Abstract
Immunoglobulin variable genes undergo several unusual genetic modifications to generate diversity, such as gene rearrangement, gene conversion, somatic hypermutation, and heavy chain class switch recombination. In view of these specialized processes, we examined the possibility that variable genes have intrinsic characteristics that allow them to be processed differently in the course of basic DNA transactions as well. This hypothesis was studied in an experimental system to gauge the relative efficiency of a DNA repair pathway, nucleotide excision repair, on a variable gene and a housekeeping gene. DNA damage was induced by ultraviolet light in murine hybridoma B cells, and repair was measured over time by an alkaline Southern blot technique, which detected removal of cyclobutane pyrimidine dimers. The rate of DNA repair in a rearranged variable gene, VHS107, was compared to that in the dihydrofolate reductase gene. Although both genes were actively transcribed, the VHS107 gene was repaired less efficiently than the dihydrofolate reductase gene. These results suggest that variable genes have inherent properties that affect the efficiency of nucleotide excision repair.
Keywords: Nucleotide excision repair, Transcription, Immunoglobulin gene, Dihydrofolate reductase gene
1. Introduction
Immunoglobulin variable (V) genes, which are located on three chromosomal loci, are unique in that they undergo unprecedented diversity by several mechanisms. (1) V genes are formed by joining several gene segments encoding V, diversity (D), and joining (J) regions by non-homologous recombination. (2) In some species, V genes undergo further diversification by frequent gene conversions between related genes using homologous recombination. (3) Following rearrangement, V genes are modified by the massive introduction of nucleotide substitutions during the process of somatic hypermutation. (4) Finally, V genes in the immunoglobulin heavy chain locus are recombined next to different constant genes by non-homologous recombination. All of these modifications suggest that the V gene loci are inherently unstable. We considered the possibility that these characteristics may also affect basic molecular processes; for example, DNA repair in the loci may be altered.
Of the several types of DNA repair, nucleotide excision repair was studied because dicyclobutane pyrimidine dimers can be readily introduced by ultraviolet (UV) light, and their removal can be monitored in specific genes (Bohr et al., 1985). This pathway does not directly contribute to any of the V gene diversification mechanisms discussed above, since V-D-J joining, hypermutation, and class switch recombination were normal in animals deficient for repair factors XPA (Winter et al., 1998), XPB (Kim et al., 1997), XPC (Shen et al., 1997), XPD (Wagner et al., 1996), XPF (Tian et al., 2004a), XPG (Tian et al., 2004b), CSA (Kim et al., 1997), and CSB (Jacobs et al., 1998), with the exception of ERCC1 (Schrader et al., 2004). However, it was used as an experimental tool because the nucleotide excision repair capacity in a specific genomic region may reflect unusual aspects of the locus. For this study, hybridoma B cells were irradiated with UV light, and the rate of repair of pyrimidine dimers was measured in a V gene and in a housekeeping gene encoding dihydrofolate reductase (DHFR).
2. Materials and methods
2.1. Cell lines and probes
Hybridoma cell lines HPCG10 and HPCG17 were made from fusion of BALB/c spleen cells to the SP2/0 cell line (Gearhart et al., 1980). Both hybridomas have a heavy (H) chain allele containing the VHS107 gene segment rearranged to D and JH1 gene segments. The hybridomas secrete anti-phosphorylcholine antibody of the IgG3 and IgG1 classes, respectively.
DNA probes were prepared by digesting plasmids with genes as follows: the DHFR probe for RNA and DNA analyses was 690 bp containing exon I and flanking sequence (Fig. 1A) from the pDsa7 plasmid (Crouse et al., 1982); the VHS107 probe for RNA analysis was 300 bp containing the coding sequence of the VH gene segment (Fig. 1B); and the JH2 probe for Southern blots was 415 bp containing the JH2 exon and flanking sequence from a PstI-BamHI digest of the unrearranged JH locus. The restriction fragments were separated on agarose gels, recovered, and labeled with [α-32P]dCTP by nick translation.
Fig. 1.
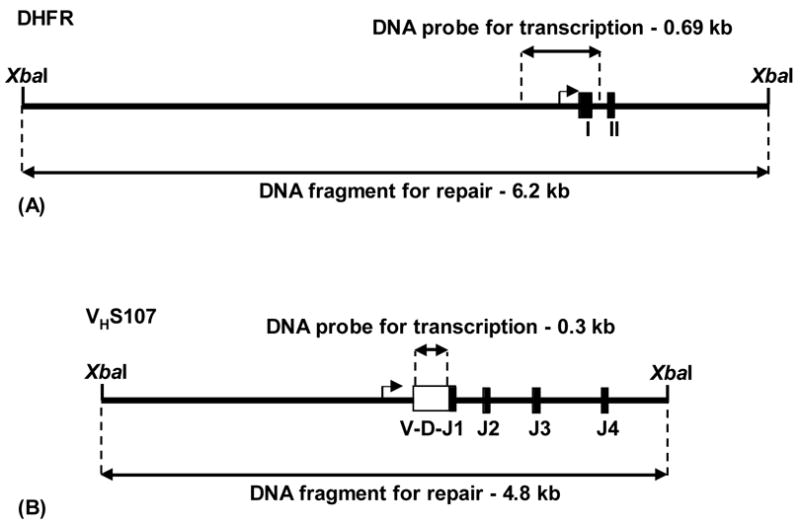
Maps of repair fragments. (A) DHFR. The 6.2 kb XbaI fragment contains exons I and II. (B) VHS107. The 4.8 kb XbaI fragment contains the VHS107 gene segment (open box) rearranged to D and JH1 segments. The fragment also contains JH2, JH3 and JH4 gene segments. Locations of probes for transcription analyses are shown. Transcription start sites are indicated by bent arrows.
2.2. Nuclear run-on RNA transcription
Nuclei were isolated from the hybridoma cells and incubated for 10 min with [α-32P]UTP, ATP, GTP and CTP according to Celano et al. (1989). Labeled RNA was hybridized to nitrocellulose slot blots containing 0.3 pmol (0.3 μg) of the DHFR probe and 0.5 pmol (0.1 μg) of the VHS107 probe. RNA synthesis was quantified after 2 days exposure to a PhosphorImager screen, and values were corrected to correspond to equal nanomoles of each probe.
2.3. Gene-specific DNA repair assay
The assay described by Bohr et al. (1985) was modified as follows. Hybridoma cells were irradiated with 40 Joules (J) per meter (m)2 of UV-C light, and allowed to repair for 0–4 h. Aliquots were taken every hour, and DNA was isolated and digested with XbaI to generate a 6.2 kb fragment for DHFR and a 4.8 kb fragment for VHS107 (Fig. 1). 20 μg of digested DNA was then treated with or without T4 denV endonuclease. DNA fragments were separated by electrophoresis on a 0.75% alkaline agarose gel, and blotted to a nylon membrane. Blots were hybridized with either DHFR or JH2 probes that were labeled with [α-32P]dCTP, and exposed and quantified by a PhosphorImager. The blot was then stripped and re-hybridized with the corresponding probe. The average number of dimers per fragment at each repair point was determined by the Poisson expression, -ln[T4 denV-treated/untreated]. For example, at 0 h, if the intensity of the treated band was 288, and the intensity of the untreated band was 784, the ratio would be 0.3673. The –ln of this ratio is 1.00, which means there is 1 dimer in the fragment. The percent of DNA repair was calculated as 1-(dimers at each time point/dimers at 0 h). For example, if there was 1 dimer at 0 h after irradiation, and 0.81 dimers after 4 h of repair, the ratio would be 0.81/1, and the percent repair would be 1–0.81 or 19%.
Repair rates were also calculated based on the increase in the fraction of undamaged DNA. The fraction that is free of dimers was calculated by dividing the intensity of the treated band by the intensity of the untreated band. The slope of the increase with time was then calculated, and percent repair was graphed.
3. Results
3.1. Transcription of VHS107 and DHFR genes in hybridoma cells
Two hybridoma cell lines, HPCG10 and HPCG17, with the same rearrangement of the VHS107 gene segment to JH1 (Gearhart et al., 1980), were used to measure transcription by the nuclear run-on technique. Labeled RNA from nuclei was hybridized to DNA probes on nitrocellulose slot blots, as shown in Fig. 2A. For both HPCG10 and HPCG17 cells, two experiments were performed, and the results of duplicate blots were averaged. The results in Fig. 2B show that the VHS107 gene was transcribed two-fold more than the DHFR gene, which was expected since the cells secrete large quantities of immunoglobulin.
Fig. 2.
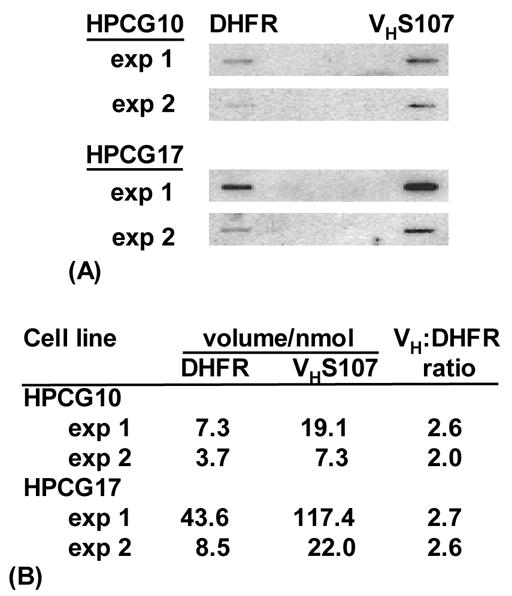
Nuclear run-on analysis of RNA in two hybridoma lines. (A) 32P-labeled nascent RNA transcripts from nuclei were hybridized to membranes containing the DHFR or VHS107 probes depicted in Fig. 1. Two experiments for HPCG10 and HPCG17 cells are shown. (B) Hybridization intensity was normalized to equal nanomoles of each probe.
3.2. Repair of UV damage in the VHS107 gene compared to the DHFR gene
We used a Southern blot technique to measure the repair of dicyclobutane pyrimidine dimers in specific genes. At a dose of 40 J/m2, about 1 dimer per 5 kb is generated. However, at this dose, the hybridoma cells underwent rapid cell death after 4 h, so cells were analyzed from 0 to 4 h, when the viability was greater than 50% (not shown), and DNA repair is underway (Mellon et al., 1986). For the repair assay, DNA was extracted from irradiated cells, and digested with a restriction enzyme. The DNA was then treated with T4 denV endonuclease, which cleaves at sites of pyrimidine dimers. As the damage is repaired over time in cells by enzymes in the nucleotide excision repair pathway, the extracted DNA will contain fewer pyrimidine dimers, and therefore have less cleavage by the endonuclease. DNA fragments were then separated on an alkaline gel, and transferred to a membrane by Southern blotting. Fragments containing the full-length restriction size were quantified by hybridization to double-strand probes. The XbaI restriction enzyme was chosen, since it yields fragments of similar size containing the 5′ ends of DHFR and VHS107 genes (Fig. 1). Although the fragments contain both 5′ intron and exon sequences, DNA repair for the DHFR gene has been reported to be constant throughout this region (Bohr et al., 1986). Although the VHS107 gene sustained slightly more initial damage (0.18 dimers per kb) than the DHFR gene (0.15 dimers per kb), we do not think this difference is significant.
Repair was determined by measuring the increasing intensity of the probe hybridized to the restriction fragments with time, and compared to DNA without T4 denV cleavage. Representative blots are shown in Fig. 3A and B; and the amount of dimers over time is listed in Table 1. HPCG10 cells had two rearranged H chain alleles that were detected by the JH2 probe, which were a 4.8 kb fragment containing the VHS107 gene rearranged to JH1, and a 8.5 kb fragment containing the VH81X gene rearranged to JH1. Only repair in the VHS107 gene was analyzed, and compared to the repair in the HPCG17 line, which had a single rearranged VHS107 chain allele. To assess the reproducibility of loading DNA into the lanes, controls containing undamaged DNA treated with or without the endonuclease were included. The intensities of fragments in the control samples were equal (not shown). Thus, although the amount of DNA between each time point varied, the amount of DNA loaded within a time point was constant, since the sample was divided in half, and treated with or without the endonuclease.
Fig. 3.
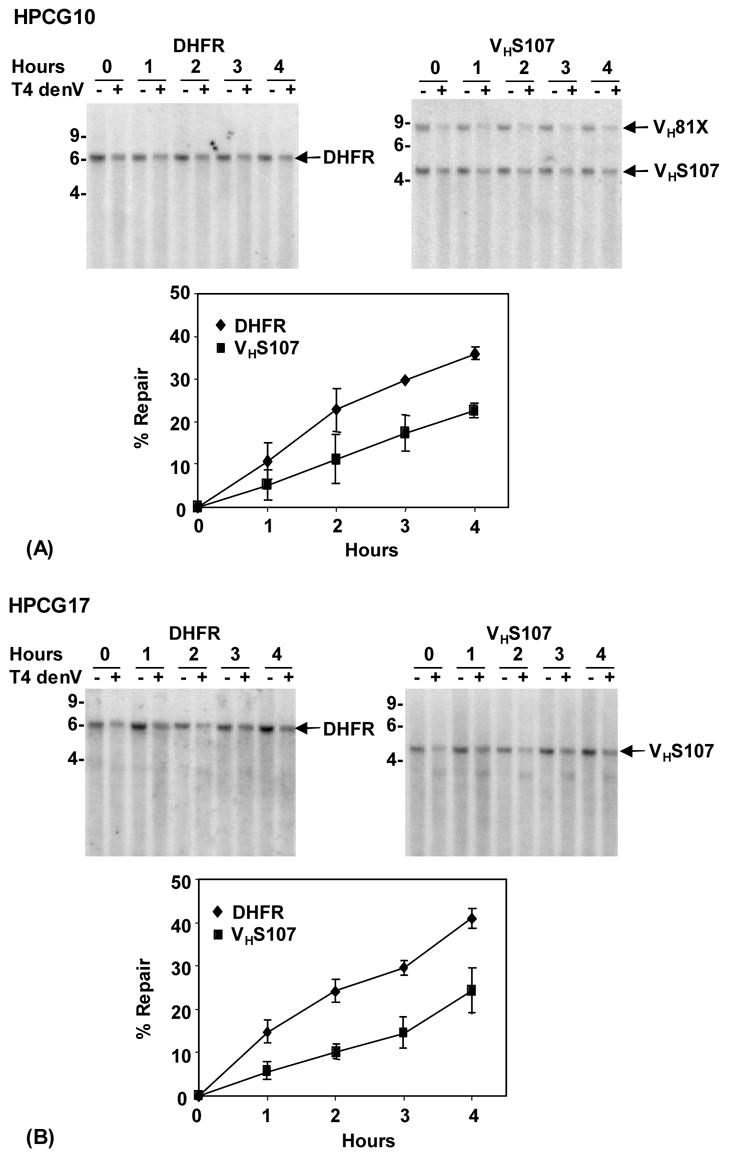
Autoradiograms and analysis of DNA repair of the DHFR and VHS107 genes. (A) Repair in HPCG10 cells. XbaI-digested DNA from different time points was treated with and without T4 denV endonuclease. A representative Southern blot is shown; size in kb is shown at the left of the blots. The blot was first hybridized to the DHFR probe, and then stripped and re-hybridized to a JH2 probe. Data was obtained from densitometric scans, which were calculated in Table 1, and graphed as the average value of two separate biological experiments. (B) Repair in HPCG17 cells. Blots were prepared, hybridized and scanned as above. The results from three separate experiments are graphed.
Table 1.
DNA repair in the DHFR and VHS107 loci
| Cell line | Locus | Time (h) after UV radiation | Dimer frequencya (40 J/m2) in exp:
|
Mean dimer frequency (dimer-free fractionb) | % Repair | ||
|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | |||||
| HPCG10c | DHFR | 0 | 0.86 | 0.82 | 0.84 (0.43) | 0 | |
| 1 | 0.74 | 0.76 | 0.75 (0.47) | 11 | |||
| 2 | 0.63 | 0.67 | 0.65 (0.53) | 23 | |||
| 3 | 0.60 | 0.59 | 0.59 (0.55) | 30 | |||
| 4 | 0.54 | 0.53 | 0.54 (0.58) | 36 | |||
| VHS107 | 0 | 0.95 | 0.76 | 0.85 (0.43) | 0 | ||
| 1 | 0.88 | 0.73 | 0.81 (0.45) | 5 | |||
| 2 | 0.82 | 0.70 | 0.76 (0.47) | 11 | |||
| 3 | 0.77 | 0.65 | 0.71 (0.49) | 17 | |||
| 4 | 0.74 | 0.58 | 0.66 (0.52) | 23 | |||
| HPCG17d | DHFR | 0 | 0.84 | 1.01 | 1.24 | 1.02 (0.36) | 0 |
| 1 | 0.73 | 0.87 | 1.03 | 0.88 (0.42) | 15 | ||
| 2 | 0.62 | 0.79 | 0.93 | 0.78 (0.46) | 24 | ||
| 3 | 0.59 | 0.73 | 0.85 | 0.72 (0.49) | 30 | ||
| 4 | 0.51 | 0.61 | 0.70 | 0.61 (0.55) | 41 | ||
| VHS107 | 0 | 0.94 | 0.86 | 0.96 | 0.92 (0.40) | 0 | |
| 0.86 | 0.83 | 0.90 | 0.86 (0.42) | 6 | |||
| 2 | 0.84 | 0.78 | 0.84 | 0.82 (0.44) | 10 | ||
| 3 | 0.82 | 0.70 | 0.83 | 0.78 (0.46) | 15 | ||
| 4 | 0.76 | 0.63 | 0.68 | 0.69 (0.50) | 24 | ||
Average number of dimers in each of the 6.2 kb DHFR and 4.8 kb VHS107 fragments.
The fraction of fragments that was free of dimers was calculated for each time point by dividing the intensity of a T4 denV-treated band by the intensity of the corresponding untreated band.
Experiment 1 values were derived from one Southern blot, and experiment 2 values were averaged from 3 blots.
Experiment 1 values were derived from one Southern blot; experiment 2 values were averaged from 2 blots; and experiment 3 values were averaged from 2 blots.
Repair of damage was examined in two separate biological experiments for HPCG10 cells, and three experiments for HPCG17 cells. For each experiment, several blots were performed, and the results from the densitometric scans were averaged. The results from Table I are graphed in Fig. 3A and B, and show that the repair rate was faster in the DHFR gene than the VHS107 fragment during the first 2 h, and then the rates were similar for the last 2 h. Thus, after 4 h, about 40% of the dimers were removed from the DHFR fragment compared to 24% in the VHS107 fragment. Repair rates were also calculated based on the increase in the fraction of dimer-free DNA listed in Table 1, and the graphs were similar (not shown).
4. Discussion
Immunoglobulin genes undergo genetic modifications that may reflect the inherent instability of the loci. This instability may also affect basic molecular transactions such as DNA repair, which employs complexes of different proteins to remove damage. We examined the nucleotide excision repair pathway, because lesions can be externally introduced by UV light into the genome at a constant frequency, and repair can be measured in specific genes using a Southern assay. This repair pathway includes a subpathway which is linked to transcription. Thus, transcribed genes are repaired faster than nontranscribed genes (Bohr et al., 1985), because transcription complexes stall at pyrimidine dimers, and repair is initiated to remove the lesions. Hybridoma B cell lines were used because they have transcribed V genes, and repair could be compared to other transcribed genes. The housekeeping DHFR gene was chosen as a control since it is transcribed in all cells, is not amplified in normal cells (Paulson et al., 1998), and is repaired at the same rate as other transcribed genes. For example, greater than 60% of induced pyrimidine dimers were removed after 8 h in DHFR (Bohr et al., 1985; Mellon et al., 1986), hypoxanthine phosphoribosyl-transferase (Vrieling et al., 1991), β-actin (Kantor et al., 1990), adenosine deaminase (Venema et al., 1991), and c-abl (Madhani et al., 1986) genes in fibroblast lines from mouse and human, and in Chinese hamster ovary cells. In contrast, repair of nontranscribed genes and bulk DNA containing vast tracts of nontranscribed regions was slower, with only 20–40% of lesions removed after 8 h.
The results reported here indicate that repair in rearranged VHS107 genes from two cell lines was slower than in DHFR genes. During the first 2 h of repair, about 10% of dimers were removed from VHS107 fragments compared to 23% removal from DHFR fragments, and during the last 2 h, the rates were similar. This suggests that repair factors are recruited less efficiently to the immunoglobulin locus after damage compared to the DHFR locus. It is possible that the V gene initially had more undetected dimers than the DHFR gene, which would affect the rate of repair. However, it has been shown that the efficiency of formation of pyrimidine dimers by UV light is similar in both a transcribed gene and bulk DNA (Bohr et al., 1985), so it is likely that both the genes studied here had similar amounts of induced damage. Our repair data on V genes are consistent with a previous report using the same assay with murine splenic B cells that were irradiated and stimulated in vitro with lipopolysaccharide (Beecham et al., 1994). In that study, there was less removal of pyrimidine dimers from the immunoglobulin Switchα-Cα and Jκ-Cκ regions compared to c-abl and Pvt1 (plasmacytoma variant translocation) genes. Thus, the repair defect appears to extend 3′ of V genes, perhaps due to the local microenvironment of the rearranged genes. Using a different technique of PCR amplification to detect removal of UV-induced dimers in human tonsil B cells, Braun and colleagues (Fairhurst et al., 1996) also found that rearranged VH genes were repaired more slowly than unrearranged VH genes.
Inefficient nucleotide excision repair might be caused by limited access of large repair complexes to the immunoglobulin loci. Histone modifications or proteins bound to DNA, which have been associated with immunoglobulin genes (Li et al., 2004; Odegard et al., 2005; Sen and Oltz, 2006; Wang et al., 2006) may impede the access of nucleotide excision repair enzymes to sites of base damage. It will be informative to examine other DNA repair pathways, including base excision and mismatch repair, to see if these processes are also compromised.
Acknowledgments
We thank David Wilson and Stella Martomo for helpful discussions on DNA repair. This research was supported by the Intramural Research Program of the NIH, National Institute on Aging.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Beecham EJ, Jones GM, Link C, Huppi K, Potter M, Mushinski JF, Bohr VA. DNA repair defects associated with chromosomal translocation breaksite regions. Mol Cell Biol. 1994;14:1204–1212. doi: 10.1128/mcb.14.2.1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohr VA, Smith CA, Okumoto DS, Hanawalt PC. DNA repair in an active gene: removal of pyrimidine dimers from the DHFR gene of CHO cells is much more efficient than in the genome overall. Cell. 1985;40:359–369. doi: 10.1016/0092-8674(85)90150-3. [DOI] [PubMed] [Google Scholar]
- Bohr VS, Okumoto DS, Ho L, Hanawalt PC. Characterization of a DNA repair domain containing the dihydrofolate reductase gene in Chinese hamster ovary cells. J Biol Chem. 1986;261:16666–16672. [PubMed] [Google Scholar]
- Celano P, Baylin SB, Casero RA., Jr Polyamines differentially modulate the transcription of growth-associated genes in human colon carcinoma cells. J Biol Chem. 1989;264:8922–8927. [PubMed] [Google Scholar]
- Crouse GF, Simonsen CC, McEwan RN, Schimke RT. Structure of amplified normal and variant dihydrofolate reductase genes in mouse sarcoma S180 cells. J Biol Chem. 1982;257:7887–7897. [PubMed] [Google Scholar]
- Fairhurst RM, Valles-Ayoub Y, Neshat M, Braun J. A DNA repair abnormality specific for rearranged immunoglobulin variable genes in germinal center B cells. Mol Immunol. 1996;33:231–244. doi: 10.1016/0161-5890(95)00145-x. [DOI] [PubMed] [Google Scholar]
- Gearhart PJ, Johnson ND, Douglas R, Hood L. IgG antibodies to phosphorylcholine exhibit more diversity than their IgM counterparts. Nature. 1980;291:29–34. doi: 10.1038/291029a0. [DOI] [PubMed] [Google Scholar]
- Jacobs H, Fukita Y, van der Horst GTJ, de Boer J, Weeda G, Essers J, de Wind N, Engelward BP, Samson L, Verbeek S, de Murcia JM, de Murcia G, te Riele H, Rajewsky K. Hypermutation of immunoglobulin genes in memory B cells of DNA-repair-deficient mice. J Exp Med. 1998;187:1735–1743. doi: 10.1084/jem.187.11.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantor GJ, Barsalou LS, Hanawalt PC. Selective repair of specific chromatin domains in UV-irradiated cells from xeroderma pigmentosum complementation group C. Mutat Res. 1990;235:171–180. doi: 10.1016/0921-8777(90)90071-c. [DOI] [PubMed] [Google Scholar]
- Kim K, Kage K, Matsuda F, Lefranc MP, Storb U. B lymphocytes of xeroderma pigmentosum or Cockayne syndrome patients with inherited defects in nucleotide excision repair are fully capable of somatic hypermutation of immunoglobulin genes. J Exp Med. 1997;186:413–419. doi: 10.1084/jem.186.3.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Luo Z, Scharff MD. Differential regulation of histone acetylation and generation of mutations in switch regions is associated with Ig class switching. Proc Natl Acad Sci USA. 2004;101:15428–15433. doi: 10.1073/pnas.0406827101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhani HD, Bohr VA, Hanawalt PC. Differential DNA repair in transcriptionally active and inactive proto-oncogenes: c-abl and c-mos. Cell. 1986;45:417–423. doi: 10.1016/0092-8674(86)90327-2. [DOI] [PubMed] [Google Scholar]
- Mellon I, Bohr VA, Smith CA, Hanawalt PC. Preferential repair of an active gene in human cells. Proc Natl Acad Sci USA. 1986;83:8878–8882. doi: 10.1073/pnas.83.23.8878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odegard VH, Kim ST, Anderson SM, Shlomchik MJ, Schatz DG. Histone modifications associated with somatic hypermutation. Immunity. 2005;23:101–110. doi: 10.1016/j.immuni.2005.05.007. [DOI] [PubMed] [Google Scholar]
- Paulson TG, Almasan A, Brody LL, Wahl GM. Gene amplification in a p53-deficient cell line requires cell cycle progression under conditions that generate DNA breakage. Mol Cell Biol. 1998;18:3089–3100. doi: 10.1128/mcb.18.5.3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader CE, Vardo J, Linehan E, Twarog MZ, Niedernhofer LJ, Hoeijmakers JHJ, Stavnezer J. Deletion of the nucleotide excision repair gene Ercc1 reduces immunoglobulin class switching and alters mutations near switch recombination junctions. J Exp Med. 2004;200:321–330. doi: 10.1084/jem.20040052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen R, Oltz E. Genetic and epigenetic regulation of IgH gene assembly. Curr Opinion Immunol. 2006;18:237–242. doi: 10.1016/j.coi.2006.03.008. [DOI] [PubMed] [Google Scholar]
- Shen HM, Cheo DL, Friedberg E, Storb U. The inactivation of the XP-C gene does not affect somatic hypermutation or class switch recombination of immunoglobulin genes. Mol Immunol. 1997;34:527–533. doi: 10.1016/s0161-5890(97)00064-3. [DOI] [PubMed] [Google Scholar]
- Tian M, Shinkura R, Shinkura N, Alt FW. Growth retardation, early death, and DNA repair defects in mice deficient for the nucleotide excision repair enzyme XPF. Mol Cell Biol. 2004a;24:1200–1205. doi: 10.1128/MCB.24.3.1200-1205.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian M, Jones DA, Smith M, Shinkura R, Alt FW. Deficiency in the nuclease activity of xeroderma pigmentosum G in mice leads to hypersensitivity to UV irradiation. Mol Cell Biol. 2004b;24:2237–2242. doi: 10.1128/MCB.24.6.2237-2242.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venema J, van Hoffen A, Karcagi V, Natarajan AT, van Zeeland AA, Mullenders LHF. Xeroderma pigmentosum complementation group C cells remove pyrimidine dimers selectively from the transcribed strand of active genes. Mol Cell Biol. 1991;11:4128–4134. doi: 10.1128/mcb.11.8.4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrieling H, Venema J, van Rooyen ML, van Hoffen A, Menichini P, Zdzienicka MZ, Simons JWIM, Mullenders LHF, van Zeeland AA. Strand specificity of UV-induced DNA repair and mutations in the Chinese hamster HPRT gene. Nucleic Acids Res. 1991;19:2411–2415. doi: 10.1093/nar/19.9.2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner SD, Elvin JG, Norris P, McGregor JM, Neuberger MS. Somatic hypermutation of Ig genes in patients with xeroderma pigmentosum (XP-D) Int Immunol. 1996;8:701–705. doi: 10.1093/intimm/8.5.701. [DOI] [PubMed] [Google Scholar]
- Wang L, Whang N, Wuerffel R, Kenter AL. AID-dependent histone acetylation is detected in immunoglobulin S regions. J Exp Med. 2006;203:215–226. doi: 10.1084/jem.20051774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter DB, Phung QH, Umar A, Baker SM, Tarone RE, Tanaka K, Liskay RM, Kunkel TA, Bohr VA, Gearhart PJ. Altered spectra of hypermutation in antibodies from mice deficient for the DNA mismatch repair protein PMS2. Proc Natl Acad Sci USA. 1998;95:6953–6958. doi: 10.1073/pnas.95.12.6953. [DOI] [PMC free article] [PubMed] [Google Scholar]


