Toime L, Fearnley IM. J Biol Chem. 2003;278:19483–19491. doi: 10.1074/jbc.M208959200. [DOI] [PubMed] [Google Scholar]