Abstract
Background
The human gastric pathogen Helicobacter pylori is co-evolved with its host and therefore, origins and expansion of multiple populations and sub populations of H. pylori mirror ancient human migrations. Ancestral origins of H. pylori in the vast Indian subcontinent are debatable. It is not clear how different waves of human migrations in South Asia shaped the population structure of H. pylori. We tried to address these issues through mapping genetic origins of present day H. pylori in India and their genomic comparison with hundreds of isolates from different geographic regions.
Results
We attempted to dissect genetic identity of strains by multilocus sequence typing (MLST) of the 7 housekeeping genes (atpA, efp, ureI, ppa, mutY, trpC, yphC) and phylogeographic analysis of haplotypes using MEGA and NETWORK software while incorporating DNA sequences and genotyping data of whole cag pathogenicity-islands (cagPAI). The distribution of cagPAI genes within these strains was analyzed by using PCR and the geographic type of cagA phosphorylation motif EPIYA was determined by gene sequencing. All the isolates analyzed revealed European ancestry and belonged to H. pylori sub-population, hpEurope. The cagPAI harbored by Indian strains revealed European features upon PCR based analysis and whole PAI sequencing.
Conclusion
These observations suggest that H. pylori strains in India share ancestral origins with their European counterparts. Further, non-existence of other sub-populations such as hpAfrica and hpEastAsia, at least in our collection of isolates, suggest that the hpEurope strains enjoyed a special fitness advantage in Indian stomachs to out-compete any endogenous strains. These results also might support hypotheses related to gene flow in India through Indo-Aryans and arrival of Neolithic practices and languages from the Fertile Crescent.
Background
Analysis of genetic diversity in microorganisms normally reflects patterns of their own evolution although it is very rare that this can portray their hosts' evolution. Co-evolution between host and pathogens can be explained only if pathogens are not horizontally transmitted, and this supports a possible phylogenetic and evolutionary parallel of the host and pathogens. Sadly, in many cases frequent horizontal transmission separates the evolution of the bacterium from that of the host. However, for some pathogens, such as H. pylori [1-3], and JC viruses [4], transmission is faithfully restricted to families within specific communities. This phenomenon has in recent times provided evidence regarding patterns of human migration [2,4,5] in different continents.
The human gastric pathogen H. pylori is presumed to have co-evolved with its host [6] and established itself in the human stomach possibly millions of years ago [7]. It has been recognized recently as a reliable biological marker of host-pathogen co-evolution and ancient human migration based on sequence variation in select gene loci. H. pylori are genetically diverse to the extreme, providing about 1,400 informative sites within 3.5 to 4.5 kb of sequence from housekeeping genes, and their global genetic structure based on such sequence-haplotypes parallels that of humans [2]. Moreover, epidemiological studies have shown that transmission occurs predominantly within families [8-11]. H. pylori therefore, could provide a window into human origins and migration [1,3] and the impact of religions and social systems on stratification of human ethnic groups [12].
A landmark study based on PCR based DNA motif analysis proposed that H. pylori jumped recently from animals to humans and, therefore, the acquisition of H. pylori by humans may be a recent phenomenon [13]. This study has been the basis for the idea of 'H. pylori free New World' [13]. However, several independent studies based on large-scale analyses of candidate gene polymorphisms contrasted the idea of recent acquisition and suggest that H. pylori might have co-evolved with humans [1,6,14].
Using the same set of Peruvian isolates described earlier by Kersulyte et al. [13], Devi et al. [3], from our group have suggested that the genetic make up of south American isolates could be an admixture of ancestral and modern lineages of H. pylori. They clearly highlighted presence of ancestral H. pylori in Peruvians that possibly survived influxes of Spanish strains from Iberian expansions in Peru about 500 years ago. Also, according to this study, the survival advantage of indigenous strains was possibly due to the acquisition of western type cagPAIs from newly arrived Spanish strains.
Previous genotyping studies on Indian isolates have largely targeted molecular epidemiological issues. However, Wirth et al. [12], for the first time, using H. pylori genotypes, addressed issues such as impact of two different religions and societal systems on stratification of human ethnic groups [12] in the remotest north eastern Ladakh area of India. In view of intriguing ideas on ancient origin of H. pylori, and the fact that ancient origins and arrival of H. pylori are hardly known in the context of the vast South Asian continent, additional evidences based on strains from different geographical regions of Asia are clearly needed.
In this study, we attempted to unravel population genetic structure and gene pool diversity of Indian isolates of H. pylori from culturally and linguistically diverse ethnic Indians. The main objective behinds the study has been to explore genetic features of the strains that might explain their ancestral origin and might help reconstruct different waves of pre-historic human migration in India. We also looked if it is possible to link some of the native strains to their ancestors in West Asia, Eurasia or Europe.
Results
DNA isolates, diagnostic PCR and epidemiological genotyping
DNA quality and purity was confirmed by agarose gel electrophoresis and diagnostic PCRs revealed presence of cagA, iceA, vacA, glmM, babB and oipA genes in all the Indian isolates we tested. The molecular epidemiological features of all the 63 strains we analyzed have been elaborated in Figure 1. Our isolates were quite diverse with respect to the plasticity region ORFs that we analyzed and no specific signature was seen dominant as regards to the arrangement or rearrangement of these ORFs. This validated that all the isolates that we looked at were in fact independent and did not represent any derivatives of clonal evolution.
Figure 1.
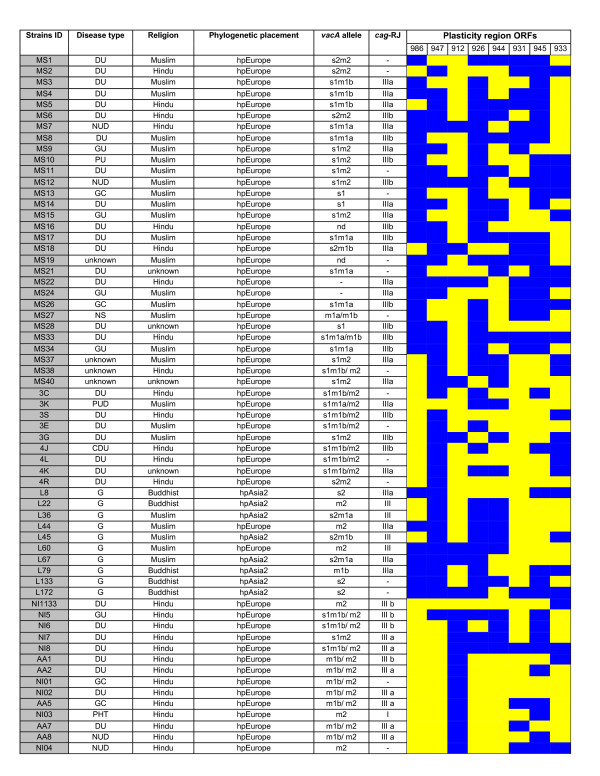
Detailed characteristics of Indian H. pylori isolates used in the study. [Yellow, region amplified or present; Blue, region absent or rearranged; -, region failed to amplify].
Specific primers amplifying different alleles (see methods section) were used to analyze the vacA allelic diversity. The sizes of the amplified products for vacA s1 and vacA s2 were 259 bp and 286 bp respectively. Of the 63 isolates analyzed, the s1 allele was detected in 33 (52.3%) and the s2 allele type was detected in 11 (17.4%) strains. The m1 variant was detected in 34 (53.9%) and the m2 variant in 37 (58.7%). The highly toxigenic vacA allele combination s1m1 was found to be dominant (33.3%) as compared to other vacA allele types. The vacA genotype s1m2 was detected in 9 isolates (14.2%) whereas vacA s2m1 and vacA s2m2 genotypes were detected in 4 isolates (6.3%) each. Not all the isolates yielded full vacA amplicons, as regions of vacA gene, in particular, the signal region posed difficulty in amplification. This is a very common phenomenon observed in H. pylori owing to frequent recombination. The vacA alleles have been shown to differ in frequency and type among East Asian isolates [15], for instance, s1c is the predominant signal sequence allele among East Asian isolates [16]. Typically, the vacA s1c was found to be completely absent in the Indian isolates.
Multilocus sequence analysis
We report that almost all of the H. pylori strains from India share significant homology to the members of sub-population hpEurope. A total of 33 MLST profiles based on DNA sequence of a concatenated multigene comprising of 7 individual gene loci (atpA, efp, mutY, ppa, trpC, ureI and yphC) were generated from Indian isolates. Data comprising of these MLST profiles were subjected to comparative genomic analysis with ~400 other H. pylori sequences from different geographical and ethnic groups [11]. Such analyses upon construction of a neighbor-joining tree in MEGA 3.1 software using Kimura-2 parameter revealed clear geographic distribution of various H. pylori populations and sub-populations, essentially in accordance with the previous results [1,3,17]. All the Indian isolates from North and South India and 2 of them from Ladakh clustered under hpEurope. Seventeen Ladakhi isolates clustered tightly to form a separate branch, hpAsia2. Results of MLST analysis in MEGA3.1 were successfully reproduced using NETWORK based phylogeny, which revealed similar acquaintances for H. pylori in India. Mirroring the spread of human populations from Africa, our network analysis suggests the co-evolution of H. pylori with Homo sapiens, as also suggested recently [6]. Both the domains of the Network tree based on 650 (data not shown) and 665 (Figure 2, left) mutating positions clearly separated African from non-African sequences. The second domain seemed to harbor higher phylogenetic information, since the resulting graph is more clearly structured, with a more accurate separation among European, Amerindian, Asian and Australasian lineages. The Indian H. pylori sequences were clustered within the European portion of the network, wherein the first domain identifies a separate branch, encompassing the majority of the Ladakhi samples, as a distinct sub-population of hpAsia2 within the European variability, and remarking the isolation of the human host population. However, many of the Ladakhi Muslim samples clustered in hpEurope and revealed a significant sequence similarity to the mainland Indian samples. These results are in agreement with previous studies on the hypervariable region of human mitochondrial DNA that showed the common origin of European and Indian populations [18] and the relative homogeneity of Indian populations regardless of their ethnic and linguistic affiliation [19].
Figure 2.
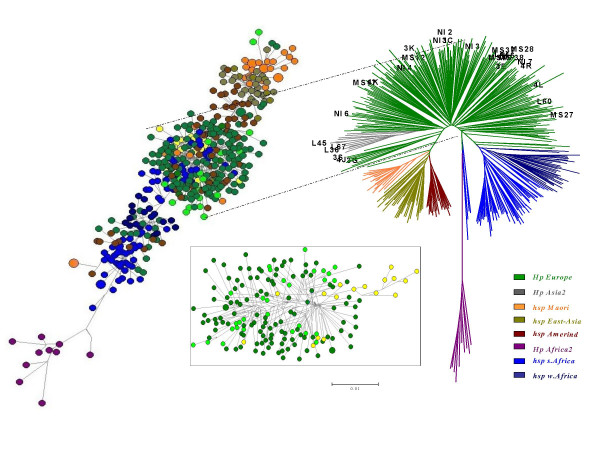
Neighbor joining tree (Kimura 2-parameter) (right) showing the global population structure of H. pylori wherein Indian isolates are highlighted. The phylogenetic tree was based on a total of 23 sequence records of South and North Indian isolates while incorporating ~400 other sequence records from pubMLST database representing different H. pylori populations and sub populations in the world. The population genetic structure was investigated by determining the multilocus haplotypes based on concatenated sequences of seven unlinked housekeeping genes that are scattered around the H. pylori chromosome. Individual isolates were assigned to bacterial populations called hpEastAsia (sub-populations: hspEAsia, hspMaori, hspAmerind), hpEurope, hpAfrica1 (hspSAfrica, hspWAfrica), hpAsia2 and hpAfrica2 [11]. Representatives from each of these (sub)-populations were chosen for subsequent analysis of the cagPAI. Isolates from the population hpAfrica2 do not contain cagPAI. Phylogenetic relationships were also estimated through NETWORK analysis (left) based on 665 mutating positions that revealed the co-evolution of the H. pylori genome. The Ladakhi (yellow) and other Indian (light green) lineages were more clearly discerned within the European (dark green) cluster (centre box), when analyses based on the remaining 650 mutating positions were performed. For the Neighbor-joining tree (right), the bootstrap values of the interior branches as calculated in MEGA, were significantly high to indicate the correct topology of the branches within the clades.
Analysis of the cagPAI and its Right Junction (RJ) motifs
Overlapping primer amplification to span entire cagPAI worked reproducibly with our isolates; Figure 3(A) reveals complete PCR output for the ~38 kb cagPAI region in 5 representative strains MS38, MS40, 3K, 4K and 3C. All the constituent genes of the PAI were successfully amplified for all the Indian isolates studied. To get more insights into composition and arrangement of the gene loci within the PAI, complete sequencing of the cagPAI of isolate 3K was performed. This isolate was from a patient with peptic ulcer disease (PUD) from South India. The size of complete cagPAI of this isolate was 36,876 bp with a G+C content of 35.9. The sequence composition and gene order in the cagPAI of 3K was compared to those of the three completely sequenced strains 26695, J99 and HPAG1 which revealed some minor differences such as fused HP0521 and HP0522 genes due to the deletion of a single nucleotide at the 3' end of HP0521. Similarly single or dinucleotide differences were observed in the cagX (HP0528), cagN (HP0538) and cagE (HP0544) and most of these insertions and deletions were observed in the intergenic regions. Broadly, the cagPAI genes were very conserved as regards to the amino acid sequences when compared with at least 15 different publicly available cagPAI sequences.
Figure 3.
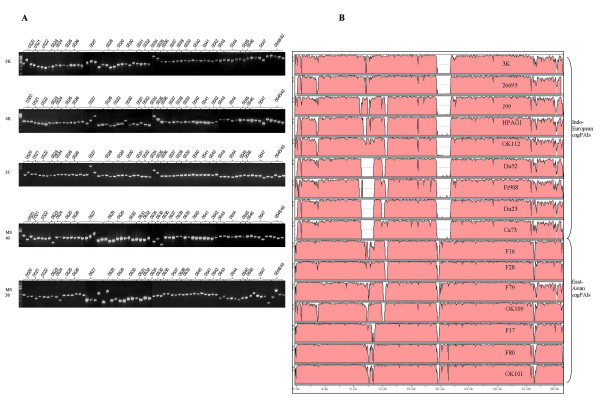
Comparative genomic analysis of the cagPAIs from Indian isolates. A) PCR based analysis of the complete cagPAI of 5 representative hpEurope Isolates: 3K, 4K, 3C, MS40 and MS38 from India. Overlapping PCR primers amplified the whole cagPAI indicating the intactness of the PAI in these isolates. B) Global pair-wise alignments of whole cagPAI sequences of different H. pylori isolates were generated by VISTA using default parameters [47]. The OK129 genome was taken as the base sequence (not shown) and rest of the sequences were aligned against it. The X-axis denotes length of the sequence under consideration and the Y-axis conveys homology in % with the base genome sequence). The Indian hpEurope isolate, 3K was aligned with other whole cagPAI sequences from GenBank along with the cagPAIs of HP 26695, HPJ99 and HPAG1. The accession numbers for the public domain sequences of the cagPAIs from Europe [9] and Japan [49] that we used in our analyses, were as follows – Ca73 (AY330638 and AY330639), Du23:2 (AY330643 and AY330644), Du52:2 (AY330640, AY330641 and AY330642), F80 (AB120421), OK112 (AB120425), F16 (AB120416), F17 (AB120417), F28 (AB120418), F79 (AB120420), OK101 (AB120422), OK109 (AB120424). Sequence of the French isolate, Fr 908 was determined in this study (EF195721). While the cagPAI sequence of the Indian isolate 3K (hpEurope) was found to be genetically highly similar to and aligning closely with the 26695 sequence, it also revealed significant sequence similarities with other isolates of European origins (that harbor Western type of cag EPIYA sequences) such as HPAG1, OK112, Du52, Du23, Ca73, J99 and Fr908. It was however largely unrelated to the East Asian like isolates (mainly harboring Asian type cag EPIYA sequences) such as F16, F28, F79, OK109, F17, OK101 and F80.
cag-RJ (the extreme right junction of the cagPAI, between 3' end of the cagA gene and the start of the glutamate racemase – glr) was studied for our 63 isolates where 99% isolates harbored type III motif. A total of 47 of 63 strains (75%) gave positive PCR results for cag-RJ (Figure 1). The type III motif was found in 27 of 39 South Indian isolates and 20 of 24 North Indian isolates. It is noteworthy that cag-RJ typeIII motifs are genetically close to European type I motifs probably due to an ancient insertion event, followed by recombinational scrambling among type I and III lineages [13]. We did not find in our Indian isolates any type II motifs, which constitute a signature characteristic of East Asian gene pool.
Genetic relationship of Indian isolates based on cagA and whole cagPAI sequences
A full-length cagPAI sequence based alignment was constructed using the Indo-European type 3K and Afro-European type Fr908 (French patient isolate) sequences determined in this study, along with 15 different whole cagPAI sequence from GenBank: Ca73, Du23: 2, Du52: 2, F16, F17, F28, F79, F80, OK101, OK109, OK112, OK129, 26695, J99 and HPAG1. Our South Indian isolate, 3K, was found to be aligning with the Western cagPAI sequences (Figure 3B).
We examined relatedness of the cagA gene sequences of tribal isolates from India to the mainstream Indian isolates and the European isolates by analyzing a 219 bp informative fragment near the 5' end of cagA which usually distinguishes the European and the East Asian strains [20]. Comparative sequence analysis was used to construct phylogenetic relationship in MEGA3.1. All the sequence records corresponding to the isolates of Santhal and Oraon tribals revealed homologies to the main stream Indian strains from Hyderabad, Lucknow and Bengal and also to all the representative European strains. These tribal isolates did not cluster with East Asian strains (Figure 4).
Figure 4.
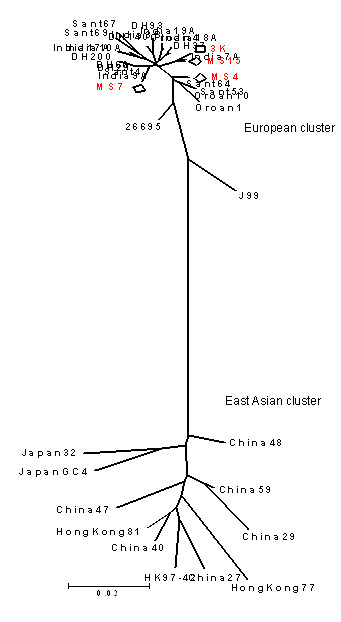
Phylogenetic tree based on the 5' end sequence of the cagA (an informative 219 bp segment of cagA was used to align sequences from unrelated isolates) suggests possible common origins for isolates from ethnic Indians and the tribal. Representative Indian genotypes (3K, MS4, Ms7 and MS15) based on this 219 bp sequence clustered tightly with previously determined genotypes of strains obtained from ethnic Bengalis [India3B (AF202219), India7A (AF202220), India9A (AF202221), India10A (AF202222), India17A (AF202223), India18A (AF202224), India19A (AF202225), DH140 (AY169293), DH200 (AY169294), DH29 (AY169295), DH37 (AY169296), DH60 (AY169297), DH93 (AY169298)] and Santhal and Oraon tribals [Sant4 (AY162446), Sant53 (AY162447), Sant64 (AY162448), Sant67 (AY162449), Sant69 (AY162450), Oraon1 (AY162451), Oraon10 (AY162452), Oraon4 (AY162453)] [20]. All the East Asian strains [China27 (AJ252979), China29 (AJ252980), China40 (AJ252982), China48 (AJ252983), China47 (AJ252985), China59 (AJ252986), Hongkong77 (AF198485), Hongkong81 (AF198486), Hongkong97-42 (AF239733), Japan GC4 (AF198484), Japan32 (AJ239726)], however, clustered together and formed a separate cluster.
This makes it clear that the cagPAI of Indian strains is a completely evolved one and probably was acquired from a European source, well before the arrival of H. pylori in India. This is also evident from the fact that the Indian strains, though of a European descent, do not share characteristic features of Asian cagPAIs.
Discussion
Although the Indian peninsula has seen many different waves of population migration [21], the Paleolithic archaeological evidence is not clearly visible to understand peopling of this country [22]. Nonetheless, the Indus Valley and Harappan civilizations portray footprints of Neolithic period [23] suggestive of the arrival of Indo-European speakers who established the caste system, an anthropologically significant prehistoric event [24,25]. The cultural and historical importance of the arrival and settlement of the Indo-Aryans is undisputed, but it is not clear if this was established through 'replacement of the existing people by outsiders' [22] or did the 'people already in India changed their habits and cultures?' [22]. Such questions have never been addressed in an unambiguous manner, even though the potential of polymorphic DNA markers in reconstruction of human migration and phylogeography [26,27] has long been appreciated. It appears that even carefully planned geographic genomics studies remained largely speculative due to the lack of a universal 'gold standard' as the classical mitochondrial DNA markers offer too few informative polymorphisms and the newly developed Y – chromosome markers are even less polymorphic than mitochondrial hypervariable regions [2]. Lately, new genetic models were successfully harnessed based on parasites and pathogens that probably accompanied their human host during evolution and much of the human history including migrations and expansions [2,4,5] in different continents. Such approaches constitute an attractive alternative to reconstruct human origins and spreads, population dynamics and bottlenecks, wars and displacements, farming and plagues etc.
Our study was aimed at tracking ancient origins of the Indian H. pylori through a two-pronged approach to i) substantiate European link of the pathogen in India and ii) to prove that the pathogenicity island was also of European origin and this PAI has not been a 'recent' addition to the genome of Indian H. pylori. Our analyses, based on MLST and comprehensive genotyping of the cagPAI, linked about 100% of the Indian isolates to H. pylori sub-population hpEurope. This perhaps conveys the message that H. pylori was most probably introduced to the Indian subcontinent by ancient Indo-European nomadic people and our findings, therefore, are consistent with the idea of a possible gene flow into India with the arrival of Indo-Aryans.
Overall, based on the MLST data (Figure 2) and the cagPAI patterns (Figure 3), we suggest that H. pylori might have arrived in India probably at the same time when Indo-European language speaking people crossed into India (~4000–10,000 years before present). Alternatively, the unquestionable common origin of Indian strains with the European ones could be actually more ancient, following the upper Paleolithic spread of Homo sapiens in Eurasia, as suggested by mtDNA variability [18], and our data on H. pylori MLST do not rule out this possibility.
Present day India represents a 'genetic playground' with tremendous diversity of cultures, and languages. However, the people are largely stratified as tribals and nontribals [25]. Four main language families are spoken, the largest being, Indo European (IE), which is prevalent in North, and the second largest Dravidian (DR) group represents languages spoken in the South [28]. The other two language groups include Tibeto-Burman (TB) of the Sino-Tibetan and the Austro-Asiatic (AA) families, largely spoken in far North and the North-east India. While most of the IE speakers belong to castes, the majority of the tribal communities (>450) speak about 750 different dialects that fit within any one of the other three language families (DR, TB, AA) [25,28]. Such an enormous cultural diversity might argue for many different populations and sub-populations of H. pylori. But until now, and including this study, H. pylori with genetic features of hpEurope have only been reported from India [29,30]. Even the newly described sub-population hpAsia2 from Ladakh is also a variant of hpEurope and many Ladakhi strains that we looked at in this study, clustered with European H. pylori clade (Figure 2). Also, the cagA sequences from H. pylori belonging to tribal Oraon and Santhals were indistinguishable from those of the mainstream Indians and Europeans (Figure 4), indicating sweeping spread of a single H. pylori genotype across the Indian peninsula. Moreover, we did not document presence of any other H. pylori populations and sub-populations such as hspAmerind, hspMaori, hpAfrica and hpEast Asia in the limited, but representative culture collection that we looked at. However, the visible footprints of other migrations into India such as from the North Eastern corridor and the presence of phenotypic features resembling to Africans in the South, make it unwise to presume an 'H. pylori free India' at the time of arrival of Indo-European speaking invaders. This issue and the fact that H. pylori's first association with humans traces back to millions of years before present, in Africa [6,17], it is more realistic to hypothesize that H. pylori of African and Asian gene pool might have already been present in India. The predominance of a single H. pylori population might therefore, point to a distinct survival advantage conferred by a fully functional (western type) cagPAI. This analogy is consistent with the scenario we previously reported [3] for the South American, Amerindian strains, which were presumably out competed by their Spanish counterparts arriving with an intact and functional western cagPAI.
Finally, it is possible that phylogeny based on highly recombining gene loci [15,29,31-35] may not be completely foolproof to extract inheritance from different ancestral populations, especially when we use tools such as MEGA 3.0 [36], which do not support admixture analysis. Moreover, phylogenetic methods based on bifurcating trees, such as Neighbor joining analysis, may not be fully appropriate for analysis at the intra-species level [37,38], especially in case of hypervariable genomic regions, where multiple homoplasy due to reversions, recurrent mutations etc., or polytomy may sometimes confound the phylogenetic interpretation. However, the housekeeping genes used here are selectively neutral and uniform as compared to virulence associated loci such as the flagellins and vacA [10], and therefore, recombinant and hybrid alleles that blur lineage inferences, could be a rare occurrence and not a routine. Partly in view of this assumption and due to our previous experiences on dissecting complex ancestry of native Peruvian isolates using phylogenetic methods [3] we did not attempt admixture analysis with complicated Bayesian statistics. However, to ensure that our conclusions did not represent shortcomings of a single method, we adopted an integrated phylogenetic approach combining MEGA/NETWORK based analyses and genotyping strategy based on full cagPAI and its left and right end sequences. Interestingly, these approaches unambiguously show the Indian H. pylori genotypes scattered among the European ones. Although this would be consistent with gene flow into India with the Indo-Aryans, or even more ancient origins following the Paleolithic expansion of humans in Eurasia, but also consistent with another scenario: migration from India to Europe. However, the later scenario becomes insignificant due to the unavailability of supporting archeological, linguistic and historical data. Nonetheless, an understanding of the time-scale would be helpful for choosing between such explanations, with the estimation of divergence times between the H. pylori sequences in the different human populations. These issues therefore need to be addressed in future.
Conclusion
In summary, we found significant overlap among genetic identities of Indian and European H. pylori based on core and flexible genome markers. This remarkable genetic similarity points to their possible common genetic origins and could therefore be potentially useful in understanding entry, survival, spread and adaptation of H. pylori in Indian stomachs. Also, this study is consistent with the hypothesis of co-evolution of H. pylori with H. sapiens and therefore, could form a reliable foundation to test and reconstruct gene flow into India with the arrival of Indo-Aryans or otherwise.
Methods
Bacterial strains, genomic DNA and diagnostic PCR
All the strains were cultured by the Centre for Liver Research and Diagnostics, Deccan college of Medical Sciences, Hyderabad, from patient biopsies. All the biopsy material was collected with necessary ethical clearances and after obtaining informed consents. Template DNA was prepared from single colony picks as described previously [39]. Genomic DNAs of the 10 Ladakhi strains were received from Mark Achtman, Max-Planck Institute für Infektionsbiologie, Berlin, Germany. Genomic DNA was isolated from strains obtained from patients with different disease types including Duodenal Ulcer (DU); Gastric Ulcer (GU); Gastric Cancer (GC); Gastritis (G); Non Ulcer Dyspepsia (NUD); Peptic Ulcer Disease (PUD); Chronic Duodenal Ulcer (CDU); Portal Hyper Tension (PHT) etc. (Figure 1). However, in the current study, the clinical background of the individual isolates was not taken into account. The Indian isolates we looked at (n = 63) were originally from Native Indian people mainly of Aryan and Dravidian ancestry from India. PCR based analyses of genes namely cagA, glmM, babB [14] and oipA were carried out to ascertain the quality of DNA samples we used. Also these PCR assays served as amplification level controls for the analysis of insertion, deletion and substitution in the cagPAI.
MLST analysis by MEGA 3.1 and NETWORK 4.2.0
A 600 bp region each from the 7 housekeeping genes spread throughout the genome atpA, efp, ureI, ppa and mutY, trpC, yphC was amplified by PCR and sequenced for all the Indian isolates exactly as described previously [3]. Sequencing was performed on both the strands, using an ABI Prism 3100 DNA sequencer (Applied Biosystems, USA). PCR and direct sequencing were performed at least twice to determine and confirm the DNA sequences for each isolate. Consensus sequence for each of the samples was generated using Genedoc (version 2.6.002). Multiple alignments of sequenced nucleotides were carried out using Clustal X (version 1.81). Neighbor joining trees were constructed in MEGA 3.0 [36] using bootstrapping at 10000 bootstrap trials and through Kimura-2 parameters. For beginning construction of phylogenetic trees based on MLST genotyping procedures, ~400 sequences of the 7 housekeeping genes of strains belonging to different established genotypes, including 40 sequences of isolates from Ladakh were obtained from the pubMLST database [40] (courtesy, Daniel Falush). The Indian H. pylori diversity represented in the final MEGA3.0 alignment and the tree thereof comprised of a total of 63 sequences inclusive of the 10 Ladakhi sequences generated in house along with the other 9 representative Ladakhi sequences from the database. We performed on MLST sequence data a network analysis using the program Network 4.2.0.0. [38,41]. In particular, the median-joining algorithm for multistate DNA data was used [42,43]. Because of a program limitation, which cannot handle more than 1000 polymorphic sites at once, we performed the analysis separately on two halves of the sequence (encompassing respectively 650 and 665 polymorphic sites). The input file (in *.rdf format) was obtained using the commercial software DNA Alignment 1.1.2.1.
Profiling of the cagA gene, the whole cagPAI and its right junction
The 5' end of the cagA gene was amplified using primers mentioned elsewhere [44] and the amplified products were sequenced with forward and reverse primers. The consensus sequences were then translated into amino acid sequences using GeneDoc software (version 2.6.002) and were then assigned to the Western or the East Asian group based on the C or D repeats present respectively in the EPIYA motif [45]. Genetic diversity of the cagA 5' end sequences for our Indian isolates: MS15, MS7, MS4 and 3K along with 26695 and J99 were compared to the other records from GenBank [20,30,46]. A phylogenetic neighbor-joining tree was constructed by MEGA 3.1 version using these sequences (Figure 4).
PCR analyses were carried out to find the status of the cagPAI using 8 sets of primers that amplified the cagA gene, its promoter region, the cagE and cagT genes and the left end of the cagPAI [8,29,34]. We also analyzed whole cagPAI of the representative isolates from India (3K, 4K, 3C, MS40 and MS38) by PCR using overlapping primers as described by Blomstergren and colleagues [9]. The entire cagPAI sequence of a single representative Indian isolate 3K was determined. The complete cagPAI sequence was aligned by VISTA programme [47] against other PAI sequences belonging to strains 26695, J99, HPAG1 and 13 other clinical isolates corresponding to H. pylori sub-populations hpEurope, hpEast Asia and hpAfrica1 (Figure 3B).
Chromosomal rearrangements are known to give rise to 5 types of insertion-deletion and substitution motifs in the region between the right end of cagA gene and the glutamate racemase (glr) gene (cag-RJ). We assessed these rearrangement profiles for all of the Indian isolates by PCR as described earlier by Kersulyte and colleagues [13].
Analysis of the chromosomal plasticity region cluster
Chromosomal plasticity region ORF's were assessed for all the 63 Indian isolates by PCR based typing to ensure that all the strains that we looked at were independent and non-clonal by descent. The PCR primers and the procedures used for evaluating the presence of the plasticity region ORF's (JHP912, HP986, JHP947, JHP926, JHP944, JHP931, JHP945 and JHP933) have been descried previously [48].
Nucleotide sequence accession numbers
The nucleotide sequences of the 7 housekeeping genes for the 23 representative Indian isolates have been deposited in the GenBank [Accession numbers, GenBank: DQ504165–DQ504183 and DQ927245–DQ927248 (atpA), DQ504184–DQ504202 and DQ927249–DQ927252 (efp), DQ504203–DQ504221 and DA927253–DA927256 (mutY), DQ504222–DQ504240 and DQ927257–DQ927260 (ppa), DQ504241–DQ504259 and DQ927261–DQ927264 (trpC), DQ504260–DQ504278 and DQ927265–DQ927268 (ureI), DQ504279–DQ504297 and DQ927269–DQ927272 (yphC)]. These sequences will also be made available through the pubMLST database maintained at the Max-Planck Institute für Infektionsbiologie, Berlin, Germany. The sequence of whole cagPAIs of the representative Indian isolate 3K and the French isolate Fr908 for which the sequence was determined in our laboratory, have been deposited in Genbank under accession nos. DQ985738 and EF195721 respectively. These and other sequences can also be requested from the authors.
Authors' contributions
SMD and IA performed and analyzed MLST, all other genotyping experiments and phylogenetic analysis. SMD also helped in analysis of babB and oipA genotyping. MAA performed vacA genotyping. IA also performed H. pylori isolation and culture. YA carried out in silico analysis of the cagPAI sequences. PF performed Network analysis on MLST data and contributed to manuscript writing. LAS and FM provided expert clinical and epidemiological support and contributed to discussions and manuscript writing. NA planned and supervised the study, edited the final draft of the manuscript and provided overall leadership. All the authors read and approved the final manuscript.
Acknowledgments
Acknowledgements
We thank Director of the Centre for DNA Fingerprinting and Diagnostics (CDFD), Hyderabad for support and guidance. Our thanks are due to various collaborators in India and abroad, who contributed to our H. pylori DNA collections. We are grateful to Daniel Falush and Mark Achtman (pubMLST.org) for international MLST data and advice. We are grateful to Seyed E. Hasnain (University of Hyderabad) for his guidance and to Chris Tyler-Smith (Sanger Centre, UK) for his critical comments on our raw data. We are also thankful to the International Society for Genomic and Evolutionary Microbiology (ISOGEM) for supporting and endorsing the study. Financial support from the Department of Biotechnology, Government of India to NA (grant ref. BT/PR2473/Med/13/106/2001) is gratefully acknowledged. Help provided by our laboratory support staff, namely, Shaikh Zamir, B Krishnamurthy and Wasim Ahmad is thankfully appreciated. NA is the Corresponding Fellow of the European Helicobacter Study Group.
Contributor Information
S Manjulata Devi, Email: manju@cdfd.org.in.
Irshad Ahmed, Email: sirfirshadahmed@gmail.com.
Paolo Francalacci, Email: pfrancalacci@uniss.it.
M Abid Hussain, Email: abid@cdfd.org.in.
Yusuf Akhter, Email: yusuf.akhter@gmail.com.
Ayesha Alvi, Email: ayesha@cdfd.org.in.
Leonardo A Sechi, Email: sechila@uniss.it.
Francis Mégraud, Email: Francis.Megraud@chu-bordeaux.fr.
Niyaz Ahmed, Email: niyaz.cdfd@gmail.com.
References
- Falush D, Wirth T, Linz B, Pritchard JK, Stephens M, Kidd M, Blaser MJ, Graham DY, Vacher S, Perez-Perez GI, Yamaoka Y, Mégraud F, Otto K, Reichard U, Katzowitsch E, Wang X, Achtman M, Suerbaum S. Traces of human migrations in Helicobacter pylori populations. Science. 2003;299:1582–1585. doi: 10.1126/science.1080857. [DOI] [PubMed] [Google Scholar]
- Wirth T, Meyer A, Achtman M. Deciphering host migrations and origins by means of their microbes. Mol Ecol. 2005;14:3289–3306. doi: 10.1111/j.1365-294X.2005.02687.x. [DOI] [PubMed] [Google Scholar]
- Devi SM, Ahmed I, Khan AA, Rahman SA, Alvi A, Sechi LA, Ahmed N. Genomes of Helicobacter pylori from native Peruvians suggest admixture of ancestral and modern lineages and reveal a western type cag-pathogenicity island. BMC Genomics. 2006;7:191. doi: 10.1186/1471-2164-7-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavesi A. Utility of JC polyomavirus in tracing the pattern of human migrations dating to prehistoric times. J Gen Virol. 2005;86:1315–1326. doi: 10.1099/vir.0.80650-0. [DOI] [PubMed] [Google Scholar]
- Holmes EC. The phylogeography of human viruses. Mol Ecol. 2004;13:745–756. doi: 10.1046/j.1365-294X.2003.02051.x. [DOI] [PubMed] [Google Scholar]
- Linz B, Balloux F, Moodley Y, Manica A, Liu H, Roumagnac P, Falush D, Stamer C, Prugnolle F, van der Merwe SW, Yamaoka Y, Graham DY, Perez-Trallero E, Wadstrom T, Suerbaum S, Achtman M. An African origin for the intimate association between humans and Helicobacter pylori. Nature. 2007;445:915–918. doi: 10.1038/nature05562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covacci A, Telford JL, Giudice GD, Parsonnet J, Rappuoli R. Helicobacter pylori virulence and genetic geography. Science. 1999;284:1328–1333. doi: 10.1126/science.284.5418.1328. [DOI] [PubMed] [Google Scholar]
- Ikenoue T, Maeda S, Gura KO, Akanuma M, Mitsuno Y, Imai Y, Yoshida H, Shiratori Y, Omata M. Determination of Helicobacter pylori virulence by simple gene analysis of the cag pathogenicity island. Clin Diag Lab Immunol. 2001;8:181–186. doi: 10.1128/CDLI.8.1.181-186.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomstergren A, Lundin A, Nilsson C, Engstrand L, Lundeberg J. Comparative analysis of the complete cag pathogenicity island sequence in four Helicobacter pylori isolates. Gene. 2004;328:85–93. doi: 10.1016/j.gene.2003.11.029. [DOI] [PubMed] [Google Scholar]
- Achtman M, Azuma T, Berg DE, Ito Y, Morelli G, Pan ZJ, Suerbaum S, Thompson S, van der Ende A, van Doorn LJ. Recombination and clonal groupings within Helicobacter pylori from different geographical regions. Mol Microbiol. 1999;32:459–470. doi: 10.1046/j.1365-2958.1999.01382.x. [DOI] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirth T, Wang X, Linz B, Novick RP, Lum JK, Blaser M, Morelli G, Falush D, Achtman M. Distinguishing human ethnic groups by means of sequences from Helicobacter pylori: lessons from Ladakh. Proc Natl Acad Sci USA. 2004;101:4746–4751. doi: 10.1073/pnas.0306629101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersulyte D, Mukhopadhyay AK, Velapatino B, Su WW, Pan ZJ, Garcia C, Hernandez V, Valdez Y, Mistry RS, Gilman RH, Yuan Y, Gao H, Alarcon T, Lopez-Brea M, Nair GB, Chowdhury A, Datta S, Shirai M, Nakazawa T, Ally R, Segal I, Wong BCY, Lam SK, Olfat F, Boren T, Engstrand L, Torres O, Schneider R, Thomas JE, Czinn S, Berg DE. Differences in genotypes of Helicobacter pylori from different human populations. J Bacteriol. 2000;182:3210–3218. doi: 10.1128/JB.182.11.3210-3218.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghose C, Perez-Perez GI, Bello MGD, Pride DT, Bravi CM, Blaser MJ. East Asian genotypes of Helicobacter pylori strains in Amerindians provide evidence for its ancient human carriage. Proc Natl Acad Sci USA. 2002;99:15107–15111. doi: 10.1073/pnas.242574599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll IM, Ahmed N, Beesley SM, Khan AA, Ghousunnissa S, O'Morain CA, Smyth CJ. Fine-structure molecular typing of Irish Helicobacter pylori isolates and their genetic relatedness to strains from four different continents. J Clin Microbiol. 2003;41:5755–5759. doi: 10.1128/JCM.41.12.5755-5759.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doorn VLJ, Figueiredo C, Mégraud F, Pena S, Midolo P, Queiroz DM, Carneiro F, Vanderborght B, Pegado MD, Sanna R, De Boer W, Schneeberger PM, Correa P, Nq EK, Atherton J, Blaser MJ, Quint WG. Geographic distribution of vacA allelic types of Helicobacter pylori. Gastroenterology. 1999;116:823–830. doi: 10.1016/S0016-5085(99)70065-X. [DOI] [PubMed] [Google Scholar]
- Gressmann H, Linz B, Ghai R, Pleissner KP, Schlapbach R, Yamaoka Y, Kraft C, Suerbaum S, Meyer TF, Achtman M. Gain and loss of multiple genes during the evolution of Helicobacter pylori. PLoS Genet. 2005;1:e43. doi: 10.1371/journal.pgen.0010043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kivisild T, Bamshad MJ, Kaldma K, Metspalu M, Metspalu E, Reidla M, Laos S, Parik J, Watkins WS, Dixon ME, Papiha SS, Mastana SS, Mir MR, Ferak V, Villems R. Deep common ancestry of Indian and western-Eurasian mitochondrial DNA lineages. Curr Biol. 1999;9:1331–1334. doi: 10.1016/S0960-9822(00)80057-3. [DOI] [PubMed] [Google Scholar]
- Sharma S, Saha A, Rai E, Bhat A, Bamezai R. Human mtDNA hypervariable regions, HVR I and II, hint at deep common maternal founder and subsequent maternal gene flow in Indian population groups. J Hum Genet. 2005;50:497–506. doi: 10.1007/s10038-005-0284-2. [DOI] [PubMed] [Google Scholar]
- Datta S, Chattopadhyay S, Nair GB, Mukhopadhyay AK, Hembram J, Berg DE, Saha DR, Khan A, Santra A, Bhattacharya SK, Chowdhury A. Virulence genes and neutral DNA markers of Helicobacter pylori isolates from different ethnic communities of West Bengal, India. J Clin Microbiol. 2003;41:3737–3743. doi: 10.1128/JCM.41.8.3737-3743.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underhill PA, Jin L, Zemans R, Oefner PJ, Cavalli-Sforza LL. A pre-Columbian Y chromosome-specific transition and its implications for human evolutionary history. Proc Natl Acad Sci USA. 1996;93:196–200. doi: 10.1073/pnas.93.1.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho-Silva DR, Zerjal T, Tyler-Smith C. Ancient Indian roots? J Biosci. 2006;31:1–2. doi: 10.1007/BF02705228. [DOI] [PubMed] [Google Scholar]
- Kenoyer JM. Ancient cities of the Indus valley civilization. Karachi: Oxford University Press; 1998. [Google Scholar]
- Bamshad M, Kivisild T, Watkins WS, Dixon ME, Ricker CE, Rao BB, Naidu JM, Prasad BV, Reddy PG, Rasanayagam A, Papiha SS, Villems R, Redd AJ, Hammer MF, Nguyen SV, Carroll ML, Batzer MA, Jorde LB. Genetic evidence on the origins of Indian caste populations. Genome Res. 2001;11:994–1004. doi: 10.1101/gr.GR-1733RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu A, Mukherjee N, Roy S, Sengupta S, Banerjee S, Chakraborty M, Dey B, Roy M, Roy B, Bhattacharyya NP, Roychoudhury S, Majumder PP. Ethnic India: a genomic view, with special reference to peopling and structure. Genome Res. 2003;13:2277–2290. doi: 10.1101/gr.1413403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalli-Sforza LL. The DNA revolution in population genetics. TIG. 1998;14:60–65. doi: 10.1016/s0168-9525(97)01327-9. [DOI] [PubMed] [Google Scholar]
- Cavalli-Sforza LL, Feldman MW. The application of molecular genetic approaches to the study of human evolution. Nat Genet. 2003;33:266–275. doi: 10.1038/ng1113. [DOI] [PubMed] [Google Scholar]
- Sahoo S, Singh A, Himabindu G, Banerjee J, Sitalaximi T, Gaikwad S, Trivedi R, Endicott P, Kivisild T, Metspalu M, Villems R, Kashyap VK. A prehistory of Indian Y chromosomes: Evaluating demic diffusion scenarios. Proc Natl Acad Sci USA. 2005;103:843–848. doi: 10.1073/pnas.0507714103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauser F, Khan AA, Hussain MA, Carroll IM, Ahmad N, Tiwari S, Shouche Y, Das B, Alam M, Ali SM, Habibullah CM, Sierra R, Megraud F, Sechi LA, Ahmed N. The cag pathogenicity island of Helicobacter pylori is disrupted in the majority of patient isolates from different human populations. J Clin Microbiol. 2004;42:5302–5308. doi: 10.1128/JCM.42.11.5302-5308.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay AK, Kersulyte D, Jeong J, Datta S, Ito Y, Chowdhury A, Chowdhury S, Santra A, Bhattacharya SK, Azuma T, Nair GB, Berg DE. Distinctiveness of genotypes of Helicobacter pylori in Calcutta, India. J Bacteriol. 2000;182:3219–3227. doi: 10.1128/JB.182.11.3219-3227.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed N, Khan AA, Alvi A, Tiwari S, Jyothirmayee CS, Kauser F, Ali M, Habibullah CM. Genomic analysis of Helicobacter pylori from Andhra Pradesh, south India: molecular evidence for three major genetic clusters. Curr Sci. 2003;85:101–108. [Google Scholar]
- Carroll IM, Ahmed N, Beesley SM, Khan AA, Ghousunnissa S, O'Morain CA, Habibullah CM, Smyth CJ. Microevolution between paired antral and paired antral and corpus Helicobacter pylori isolates recovered from individual patients. J Med Microbiol. 2004;53:669–677. doi: 10.1099/jmm.0.05440-0. [DOI] [PubMed] [Google Scholar]
- Kauser F, Hussain MA, Ahmed I, Ahmad N, Habeeb A, Khan AA, Ahmed N. Comparing genomes of Helicobacter pylori strains from the high altitude desert of Ladakh, India. J Clin Microbiol. 2005;43:1538–1545. doi: 10.1128/JCM.43.4.1538-1545.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prouzet-Mauleon V, Hussain MA, Lamouliatte H, Kauser F, Megraud F, Ahmed N. Pathogen evolution in vivo: genome dynamics of two isolates obtained nine years apart from a duodenal ulcer patient infected with a single Helicobacter pylori strain. J Clin Microbiol. 2005;43:4237–4241. doi: 10.1128/JCM.43.8.4237-4241.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ando T, Peek RM, Pride D, Levine SM, Takata T, Lee YC, Kusugami K, van der Ende A, Kuipers EJ, Kusters JG, Blaser MJ. Polymorphisms of Helicobacter pylori HP0638 reflect geographic origin and correlate with cagA status. J Clin Microbiol. 2002;40:239–246. doi: 10.1128/JCM.40.1.239-246.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Nei M. Integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinfor. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- Herrnstadt C, Elson JL, Fahy E, Preston G, Turnbull DM, Anderson C, Ghosh SS, Olefsky JM, Beal MF, Davis RE, Howell N. Reduced-median-network analysis of complete mitochondrial DNA coding-region sequences for the major African, Asian, and European haplogroups. Am J Hum Genet. 2002;70:1152–1171. doi: 10.1086/339933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posada D, Crandall KA. Intraspecific gene genealogies: trees grafting into networks. Trends Ecol Evol. 2001;16:37–45. doi: 10.1016/S0169-5347(00)02026-7. [DOI] [PubMed] [Google Scholar]
- Kauser F, Hussain MA, Ahmed I, Srinivas S, Devi SM, Majeed AA, Rao KR, Khan AA, Sechi LA, Ahmed N. Comparative genomics of Helicobacter pylori isolates recovered from ulcer disease patients in England. BMC Microbiol. 2005;5:32. doi: 10.1186/1471-2180-5-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 'pubMLST database' http://www.pubmlst.org
- 'Network package' http://www.fluxus-engineering.com
- Bandelt H-J, Forster P, Sykes BC, Richards MB. Mitochondrial portraits of human populations. Genetics. 1995;141:743–753. doi: 10.1093/genetics/141.2.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandelt H-J, Forster P, Röhl A. Median-joining networks for inferring intraspecific phylogenies using median networks. Mol Biol Evol. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- Yamaoka Y, Orito E, Mizokami M, Gutierrez O, Saitou N, Kodama T, Osato MS, Kim JG, Ramirez FC, Mahachai V, Graham DY. Helicobacter pylori in north and south America before Columbus. FEBS Lett. 2002;517:180–184. doi: 10.1016/S0014-5793(02)02617-0. [DOI] [PubMed] [Google Scholar]
- Hatakeyama M. Oncogenic mechanisms of the Helicobacter pylori CagA protein. Nat Rev Cancer. 2004;4:688–694. doi: 10.1038/nrc1433. [DOI] [PubMed] [Google Scholar]
- Rahman M, Mukhopadhyay AK, Nahar S, Datta S, Ahmad MM, Sarker S, Masud IM, Engstrand L, Albert MJ, Nair GB, Berg DE. DNA-Level characterization of Helicobacter pylori strains from patients with overt disease and with benign infections in Bangladesh. J Clin Microbiol. 2003;41:2008–2014. doi: 10.1128/JCM.41.5.2008-2014.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I. VISTA: computational tools for comparative genomics. Nucleic Acids Res. 2004;32:W273–W279. doi: 10.1093/nar/gkh458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Occhialini A, Marais A, Alm R, Akanuma M, Mitsuno Y, Imai Y, Yoshida H, Shiratori Y, Omata M. Distribution of open reading frames of plasticity region of strain J99 in Helicobacter pylori strains isolated from gastric carcinoma and gastritis patients in Costa Rica. Infect Immun. 2000;68:6240–6249. doi: 10.1128/IAI.68.11.6240-6249.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azuma T, Yamakawa A, Yamazaki S, Ohtani M, Ito Y, Muramatsu A, Suto H, Yamazaki Y, Keida Y, Higashi H, Hatakeyama M. Distinct diversity of the cag pathogenicity island among Helicobacter pylori strains in Japan. J Clin Microbiol. 2004;42:2508–2517. doi: 10.1128/JCM.42.6.2508-2517.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]


