Abstract
Purpose
A previous study demonstrated that CTCF (CCCTC binding factor) regulates homeobox Pax6 gene expression in early embryonic stages and plays a dominant role in eye development. The purpose of the present study was to explore further the mechanism of CTCF controlling Pax6 gene expression in human retinoblastoma (Rb) cells and in the development of chicken and mouse retinas.
Methods
Northern and Western analyses were used to detect expressions of CTCF and Pax6 in Rb cells. Pax6 transcription reporter and deletion mutants were used to study the regulatory interaction between CTCF and Pax6 in Rb cells and in the retina of chicken embryos. CTCF transgenic chicken embryos and mice were established by lipofection and microinjection of linearized cytomegalovirus (CMV)-CTCF construct into fertilized eggs and mouse oocytes, respectively. Injected oocytes were implanted in the uterus of foster mothers through microinjection into the ovarian duct. The expression of CTCF and Pax6 was determined in embryo sections by immunochemistry.
Results
Stimulation of Rb cells with 10% FBS resulted in an increase in CTCF expression and a decrease in Pax6 expression. To study the regulatory mechanism, the Pax6 reporter and its deletion mutant activities were determined in transfected Rb cells and chicken embryonic retinas, revealing that CTCF interacts with the Pax6 gene in Rb cells through transcription control in the 5′-flanking region upstream from the Pax6 P0 promoter. Overexpression of CTCF in Rb cells suppressed Pax6 reporter activity and downregulated endogenous Pax6 expression. In contrast, downregulation of CTCF expression by knockdown of CTCF mRNA using specific small interfering (si)RNA markedly enhanced Pax6 expression in Rb cells. Further study in CTCF transgenic mouse embryos verified that overexpression of CTCF suppressed Pax6 gene expression in the retina.
Conclusions
CTCF plays an important role in regulating Pax6 expression in Rb cells and in the developmental retina, and the regulation of Pax6 gene expression by CTCF in the retina is through transcriptional regulation.
A homeobox transcription factor, Pax6, is one of the nine members of the Pax gene family in vertebrates.1 PAX6 plays essential roles in the development of ocular structures, including the cornea, iris, lens, and retina.2–5 Overexpression of Pax6 results in ectopic compound eyes in Drosophila and well-formed ectopic lenses in Xenopus laevis.6,7 Mutation of Pax6 can cause small eye (Sey) defects in mice and aniridia in the human eye.8–10 In addition, homozygous mutation of the Pax6 gene results in missing eye structures in Drosophila.11 Expression of the Pax6 gene is regulated via two promoters, P0 and P1.12–14 The P0 promoter–initiating transcriptions predominate in the cornea, conjunctival epithelia, lens placode, and retina, and the P1 promoter initiates transcriptions mainly in the lens placode, optic vesicle, and central neural system.14 An ectoderm enhancer (EE), found approximately −3.5 kb upstream of the P0 promoter, plays a key role in enhancing Pax6 transcription.15 There is also a regulatory element in intron 4 of the Pax6 gene to direct expression of P0 and P1 promoters in amacrine cells, ciliary body, and iris.14 However, it is still not clear how the tissue-specific expression of the Pax6 gene is regulated during embryonic eye development. Results of recent studies in our laboratory indicate that the transcription of the Pax6 gene is regulated by CTCF, a nuclear protein and transcription regulator.16,17
CTCF is a zinc finger (ZF) phosphoprotein that interacts with targeting DNA sequences by its DNA-binding domain containing 11 ZFs.18 CTCF protein is composed of 728 amino acids and its molecular mass is 82 kDa. Message RNA encoding CTCF is composed of 4.1-kb nucleotides and contains a long open reading frame (21.8 kb). However, the reported protein size of CTCF is 130 kDa, according to SDS-PAGE. The discrepancy of CTCF protein sizes between the predicted and apparent molecular mass suggests anomalous migration of the protein in SDS-PAGE.19 On the one hand, CTCF downregulates transcriptions of several important genes including c-myc, β-globin, and chicken lysozyme genes.20 On the other hand, CTCF can upregulate expression of the amyloid protein precursor gene through transcriptional control.21 More interesting, CTCF is a major factor that mediates monoallelic expression of imprinted genes, such as IGF2 (insulin-like growth factor-2) and H19 through a DNA methylation-sensitive mechanism.22,23 In fact, CTCF regulates DNA imprinting and controls IGF2 and H19 expression from the paternal allele and maternal allele, respectively. Another recent finding suggests that CTCF is a transacting factor for X-inactivation, silencing of one of two female X chromosomes.24 Expression patterns of CTCF suggest that it may play roles in the eye, because it is not only expressed in a wide range of mature tissues, but is also highly expressed in the eye in the ciliary marginal zone, inner nuclear layer, and anterior lens epithelium.18,25
In the present study, we report Rb cells in which fetal bovine serum (FBS)–induced increases in CTCF expression effectively suppressed Pax6 expression. We used the Pax6-LacZ reporter containing the 4.2-kb DNA fragment isolated from the 5′ flank region of the Pax6 gene upstream from the P0 promoter and its deletion mutants to study in retina-derived Rb cells the interaction of CTCF and Pax6 transcription. Furthermore, we investigated regulation of Pax6 reporter activities by CTCF in Rb cells and in the retina. Overexpression of CTCF in transgenic chicken and mouse revealed the role of CTCF in controlling Pax6 gene expression through transcription regulation.
Methods
Cell Culture and Transfection
Human Rb cells were cultured in RPMI medium 1640 contained 10% FBS (Invitrogen-Gibco, Carlsbad, CA). Rb cells were incubated in a humidified incubator supplied with 95% air and 5% CO2 at 37°C. Culture medium was replaced every 2 days. The number of cells was counted by a cytometer, and cell viability was determined by trypan blue exclusion.
Full-length cDNAs encoding human CTCF were cloned into an expression vector (pcDNA4/To/A; Invitrogen-Gibco) and termed pcDNA4-CTCF and pcDNA4-Pax6, respectively. These constructs were established for overexpression of CTCF in Rb cells. The constructs and pcDNA4/To/A vector alone (serving as the control) were transfected into Rb cells by electroporation with the following protocol. Rb cells were washed twice with cold PBS and resuspended in PBS (106 cell/mL). DNA samples (5 μg each) were added into 0.8 mL cells in electroporation cuvettes with a 400-μm gap and placed into an electroporator (Bio-Rad Laboratories, Hercules, CA). Electroporation was performed by exposure of cells to the electric field (300 V) with a time constant of 0.5 ms (τ = RC). The electroporated cells were immediately transferred into 3 mL prewarmed RPMI 1640 medium containing 10% FBS and cultured in normal conditions for 10 to 12 hours. Transfected cells were washed with PBS to remove cell debris and were then cultured in the normal medium for 2 to 3 days before the experiments.
For RNA interference (RNAi) experiments, a human CTCF-specific double-stranded, small interfering (si)RNA was synthesized (Silencer siRNA Construction kit; Ambion, Austin, TX). The sense strand sequence was aaggaaugucuucuuuacacc, and the antisense strand was aagguguaaagaagacauucc. In addition, a control double strand siRNA (sense, aacauucgguagauuccucgc, and antisense, aagcgaggaaucuaccgaaug) was synthesized by the same method. The sequence homologies of siRNA primers were examined by using the NIH Blast program (National Center for Biotechnology Information, Bethesda, MD). A lipid transfection system (siPORT; Ambion) was used to introduce the siRNAs into Rb cells by the following protocol: Rb cells were rinsed twice with ice-cold PBS and resuspended in PBS with a density of 106 cell/mL. Rb cells were placed into six-well culture plates containing serum-free 1640 medium. The mixtures of siRNA and lipid reagent (siPORT; Ambion) were added to 1 mL Rb cells at a final concentration of 25 nM. The cells were incubated in normal culture conditions for 4 hours before 3 mL RPMI 1640 medium containing 10% FBS was added to each well. In our previous study,17 we found that the level of CTCF mRNA was markedly decreased within 2 to 3 days after the transfection (data not shown). All experimental procedures were performed with these cells on the third day after siRNA transfection.
Reporter Gene Constructs and Expression
A 4.29-kb DNA fragment located upstream of the Pax6 gene P0 promoter was cloned by PCR with mouse genomic DNA used as a template. Both sense (5′GGTTACACCA GAAGCACCCCAACC) and anti-sense (5′CATCCGCAGCAACTCCTCTACCATC) primers were designed for PCR experiments based on the Pax6 gene sequence obtained from GenBank (accession numbers AF098639 and AF008212; http://www.ncbi.nlm.nih.gov/Genbank; provided in the public domain by the National Center for Biotechnology Information, Bethesda, MD). The 4.29-kb DNA fragment of the Pax6 gene was cloned into a promoterless β-gal-basic vector (BD-Clontech, Redwood, CA). Deletion mutants were obtained by digestion of the 4.2-kb DNA fragment with restriction enzymes. These mutants included DNA fragments of 3.5 kb (cleaved by EcoRI at nucleotide 712), 2.3 (cleaved by SmaI at nucleotide 1897), and 1.2 kb (cleaved by SalI at nucleotide 3058). In addition, an internal-delete mutant was made by removing an internal fragment between nucleotide 2470 (cleaved by XbaI) and 3035 (cleaved by XhoI) from the 4.2-kb DNA fragment. To establish reporter constructs, these mutant fragments were individually subcloned into a promoterless β-gal-basic vector. These reporter constructs were termed P4.2, P3.5, P2.3, P1.2, and Pxba. Each of reporter constructs (5 μg each) and an internal control plasmid pRL-TK (a luciferase reporter plasmid from Promega, Madison, WI) were cotransfected into target cells by electroporation using the protocol described earlier. On day 3 after transfection, transfected cells were harvested for measuring β-galactosidase and luciferase activities. The cells were transfected with β-gal control vector, SV40 promoter (positive control), and β-gal-basic vector (negative control). In addition, we found that Rb cells have very low β-galactosidase background (data not shown).
Analysis of β-Galactosidase Activity
Rb cells were rinsed twice with ice-cold PBS and suspended in lysate buffer containing (mM): 100 KH2PO4 and 1 dithiothreitol (pH 7.8). The cells were ruptured by three freeze-thaw cycles and precleared by centrifugation at 13,000g for 5 minutes. β-galactosidase activity was tested by a luminescent β-gal system using a chemiluminescent substrate (Galacton-star; BD-Clontech). Chemiluminescent signals from metabolic products of the substrate by β-galactosidase were detected by luminometer (Sirius; Berthold Detection System, Pforzheim, Germany). Luciferase activity of cotransfected pRL-TK from the same cell lysates served as the internal control for normalizing LacZ activity. Luciferase activity was measured by a luciferase detection kit from Promega. Finally, Pax6-Lac-Z reporter activities were normalized and presented as a ratio of measured β-galactosidase activity over luciferase activity (β-galactosidase activity[b]/luciferase activity).
Transfection and X-Gal Staining of Chicken Embryos
The transfection of chicken embryos was performed according to a method published by Muramatsu et al.26 Fertilized eggs of chicken were hatched at 38°C with 100% humidity. At day 1 of incubation, DNA constructs (P4.2, P2.3, and P1.2 and blank vector β-gal basic) were individually injected onto blastoderm. For each injection of chicken embryos, DNA (3 μg) was premixed with 6 μL of a lipophilic (lipofectAMINE 2000; Invitrogen-Gibco), and the premixture was brought to a final volume of 60 μL by adding DMEM. A window (<1 cm in diameter) located at the upper blastoderm was opened in the shell of chicken eggs. Injections of lipid-DNA mixture onto the chicken blastoderm were performed by using a fine-tipped glass pipette. After injection, the window on the shell was sealed with a piece of plastic film. Injected eggs were incubated in a rocking incubator with 38°C temperature and 100% humidity for expected days. At 7 days incubation, chicken embryos were collected in PBS solution and washed three times with PBS. Chicken embryos were fixed for 60 minutes in 2% paraformaldehyde in PBS. After rinsing with PBS, chicken embryos were stained overnight at room temperature with a solution containing 1 mg/mL X-gal, 5 mM potassium ferrocyanide, 5 mM potassium ferricyanide, and 2 mM MgCl2 in PBS (pH 7.3). The stained embryos were dehydrated and embedded in paraffin. Finally, 10 μm thickness sections of chicken embryos were cut, mounted on glass slides, and deparaffinized with xylene.
To verify DNA concentrations that were injected into each embryo, we extracted tissue samples from each embryonic head for genomic DNA. After phenol-chloroform extraction, DNA samples in lysates were precipitated in −20°C isopropanol overnight. The resultant DNA pellets were washed with 70% ethanol and resuspended in TE buffer. DNA samples were purified and used as templates for semiquantitative PCR that was performed with a pair of primers specific to the μ-galactosidase coding frame. A 180-bp PCR product was yielded by the PCR experiment. For PCR experiments, 20 ng of template DNA was used for each sample and PCR reactions were performed for 25 cycles.
Northern Blot Analysis
Total RNA obtained from Rb cells was extracted with a guanidium thiocyanate procedure.27,28 Briefly, 1 × 107 cells were collected for each sample. The cells were rinsed with ice-cold PBS and lysed in 1 mL guanidium solution (5 M guanidine hydrochloride, 50 mM Tris-HCl [pH 8], 0.5% N-lauroylsarcosine, and 100 mM β-mercaptoethanol). The lysate was extracted with 50:50 phenol-chloroform three times. RNAs were precipitated in ethanol and spun down by centrifugation at 12,000 rpm for 15 minutes. RNA (20 μg) for each sample was loaded in 1% agarose gel denatured with 2.2 M formaldehyde. The fractionated RNAs were transferred onto a nylon membrane. The membrane was hybridized with [α-32P] labeled CTCF probe with a standard procedure and visualized by exposure of x-ray films at −80°C overnight. Then, the membranes were stripped by standard protocol and rehybridized with [α-32P]-labeled GAPDH probe for the internal control. A CTCF probe used in Northern blot analysis was an approximately 1.2-kb fragment from the 5′ end of human CTCF cDNA.
Western Blot Analysis
Western blot assay was performed as described previously.28 In brief, 5 × 105 cells were harvested in 0.5 mL lysate buffer containing (mM): 137 NaCl, 1.5 MgCl2, 2 EDTA, 10 sodium pyrophosphate, 25 β glycerophosphate, 10% glycerol, 1% Triton X-100, 1 Na-orthovanadate, 1 phenylmethylsulfonyl fluoride, 10 mg/mL leupeptin, and 20 Tris [pH 7.5]. Cell lysates were precleared by centrifugation at 13,000g for 20 minutes. Samples were denatured by adding an equal volume of 2× Laemmli buffer and boiling for 5 minutes. Cellular proteins were fractionated by electrophoresis in 8% SDS-PAGE gel. Cellular proteins were then transferred to polyvinylidene difluoride (PVDF) membranes that were incubated with primary antibodies including rabbit anti-CTCF antibodies (1:5000; Upstate Biotechnology, Lake Placid, NY), rabbit anti-Pax6 (1:2000; Santa Cruz Biotechnology, Santa Cruz, CA). Donkey anti-rabbit immunoglobulin G conjugated with HRP (1:1000; Santa Cruz Biotechnology) was used as the secondary antibody. Signals in the membrane were visualized with a horseradish peroxidase blot detection kit (Santa Cruz Biotechnology). All membranes were stripped by using a standard stripping protocol. The membranes were rehybridized with mouse anti-actin and goat anti-mouse IgG antibodies for internal and loading controls.
CTCF Transgenic Mouse and Embryo Section Preparation
Fertilized eggs from donor mice were injected with a MulI/NotI fragment (~3.1 kb) containing the CMV promoter and cDNA encoding full-length CTCF by using a microdispenser. CTCF-injected eggs were cultured in vitro for 2 days, and the eggs were transplanted into the ovarian duct of maternal recipients. Embryos were collected from the uterus at 7, 11, and 14 days after transplantation. Harvested embryos were fixed in 4% paraformaldehyde in PBS for histologic examinations. Tissue sections were obtained from paraffin-fixed embryo blocks by removing wax with the following protocol. Embryonic tissue samples were dehydrated by serial incubations in 50%, 70%, 85%, 95%, and 100% ethanol (30 minutes at each concentration). After 10 minutes of incubation of tissues in xylene, embryos were transferred into tubes that contained xylene-paraffin (50:50) at 60°C and kept at room temperature overnight. All mouse embryos were incubated in paraffin at 60°C for 1 hour. Finally, embryonic tissue samples were individually embedded in wax blocks that were cut into sections with a thickness of 6 μm. All animals were treated in accordance with the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research, using protocols approved and monitored by the Animal Care Committee of Harbor-UCLA REI, School of Medicine at UCLA.
Results
Regulation of CTCF and Pax6 Expression by Serum in Rb Cells
Previous studies report that CTCF is expressed in many tissue and cell types, including in the retina.25,29 We chose Rb cells in our study because human Rb cells originate from the retina and can grow continuously in a culture condition supplemented with 10% FBS. Expressions of CTCF in several cell types were examined by Western blot analysis using an anti-CTCF antibody. Expressions of CTCF protein were found in Rb cells, rabbit corneal epithelial (RCE) cells and hematopoietic myeloblastic ML-1 and FDC-P1 cells (Fig. 1A). The expression level of CTCF in eye-derived RCE and Rb cells appeared lower than in hematopoietic cells. This lower expression pattern was also observed in human corneal epithelial cells that express Pax6 proteins (data not shown). To study the regulatory relationship between CTCF and Pax6, we synchronized the Rb cells and control ML-1 cells by serum starvation for 36 hours, and then the cells were stimulated with 10% FBS (Fig. 1B). Expressions of CTCF mRNA were markedly increased in response to FBS stimulation in both cell types detected with Northern blot analysis. In contrast, Northern and Western blot analysis showed that expressions of Pax6 mRNA and protein were diminished in Rb cells, respectively, but not in ML-1 cells, suggesting that there is a regulatory correlation in Rb cells between CTCF and Pax6 expression and that Pax6 is specifically expressed only in retina-derived Rb cells, but not in hematopoietic ML-1 cells.
Figure 1.

Serum-induced expression changes of CTCF and PAX6. (A) Western blot analysis of CTCF protein expression. CTCF protein was identified in various cell types by Western blot analysis using a specific anti-CTCF antibody. β-Actin was used as an internal control for Western blot analysis. (B) Expressions of CTCF and Pax6 mRNAs, and PAX6 protein in Rb cells in response to serum stimulation. Rb cells were cultured in the normal condition, as introduced in methods and serum-starved 36 hours before 10% FBS stimulation for 6 hours. Total RNAs of these cells were harvested for Northern blot analysis of CTCF and Pax6 mRNAs and Western blot analysis were performed to detect cellular protein expression of CTCF and Pax6. Afterward, the same membranes as were used in Northern and Western blot analyses were stripped and rehybridized with GAPDH probe and β-actin antibody, respectively as the internal controls.
Transcriptional Regulation of the Pax6 P0 Promoter in Rb Cells
A 4.2-kb upstream fragment (P4.2) of the Pax6 P0 promoter was cloned from mouse genomic DNA by PCR and subcloned into a promoterless β-gal-basic vector (BD-Clontech), to generate a reporter construct (Fig. 2A). In addition, 5′ end deletion mutants (P3.5, P2.3, and P1.2) and an internal deletion mutant (Pxba) of the reporter construct were generated and subcloned into the promoterless pβ-gal-basic vector. These Pax6 P0 report mutants were used to study the potential regulative element in the 5′ end flank region of the P0 promoter (Fig. 1A). The Pax6 P0 promoter activity of these constructs was examined by transient expression in Rb cells. β-Galactosidase activities were dramatically elevated after transfection of the P4.2 pβ-gal construct, indicating that the P4.2 fragment upstream Pax6 P0 promoter is able to initiate the reporter gene expression in Rb cells (Fig. 2B). For control experiments, cells were transfected with the SV40-β-gal vector to express β-galactosidase activities (data not shown).
Figure 2.
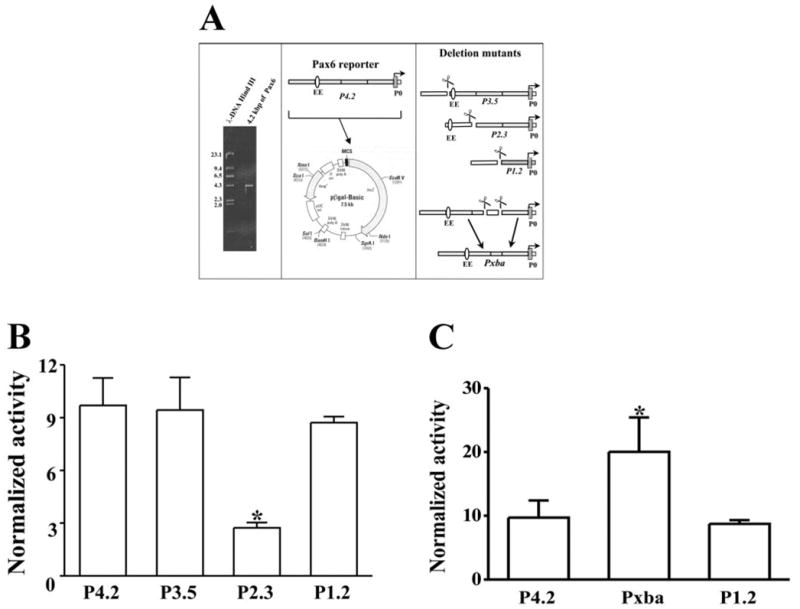
Constructions of β-gal–Pax6 reporter and deletion mutants. (A) Cloning 5′ flank region of Pax6 P0 promoter and deletion mutants. A 4.2-kb DNA fragment upstream Pax6 P0 promoter was cloned from mouse genomic DNA by PCR. A β-gal–Pax6 reporter construct was established by inserting the 4.2-kb fragment into a promoterless β-gal-basic vector. Three mutants (P3.5, P2.3, and P1.2) were made by deleting 5′ end nucleotides in the β-gal–Pax6 reporter. In addition, a mutant of P4.2 β-gal–Pax6 construct was generated by deleting an internal XbaI-XhoI fragment (~500 bp) in the region of −1.7 to −1.2 kb upstream of the P0 promoter. (B) Expression of the β-gal–Pax6 reporter and deletion mutants in Rb cells. (C) Expression of Pxba mutant lacking the potential repressor element. The β-gal–Pax6 reporter construct and its mutants were transfected into Rb cells by electroporation, with cotransfection of an internal control plasmid pRL-TK (a vector expressing luciferase driven by the HSV-TK promoter). Pax6 promoter activity was determined by measuring β-galactosidase activity with a luminescent β-gal system. The internal control luciferase activity was determined with a luminescent luciferase assay kit. The Pax6 promoter activities are presented as a ratio of measured β-galactosidase activity over luciferase activity (β-galactosidase activity[b]/luciferase activity). In each group of experiments, four DNA constructs were individually transfected. At least three individual experiments were performed for each group (n = 3). Data were plotted as the mean ± SE. *Significant difference (P < 0.05, n = 4).
Deletion mutants (P3.5, P2.3, and P1.2) of the P4.2 construct were also transfected in Rb cells, to analysis whether CTCF regulates Pax6 transcription through an element in the 5′ flanking region upstream Pax6 P0 promoter (Fig. 2B). The mutant P2.3 demonstrated a significant reduction of β-galactosidase activity in Rb cells (Fig. 2B). The further deletion of P2.3 (mutant P1.2) resulted in restoring P0 promoter activity, suggesting that there is a repressor element for CTCF to interact between −2.3 and −1.2 kb upstream from the P0 promoter, consistent with previous studies indicating that there are five repeats of CCCTC sequence in this region.15 In fact, we found only the clusters of binding sites for CTCF were involved in the Pax6 promoter inhibition (data not shown). To confirm the prediction, an additional mutant (Pxba) was generated previously by dropping an internal XbaI-XhoI fragment (~500 bp) in the region of −1.7 to −1.2 kb in the P4.2 construct. After transfecting Rb cells, the Pxba mutant expressed the strongest β-galactosidase activity (Fig. 2C).
Effect of the Repressor on P0 Promoter Activity in Developing Chicken Retina
Pax6 plays an essential role in the development of ocular structures including the retina. To determine whether the identified CTCF-interactive repressor plays functional roles in regulating Pax6 gene expression during retinal development, P4.2-β-gal construct and deletion mutants were introduced into the blastoderm on day 1 of chicken embryos (day E1). β-Galactosidase activities in the chicken embryo were determined by X-gal staining on embryonic day (E) 7. X-Gal signal was not observed by nude eyes in the whole embryo body. However, X-gal activities were found to be concentrated in the retina of chicken embryos injected with P4.2-β-gal and the mutants (day 7, Fig. 3A). In addition, X-gal activity was found in the lens epithelium, corneal epithelium, and neurons in the forebrain regions (data not shown). Moreover, X-gal blue staining was not detected in tissue sections of the liver, heart, bone, and muscle of chicken embryos, suggesting eye- and neurospecific effects of the Pax6 P0 promoter. Chicken embryos injected with P1.2 mutant demonstrated a much stronger X-gal activity compared with P4.2-β-gal–injected embryos, but there was a weaker expression of X-gal activity in P2.3 mutant–injected chicken embryos (Fig. 3A). To confirm different intensity of X-gal signal in injected embryos was not due to copy number variations in the injected DNA, we used the remaining chicken embryonic heads fixed in blocks to extract DNA samples for detecting the level of β-galactosidase gene in tissues. These DNA samples were used as templates for semiquantitative PCR experiments. As shown in Figure 3B, there were uniform injection level of β-galactosidase gene in all tested embryos, and there were at least three individual embryos were tested for each DNA construct. These results are consistent with our previous studies demonstrating that there is a CTCF-interactive repressor in the region upstream from the Pax6 P0 promoter and that the CTCF-interactive repressor is functional in vivo in chicken retinal development.
Figure 3.
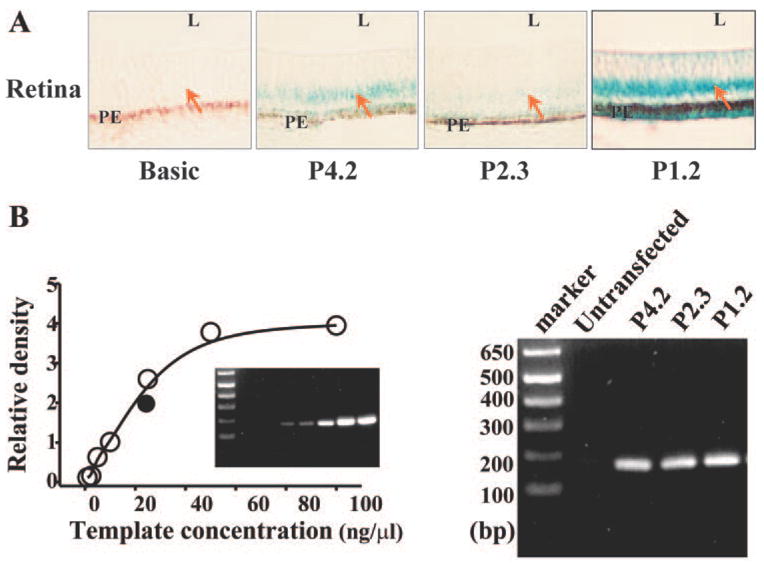
Expression of P4.2 reporter and deletion mutants in chicken retina. (A) X-gal staining of the retina of chicken embryo. P4.2 β-gal–Pax6 reporter and deletion mutants (P2.3 and P1.2) were introduced into the blastoderm of the chicken at E1. At E7, embryos were collected and stained with X-gal. Tissue sections were processed at a thickness of 10 μm. L and PE represent lens and pigment epithelial layer, respectively. (B) Copy numbers of the P4.2 reporter and mutants in transfected chicken embryos. Semi-quantitative PCR was performed with embryo DNA templates and a pair of primers specific to β-galactosidase gene sequences (180 bp apart). Chicken embryonic DNAs were isolated from the same individual embryos as shown in (A). The amount of PCR products as a function of template concentrations was plotted to generate a standard curve. DNA templates were diluted into 1, 2.5, 5, 10, 25, 50, and 100 ng/μL. There were at least six chicken embryos for each transfection experiment. Results from these experiments were very consistent.
Effect of CTCF on Pax6 P0 Promoter Activity and Pax6 Expression
To establish more firmly the role of CTCF in regulation of Pax6 P0 promoter activity, we subcloned the full-length cDNA encoding CTCF into a pcDNA4/To/A expression vector. The pcDNA4/To/A-CTCF construct that contains the CMV promoter was used for highly efficient, transient CTCF expression experiments in Rb cells. Cotransfection of pcDNA4/To/A-CTCF with P4.2-β-gal reporter significantly inhibited β-galactosidase activity in Rb cells (Fig. 4A). However, there are no changes in β-galactosidase activity in cells cotransfected with pcDNA4/To/A-CTCF and Pxba or P1.2 mutants, since the repressor element was deleted in these mutants (Fig. 4A). To study further the regulatory effect of CTCF on Pax6 expression, pcDNA4/To/A-CTCF–transfected Rb cells were analyzed by Northern and Western blot analysis. Overexpression of CTCF in Rb cells significantly increased CTCF mRNA expression and suppressed Pax6 protein expression (Fig. 4B). There were no changes in control experiments in GAPDH or β-actin expression levels. In contrast, endogenous CTCF mRNA was knocked down by RNAi technique to verify the regulatory correlation between CTCF and Pax6 expression. CTCF siRNA effectively knocked down CTCF protein expression in transfected cells up to 72 hours of detection period by Western blot analysis (Fig. 4C). Suppressed CTCF protein expression by RNAi resulted in an increase in Pax6 protein expression (Fig. 4C). The control experiments were performed by transfection of Rb cells with random and nonspecific siRNA. There were no changes in either CTCF mRNA or Pax6 protein levels in the control cells. Our results clearly indicate that, in vivo, CTCF downregulates Pax6 transcription and protein expression in Rb cells.
Figure 4.

Effect of CTCF on Pax6 P0 promoter activity and Pax6 expression in Rb cells. (A) Effect of overexpression of CTCF on activities of P4.2 reporter and deletion mutants. Full-length cDNA encoding CTCF was subcloned into pcDNA4/To/A vector (termed pcDNA-CTCF). The pcDNA-CTCF plasmid or pcDNA4/To/A vector was cotransfected into Rb cells with the P4.2 β-gal reporter and mutants (P1.2 and Pxba). The pBR-TK plasmid was also transfected as internal controls. Pax6 promoter activity was determined by measuring β-galactosidase activity with a luminescent β-gal system. Luciferase activity was determined for internal controls by using a luminescent lucif-erase assay kit. Pax6 promoter activities are presented as a ratio of measured β-galactosidase activity over luciferase activity (β-galactosidase activity[b]/luciferase activity). All three DNA constructs were transfected in each group of experiments. Each group of experiments was repeated at least three times (n = 3). Data were plotted as means with SE bars. *P < 0.05 and **P < 0.01, by comparison of those cells transfected with the blank vector and P4.2. (B) Effect of overexpression of CTCF on PAX6 expression. PcDNA-CTCF was transfected into Rb cells by electroporation. CTCF mRNA was detected 72 hours after transfection by Northern blot analysis and PAX6 expression was detected by Western blot analysis in CTCF-transfected Rb cells. Pax6 antibody (rabbit, 1:1000; Covance, Vienna, VA). The experiments were repeated by at least three independent experiments. The same membranes as used in Northern and Western blot analysis were stripped and rehybridized with GAPDH probe or β-actin antibody for internal controls. (C) Effect of CTCF knockdown on Pax6 expression. CTCF siRNA was introduced into Rb cells by lipofection for knockdown of CTCF mRNA. For control cells, a nonsense siRNA was transfected. CTCF and Pax6 expressions were detected 48 to 72 hours after siRNA transfection by Western blot analysis using anti-CTCF and anti-Pax6 antibodies (goat, 1:1000). The same membranes were stripped and rehybridized with anti-β-actin for the internal control. All results were repeated by at least three independent experiments.
Effect of CTCF on Pax6 Expression in the Retina of Transgenic Mice
The functional correlation between CTCF and Pax6 expression was further studied by introducing a CTCF transgene in the early developmental stages of the mouse embryo. The effect of overexpression of CTCF on Pax6 expression was observed in the retina of transgenic mice. Mouse embryos (from 28 mice) analyzed in our experiments were collected at E14. We found that there were significant increases in CTCF expression in the retina of CTCF transgenic mice compared with the control mice (Fig. 5A, left). Pax6 expression in retinal sections of CTCF transgenic mice was compared with that in the control retinas. In transgenic mice with high expression of CTCF, the expression of Pax6 protein was markedly decreased (Fig. 5B, right). To demonstrate further that the effect of overexpression of CTCF on Pax6 expression, retinal slides of consecutive sections were label with immunofluorescent stain using antibodies specific to CTCF and Pax6, respectively (Fig. 5B). Pax6 expression in the retinas from CTCF-overexpressing mice was markedly diminished compared with its wild-type counterpart. The results provide strong evidence in the retina of mice that there is a regulatory correlation between CTCF function and Pax6 expression in the developmental retina.
Figure 5.
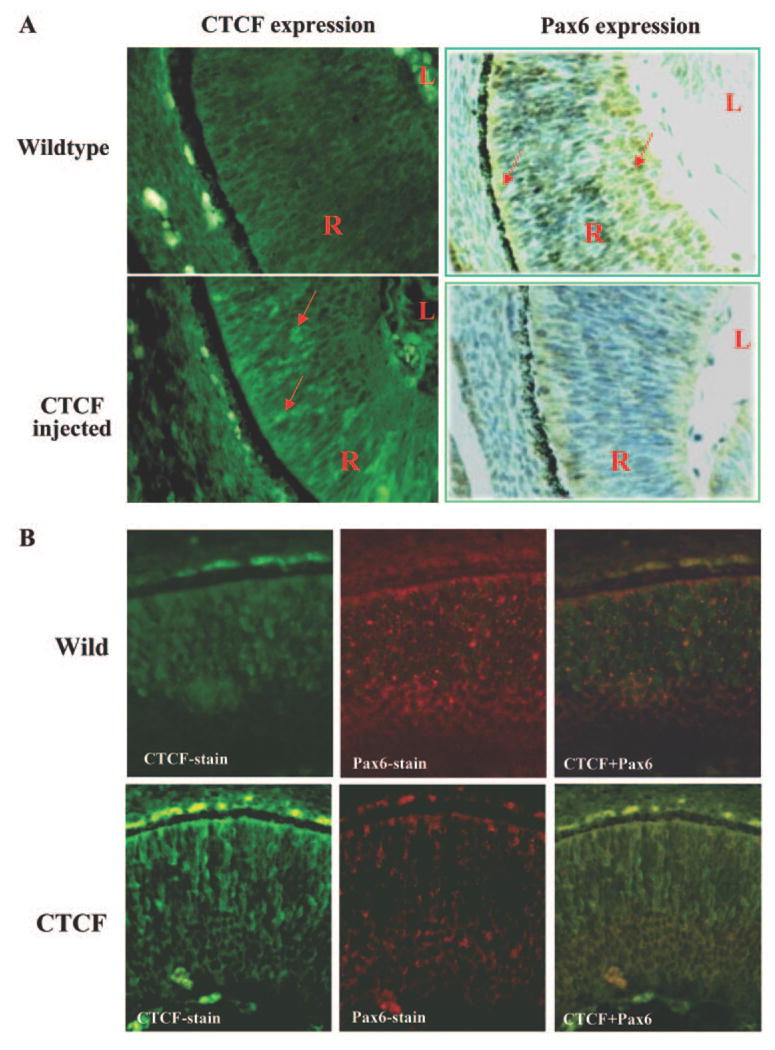
Effect of overexpression of CTCF on Pax6 expression in the mouse retina. (A) Alteration of Pax6 expression in the retina of CTCF transgenic mice. Expressions of CTCF and Pax6 proteins in mouse retinal tissue were stained with FTIC-labeled antibody and ABC kits, respectively. (B) Localization of CTCF and Pax6 expression in the retina in consecutive slide sections. Pax6 and CTCF proteins were detected with 1:100 rabbit anti-pax6 antibody (Abcam, Cambridge, MA) and 1:100 rabbit anti-CTCF, respectively. Positive staining signals were visualized by using Cy3-goat anti rabbit IgG for 1 hour at room temperature. Mouse embryos were collected at E14 and fixed in 4% paraformaldehyde-PBS. Tissues were embedded in wax and sectioned at 6 μm. CTCF in the retina of mice was detected with a light/fluorescence microscope with a 20× objective. Pax6 in the retina of mice was detected with a light/fluorescence microscope with a 20× objective. Images were photographed with a digital camera at 4.1 megapixels. Arrows: positively strained cells in the retina.
Discussion
We found that there was a physiological correlation between the expression of CTCF and Pax6 in the retina and retina-derived cells. Northern and Western blot analyses provided further evidence that CTCF mRNA and protein was indeed expressed in cultured eye-derived Rb cells and hematopoietic cells, but Pax6 protein was specifically expressed in Rb cells (Fig. 1). Expression of CTCF in Rb cells increased in response to stimulation of FBS accompanied by a decrease in Pax6 expression, suggesting functional roles of serum-containing cytokines in regulating CTCF and Pax6 activities (Fig. 1B). Additional experiments were performed to confirm the regulatory relationship between CTCF and Pax6. First, cDNA encoding full-length CTCF was introduced in Rb cells to induce overexpression of CTCF. The CTCF mRNA level in transfected cells was markedly increased, and endogenous Pax6 protein expression was suppressed (Fig. 4B). Second, synthesized CTCF siRNA 21 nucleotides in length, according to the sequence of CTCF mRNA, was introduced in Rb cells to knockdown mRNA expression of CTCF. Suppression of CTCF by knockdown of CTCF mRNA resulted in decreases in CTCF protein levels and increases in endogenous Pax6 protein expression in CTCF siRNA-transfected Rb cells (Fig. 4C). Finally, the effect of CTCF activity on Pax6 expression was studied in the retina of transgenic mice into which the CTCF transgene was injected obtained during the fertilized egg stage from donor mice. There was a marked increase in expression of CTCF protein in the developing retina of the CTCF transgenic mice compared with wild-type mice. As expected, Pax6 protein expression in the developing retina of CTCF transgenic mice was significantly decreased. The results provide firm evidence indeed that there is a functional correlation between CTCF and Pax6 expression in the retina of mice. In summary, the present study identified a novel functional role of CTCF in regulating Pax6 gene expression in the retina. This regulatory function of CTCF is responsive to stimulation of serum-containing cytokines and through controlling Pax6 P0 promoter activity. CTCF protein can interact with the regulatory element and inhibit activity of the Pax6 P0 promoter in developing retinal cells.
We found in the present study that the ZF protein CTCF in the retina involved the regulation of homeobox Pax6 expression that may be induced by a mechanism involving transcriptional inhibition of the gene. CTCF interacts with a repressor element located in the region of −1.2-kb upstream Pax6 P0 promoter.16 Our studies using P4.2-β-gal construct and deletion mutants to detect the repressor element reveal a similar result from a previous report that in the area of −3.5 to −3.3 kb upstream, the Pax6 P0 promoter contains an ectoderm enhancer (EE) for Pax6 P0 promoter activity.14 It is also consistent with our previous study about a mutant (Pxba) lacking a 500-bp fragment −1.2 kb upstream from the P0 promoter.16 Expression of mutant Pxba demonstrated a significant enhancement of Pax6 P0 activity in Rb cells because the EE element is retained and the CTCF interactive element is removed in the reporter construct (Fig. 2C). In the other group of experiments, cDNA encoding full-length CTCF was cotransfected into Rb cells with P4.2 reporter and mutants (P1.2 and Pxba). We expected that the interaction between CTCF and P0 promoter activation would be functional in Rb cells. Indeed, overexpression of CTCF suppressed Pax6 P0 promoter activity in Rb cells cotransfected with the P4.2 reporter that retains both the EE and CTCF repressor elements (Fig. 4A). In contrast, overexpression of CTCF in Rb cells cotransfected with Pxba and P1.2 mutants did not affect P0 reporter activity, because both of these mutants lack the element containing CTCF-interactive sites. These results support the notion that the inhibitory effect of CTCF overexpression on Pax6 transcription is regulated through the interaction of CTCF and Pax6 P0 promoter activity. Thus, we have for the first time found in retina-related cells that EE activity is inhibited in retina-derived cells by the further downstream repressor element in response to CTCF activation, resulting in suppression of P0 promoter activity and Pax6 transcription.
For further supporting evidence, we injected P4.2 and its mutant reporter constructs in the chicken embryo to test the effect of overexpression of CTCF on Pax6 expression in vivo in the retina. P0 promoter activity was observed in the chicken retina that was injected with the P4.2 reporter construct, but was not observed in the chicken retina injected with a P2.3 mutant reporter construct because the EE element is deleted in the P2.3 mutant. However, there was significant enhanced reporter activity in the chicken retina that was injected with a P1.2 mutant reporter because both EE and CTCF repressor elements are eliminated in this mutant. These results lead us to the conclusion that the regulatory effect of CTCF on Pax6 transcription may involve several different mechanisms including (1) the blockage of the access of enhancer region for promoters that has been shown in H19 and IGF-2 transcription—namely, boundary functions22,30,31; (2) the blockage of elongation stages in transcription; (3) the prevention of the target gene from being transcribed by removing acetyl groups from histone proteins in the chromatin; and (4) the inhibition of the transcription-initiation complex. In addition, our findings about the effect of CTCF on the suppression of Pax6 P0 promoter activity are consistent with the previous report that CTCF protein serves as a repressive transcription factor in promoters of the chicken and human c-myc genes.18,29,32,33 As mentioned earlier, CTCF may regulate Pax6 transcription through an insulation mechanism very similar to the mechanism through which CTCF blocks the access of enhancer region to promoter in H19 and IGF2 transcription, known as boundary functions.22,30,31 However, the results presented in this study provide additional evidence to support the mechanism that CTCF interacting with binding sites located between the enhancer and P0 promoter may work just as an insulator-binding site in regulating Pax6 gene expression.
Acknowledgments
Supported by National Eye Institute Grants EY12953 and EY15281 (LL) and SJU- Shanghai Second Medical University (SSMU) Grant 2003hzjj001 (ZL).
Footnotes
Disclosure: T. Li, None; Z. Lu, None; L. Lu, None
References
- 1.Underhill DA. Genetic and biochemical diversity in the Pax gene family. Biochem Cell Biol. 2000;78:629–638. [PubMed] [Google Scholar]
- 2.Walther C, Gruss P. Pax-6, a murine paired box gene, is expressed in the developing CNS. Development. 1991;113:1435–1449. doi: 10.1242/dev.113.4.1435. [DOI] [PubMed] [Google Scholar]
- 3.Martin P, Carriere C, Dozier C, et al. Characterization of a paired box- and homeobox-containing quail gene (Pax-QNR) expressed in the neuroretina. Oncogene. 1992;7:1721–1728. [PubMed] [Google Scholar]
- 4.Koroma BM, Yang JM, Sundin OH. The Pax-6 homeobox gene is expressed throughout the corneal and conjunctival epithelia. Invest Ophthalmol Vis Sci. 1997;38:108–120. [PubMed] [Google Scholar]
- 5.Davis JA, Reed RR. Role of Olf-1 and Pax-6 transcription factors in neurodevelopment. J Neurosci. 1996;16:5082–5094. doi: 10.1523/JNEUROSCI.16-16-05082.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gehring WJ. The master control gene for morphogenesis and evolution of the eye. Genes Cells. 1996;1:11–15. doi: 10.1046/j.1365-2443.1996.11011.x. [DOI] [PubMed] [Google Scholar]
- 7.Altmann CR, Chow RL, Lang RA, Hemmati-Brivanlou A. Lens induction by Pax-6 in Xenopus laevis. Dev Biol. 1997;185:119–123. doi: 10.1006/dbio.1997.8573. [DOI] [PubMed] [Google Scholar]
- 8.Hill RE, Favor J, Hogan BL, et al. Mouse small eye results from mutations in a paired-like homeobox-containing gene. Nature. 1991;354:522–525. doi: 10.1038/354522a0. [DOI] [PubMed] [Google Scholar]
- 9.Jordan T, Hanson I, Zaletayev D, et al. The human PAX6 gene is mutated in two patients with aniridia. Nat Genet. 1992;1:328–332. doi: 10.1038/ng0892-328. [DOI] [PubMed] [Google Scholar]
- 10.Hanson IM, Fletcher JM, Jordan T, et al. Mutations at the PAX6 locus are found in heterogeneous anterior segment malformations including Peters’ anomaly. Nat Genet. 1994;6:168–173. doi: 10.1038/ng0294-168. [DOI] [PubMed] [Google Scholar]
- 11.Quiring R, Walldorf U, Kloter U, Gehring WJ. Homology of the eyeless gene of Drosophila to the small eye gene in mice and aniridia in humans. Science. 1994;265:785–789. doi: 10.1126/science.7914031. [DOI] [PubMed] [Google Scholar]
- 12.Plaza S, Dozier C, Saule S. Quail Pax-6 (Pax-QNR) encodes a transcription factor able to bind and trans-activate its own promoter. Cell Growth Differ. 1993;4:1041–1050. [PubMed] [Google Scholar]
- 13.Plaza S, Dozier C, Turque N, Saule S. Quail Pax-6 (Pax-QNR) mRNAs are expressed from two promoters used differentially during retina development and neuronal differentiation. Mol Cell Biol. 1995;15:3344–3353. doi: 10.1128/mcb.15.6.3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu PX, Zhang X, Heaney S, Yoon A, Michelson AM, Maas RL. Regulation of Pax6 expression is conserved between mice and flies. Development. 1999;126:383–395. doi: 10.1242/dev.126.2.383. [DOI] [PubMed] [Google Scholar]
- 15.Williams SC, Altmann CR, Chow RL, Hemmati-Brivanlou A, Lang RA. A highly conserved lens transcriptional control element from the Pax-6 gene. Mech Dev. 1998;73:225–229. doi: 10.1016/s0925-4773(98)00057-4. [DOI] [PubMed] [Google Scholar]
- 16.Li T, Lu Z, Lu L. Regulation of eye development by transcription control of CCCTC binding factor (CTCF) J Biol Chem. 2004;279:27575–27583. doi: 10.1074/jbc.M313942200. [DOI] [PubMed] [Google Scholar]
- 17.Li T, Lu L. Epidermal growth factor-induced proliferation requires Down-regulation of Pax6 in corneal epithelial cells. J Biol Chem. 2005;280:12988–12995. doi: 10.1074/jbc.M412458200. [DOI] [PubMed] [Google Scholar]
- 18.Filippova GN, Fagerlie S, Klenova EM, et al. An exceptionally conserved transcriptional repressor, CTCF, employs different combinations of zinc fingers to bind diverged promoter sequences of avian and mammalian c-myc oncogenes. Mol Cell Biol. 1996;16:2802–2813. doi: 10.1128/mcb.16.6.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klenova EM, Nicolas RH, U S, et al. Molecular weight abnormalities of the CTCF transcription factor: CTCF migrates aberrantly in SDS-PAGE and the size of the expressed protein is affected by the UTRs and sequences within the coding region of the CTCF gene. Nucleic Acids Res. 1997;25:466–474. doi: 10.1093/nar/25.3.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Awad TA, Bigler J, Ulmer JE, et al. Negative transcriptional regulation mediated by thyroid hormone response element 144 requires binding of the multivalent factor CTCF to a novel target DNA sequence. J Biol Chem. 1999;274:27092–27098. doi: 10.1074/jbc.274.38.27092. [DOI] [PubMed] [Google Scholar]
- 21.Vostrov AA, Quitschke WW. The zinc finger protein CTCF binds to the APBbeta domain of the amyloid beta-protein precursor promoter; evidence for a role in transcriptional activation. J Biol Chem. 1997;272:33353–33359. doi: 10.1074/jbc.272.52.33353. [DOI] [PubMed] [Google Scholar]
- 22.Bell AC, Felsenfeld G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature. 2000;405:482–485. doi: 10.1038/35013100. [DOI] [PubMed] [Google Scholar]
- 23.Nakagawa H, Chadwick RB, Peltomaki P, Plass C, Nakamura Y, de La Chapelle A. Loss of imprinting of the insulin-like growth factor II gene occurs by biallelic methylation in a core region of H19-associated CTCF-binding sites in colorectal cancer. Proc Natl Acad Sci USA. 2001;98:591–596. doi: 10.1073/pnas.011528698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Percec I, Bartolomei MS. Genetics Do X chromosomes set boundaries? Science. 2002;295:287–288. doi: 10.1126/science.1068663. [DOI] [PubMed] [Google Scholar]
- 25.Filippova GN, Lindblom A, Meincke LJ, et al. A widely expressed transcription factor with multiple DNA sequence specificity, CTCF, is localized at chromosome segment 16q22.1 within one of the smallest regions of overlap for common deletions in breast and prostate cancers. Genes Chromosomes Cancer. 1998;22:26–36. [PubMed] [Google Scholar]
- 26.Muramatsu T, Mizutani Y, Ohmori Y, Okumura J. Comparison of three nonviral transfection methods for foreign gene expression in early chicken embryos in ovo. Biochem Biophys Res Commun. 1997;230:376–380. doi: 10.1006/bbrc.1996.5882. [DOI] [PubMed] [Google Scholar]
- 27.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 28.Li T, Dai W, Lu L. Ultraviolet-induced junD activation and apoptosis in myeloblastic leukemia ML-1 cells. J Biol Chem. 2002;277:32668–32676. doi: 10.1074/jbc.M203519200. [DOI] [PubMed] [Google Scholar]
- 29.Klenova EM, Nicolas RH, Paterson HF, et al. CTCF, a conserved nuclear factor required for optimal transcriptional activity of the chicken c-myc gene, is an 11-Zn-finger protein differentially expressed in multiple forms. Mol Cell Biol. 1993;13:7612–7624. doi: 10.1128/mcb.13.12.7612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hark AT, Schoenherr CJ, Katz DJ, Ingram RS, Levorse JM, Tilghman SM. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature. 2000;405:486–489. doi: 10.1038/35013106. [DOI] [PubMed] [Google Scholar]
- 31.Kanduri C, Pant V, Loukinov D, Pugacheva E, et al. Functional association of CTCF with the insulator upstream of the H19 gene is parent of origin-specific and methylation-sensitive. Curr Biol. 2000;10:853–856. doi: 10.1016/s0960-9822(00)00597-2. [DOI] [PubMed] [Google Scholar]
- 32.Lobanenkov VV, Gudvin GG. CCCTC-binding protein: a new nuclear protein factor which interaction with 5′-flanking sequence of chicken c-myc oncogene correlates with repression of the gene (in Russian) Dokl Akad Nauk SSSR. 1989;309:741–745. [PubMed] [Google Scholar]
- 33.Lobanenkov VV, Nicolas RH, Adler VV, et al. A novel sequence-specific DNA binding protein which interacts with three direct repeats of the CCCTC-motif in the 5′-flanking sequence of the chicken c-myc gene. Oncogene. 1990;5:1743–1753. [PubMed] [Google Scholar]


