Abstract
Reconstituted influenza virosomes (virus membrane envelopes) have been used previously to deliver pDNA (plasmid DNA) bound to their external surface to a variety of target cells. Although high transfection efficiencies can be obtained with these complexes in vitro, the virosome-associated DNA is readily accessible to nucleases and could therefore be prone to rapid degradation under in vivo conditions. In the present study, we show a new method for the production of DNA–virosomes resulting in complete protection of the DNA from nucleases. This method relies on the use of the short-chain phospholipid DCPC (dicaproylphosphatidylcholine) for solubilization of the viral membrane. The solubilized viral membrane components are mixed with pDNA and cationic lipid. Reconstitution of the viral envelopes and simultaneous encapsulation of pDNA is achieved by removal of the DCPC from the mixture through dialysis. Analysis by linear sucrose density-gradient centrifugation revealed that protein, phospholipid and pDNA physically associated to particles, which appeared as vesicles with spike proteins inserted in their membranes when analysed by electron microscopy. The DNA–virosomes retained the membrane fusion properties of the native influenza virus. The virosome-associated pDNA was completely protected from degradation by nucleases, providing evidence for the DNA being highly condensed and encapsulated in the lumen of the virosomes. DNA–virosomes, containing reporter gene constructs, transfected a variety of cell lines, with efficiencies approaching 90%. Transfection was completely dependent on the fusogenic properties of the viral spike protein haemagglutinin. Thus, DNA–virosomes prepared by the new procedure are highly efficient vehicles for DNA delivery, offering the advantage of complete DNA protection, which is especially important for future in vivo applications.
Keywords: gene delivery, haemagglutinin, influenza virus, plasmid DNA, virosome
Abbreviations: BHK, baby hamster kidney; CHO, Chinese hamster ovary; DCPC, dicaproylphosphatidylcholine; DODAC, dioleoyldimethyl-ammonium chloride; DOTAP, 1,2-dioleoyl-3-trimethylammonium-propane; EGFP, enhanced green fluorescent protein; excimer, excited dimer; FCS, foetal calf serum; HA, haemagglutinin; HBS, Hepes-buffered saline; MDCK, Madin–Darby canine kidney; NA, neuraminidase; pDNA, plasmid DNA; PEG, poly(ethylene glycol); pyrPC, 1-hexadecanoyl-2-(1-pyrenedecanoyl)-sn-glycero-3-phosphocholine; SPLP, stabilized plasmid–lipid particle
INTRODUCTION
Delivery vehicles for the transfer of genes to target cells can be divided into viral and non-viral systems. Viral vectors are obtained by replacement of one or more viral genes by a gene of interest and are considered to be the most efficient transducing systems to date (reviewed in [1,2]). Their efficiency relates to properties of the viral capsid proteins or membrane glycoproteins, such as the ability to bind to cellular receptors and to pass through or fuse with cellular membranes. However, the safety of viral vector systems remains a matter of major concern, and issues related to insertion mutagenesis as observed with retroviruses [3] and the induction of undesirable immune responses and inflammation [4,5] still pose major challenges.
For non-viral gene delivery, chemical approaches (e.g. cationic lipids, cationic polymers, nanoparticles) and physical methods (e.g. gene gun, electroporation) are being employed [6]. Among the chemical systems, cationic liposomes are the most extensively studied vehicles (reviewed in [7–10]). In these systems, cationic lipids condense DNA through electrostatic interactions with the negatively charged phosphate groups of the nucleic acid, thereby forming so-called lipoplexes. In the presence of an excess of cationic lipid, due to their overall positive charge, these lipoplexes bind to the negatively charged cell surface. After uptake by endocytosis, the pDNA (plasmid DNA) is released into the cytosol by an as-yet-unknown mechanism. Lipoplexes are quite efficient in transfection of target cells in culture [11]. With respect to in vivo use, liposomal delivery systems are regarded as relatively safe with regard to their immunogenicity and oncogenicity [12,13]. However, their successful application in vivo has been hampered by toxicity issues [8] and low transfection efficiencies [7,10].
Virosomes can be considered as hybrids between viral and liposomal delivery systems, combining the characteristics of cellular interactions of viral vectors with the safety of liposomal delivery systems. Virosomes are reconstituted membrane vesicles prepared from enveloped viruses, but lack the genetic material of the native virus. By virtue of the viral membrane glycoproteins embedded in their membrane, virosomes bind to cellular receptors and fuse with the target cell membrane, thus actively delivering encapsulated compounds to the cytosol of target cells.
Influenza virus has been used extensively for the generation of virosomes. The native virus enters cells through interaction of the viral envelope glycoprotein HA (haemagglutinin) with sialic acid residues on the target cell surface. This is followed by entry of the virus particles into the endocytic pathway and subsequent low pH-dependent fusion of the viral envelope with the endosomal membrane [14,15].
Virosomes derived from influenza virus exploit these properties of the viral HA and have been employed for the delivery of biologically active macromolecules, including pDNA, into cells [16–19]. Also virosomes derived from other enveloped viruses, such as Sendai virus and vesicular stomatitis virus, have been used for delivery purposes, including delivery of pDNA [20–22]. Preparation of virosomes from influenza virus or other enveloped viruses generally involves solubilization of the viral membrane, followed by removal of the nucleocapsid by ultracentrifugation. Subsequently, reconstitution of the viral membrane is accomplished by removal of the detergent either by dialysis or by adsorption to a hydrophobic resin [23–25]. However, other methods such as post-insertion of membrane proteins into pre-formed liposomes [22] and fusion of virus particles with liposomes have also been reported [26].
Previously, we reported on a method to prepare influenza virosomes suitable for pDNA delivery [16]. The cationic lipid DODAC (dioleoyldimethylammonium chloride) was incorporated in the membrane of these virosomes to enable binding of pDNA to the surface of the reconstituted virosomes. The virosome–DNA complexes thus formed were found to efficiently deliver the virosome-associated DNA to target cells in a process that was completely dependent on the membrane fusion activity of the virosomal HA [16]. Virosomes with 30 mol% DODAC relative to the viral phospholipid showed the most efficient binding of pDNA and optimal transfection of cultured cells.
However, a major drawback of virosomes with surface-bound pDNA is that the DNA is readily accessible for nucleases, especially during in vivo applications. The low degree of condensation of virosome-bound pDNA (due to a low positive-to-negative charge ratio of approx. 0.2) further contributes to vulnerability for nucleases. Increasing this charge ratio might be expected to result in a higher degree of condensation and perhaps in a better protection from nuclease activity. However, it has been shown that an increase in the charge ratio results in reduced transfection efficiency [16].
To improve protection of the pDNA, while maintaining high transfection efficiencies, we set out to develop a new method aimed at encapsulation of pDNA in the lumen of virosomes as opposed to attachment of the DNA to the virosomal surface. This new method is based on a technology we have employed successfully for the encapsulation of small interfering RNA molecules, which have a length of approx. 20 nucleotides [17]. The method relies on the use of the short-chain phospholipid DCPC (dicaproylphosphatidylcholine) for solubilization of the virus membrane. Owing to its high critical micelle concentration, DCPC can be removed by dialysis. Thus, virosome reconstitution can be performed by a gentle method, which is expected to be compatible with the presence of pDNA. We demonstrate that, using this method, pDNA can be encapsulated in virosomes with high efficiency. In the resulting virosomes the DNA is well protected from nucleases. Moreover, these virosomes efficiently transfect a variety of cell lines in a process involving the membrane fusion activity of the virosomal HA.
EXPERIMENTAL
Materials
DCPC (1,2-dihexanoyl-sn-glycero-3-phosphocholine; dicaproylphosphatidylcholine) and DOTAP (1,2-dioleoyl-3-trimethylammonium-propane) were obtained from Avanti Polar Lipids (Alabaster, AL, U.S.A.). DODAC was from Inex Pharmaceuticals (Vancouver, BC, Canada). PicoGreen reagent and pyrPC [1-hexadecanoyl-2-(1-pyrenedecanoyl)-sn-glycero-3-phosphocholine] were obtained from Molecular Probes (Leiden, The Netherlands). Octaethyleneglycol (n-dodecyl) monoether (C12E8) was purchased from Calbiochem (La Jolla, CA, U.S.A.). Influenza virus (A/Panama/2007/99) was kindly provided by Solvay Pharmaceuticals B.V. (Weesp, The Netherlands). β-Propiolactone was from Acros Organics (Geel, Belgium). The pCMV-EGFP (enhanced green fluorescent protein) plasmid was a gift from Alida Weeke-Klimp (Department of Cell Biology, Liposome Research Section, University Medical Center Groningen, Groningen, The Netherlands).
Preparation of virosomes with encapsulated pDNA
Preparation of virosomes with cationic lipid and encapsulated pDNA was based on the method for the production of virosomes with encapsulated small interfering RNA described previously [17]. Influenza virus (1.78 μmol/ml viral phospholipid) was inactivated by incubation with β-Propiolactone (1:1000, v/v) for 24 h, followed by extensive dialysis. Subsequently, the virus (1.5 μmol of phospholipid) was sedimented by ultracentrifugation at 30000 rev./min in a SW55Ti rotor for 1 h at 4 °C, and the viral pellet was resuspended in 375 μl HBS (Hepes-buffered saline; 5 mM Hepes, pH 7.4, and 0.15 M NaCl). The viral membrane was dissolved by addition of 375 μl of 200 mM DCPC in HBS, followed by incubation on ice for 30 min. The nucleocapsid was removed by ultracentrifugation at 50000 rev./min in a TLA 100.3 rotor for 30 min at 4 °C, and the supernatant containing the dissolved membrane components was used for the reconstitution of the viral membranes. The required amounts of DOTAP or DODAC were dissolved in chloroform (typically 0.808 μmol, corresponding to 35% of total phospholipid in the virosome preparation), dried under a stream of nitrogen and kept under vacuum for 2 h. pDNA (65 μg in 130 μl) was added to the dried cationic lipids, followed by a 15 min incubation. The supernatant containing the solubilized viral membrane components was added to the cationic lipid/pDNA solution and the mixture was incubated for 30 min at 20 °C. Subsequently, the mixture was dialysed in a 0.5–3 ml Slide-A-Lyser (with a molecular mass cut off of 10 kDa; Pierce Rockford, Illinois, U.S.A.) against 2 l of HBS overnight at 4 °C, followed by another 4 h of dialysis after buffer refreshment. The obtained virosome preparation was purified on a discontinuous sucrose gradient [10–50% (w/v) in HBS] by ultracentrifugation at 30000 rev./min in an SW55Ti rotor for 1.5 h at 4 °C. The virosomes were recovered from the interface of the two sucrose layers and dialysed against HBS overnight at 4 °C. Virosomes were analysed for protein content by a micro Lowry assay [27] and for pDNA content by the PicoGreen assay (described in detail below). Phosphate concentration was determined by the method of Böttcher et al. [28] and the amount of phospholipid was calculated by subtraction of the amount of phosphate present in the pDNA as determined by the PicoGreen assay.
Virosomes with pDNA attached to the surface
Virosomes with pDNA attached to the surface were prepared as described by Schoen et al. [16], with some modifications. Instead of the detergent C12E8, DCPC was used to solubilize the viral membrane, and, for the removal of DCPC, dialysis was performed as an alternative to adsorption to BioBeads SM2. We have shown previously that virosomes produced by the C12E8 and the DCPC methods are similar in morphology and demonstrate similar fusogenic properties [29]. Virosomes were prepared with either DODAC or DOTAP (30% relative to total lipid and 43% relative to viral lipid). For the binding of pDNA to the surface of the virosomes, typically 6 μg of pDNA was incubated with 20 nmol of virosomal phospholipids for 30 min as described by Schoen and coworkers [16].
PicoGreen assay
This assay was adapted from Ferrari et al. [30]. DNA–virosomes (approx. 40 ng of DNA) were diluted in 20 mM sodium acetate (pH 6) containing 0.13% Zwittergent (Zwittergent 3-14; Calbiochem, La Jolla, U.S.A.) to a final volume of 100 μl in a 96-well plate. PicoGreen (100 μl) solution [1:200, v/v in 20 mM sodium acetate (pH 6)] was added to the virosome samples, followed by incubation for 15 min. The fluorescence was read using a FL500 microplate fluorescence reader (Bio-Tek instruments) with excitation filters set at 485±20 nm and emission filters set at 530±25 nm.
Benzonase assay
Virosomes with encapsulated pDNA (2 μg DNA), virosomes with pDNA attached to the surface and naked pDNA (control) were treated with 1 unit of benzonase (Roche, Basel, Switzerland) in HBS containing 0.6 mM MgCl2 for 30 min at 37 °C. The reaction was stopped by chelation of MgCl2 using a 4× molar excess of EDTA (2.5 mM) and the DNA–virosomes were solubilized with 8 mM C12E8. For total degradation of pDNA, the DNA–virosomes and naked pDNA were treated with C12E8 prior to benzonase treatment. Total DNA was determined from samples not exposed to benzonase and only treated with detergent. pDNA was isolated using a QIAprep spin miniprep kit (QIAGEN, Venlo, The Netherlands), resolved on a 1% (w/v) agarose gel and visualized by ethidium bromide staining.
Sucrose density-gradient analysis
The virosome preparations were analysed for co-migration of phospholipid, protein and DNA on a linear sucrose density-gradient [10–60% (w/v) in HBS]. The gradients were centrifuged to equilibrium at 50000 rev./min in an SW55Ti rotor for 65 h at 4 °C. Fractions were analysed for protein, phosphate and DNA content as described above.
Electron microscopy
Virus or virosomes were dialysed against ammonium acetate buffer (75 mM ammonium acetate and 2.5 mM Hepes, pH 7.4) overnight at 4 °C. Subsequently, the virus or virosome suspensions were applied to glow-discharged 200-mesh grids covered with a Formvar film. Virosomes or virus were stained with freshly prepared 3% (w/v) ammonium molybdate (pH 7.2) and examined in a Philips CM 12 electron microscope.
Membrane fusion and fusion inactivation
For determination of their fusogenic properties, virosomes were labelled with 10 mol % pyrPC relative to the amount of viral phospholipids. When incorporated in the virosomal membrane at high concentrations, the pyrene probe forms so-called excimers (excited dimers) with a typical fluorescence emission at approx. 475 nm. Fusion of the virosomes with much larger erythrocyte ghosts used as target membranes results in dilution of the pyrene probe into the bilayer of the target. As a consequence, excimer formation is diminished and the fluorescence intensity at 475 nm decreases, which is a direct measure for fusion [31,32].
PyrPC was co-reconstituted in the virosomal membrane by including it in the mixture containing the solubilized viral membrane components prior to the dialysis step. To this end, pyrPC was dissolved in chloroform and the required amount was added to the test tube, which also contained the cationic lipid solution. The mixture was subsequently dried and virosomes were prepared as described above. Fusion of the virosomes with erythrocyte ghosts and fusion inactivation were performed as described previously [32,33]. Fusion inactivation involved exposure of the virosomes to pH 5 in the absence of target membranes, followed by neutralization of the solution.
Cells
BHK (baby hamster kidney)-21 cells were grown in BHK-21 medium (Glasgow minimal essential medium, Invitrogen, Paisley, Scotland), supplemented with 5% (v/v) FCS (foetal calf serum; Bodinco BV, Alkmaar, The Netherlands), 20 mM Hepes (Invitrogen) and 10% (v/v) tryptose phosphate broth (Invitrogen). MDCK (Madin–Darby canine kidney) cells were grown in Iscove's medium (Invitrogen) supplemented with 10% FCS. CHO (Chinese hamster ovary)-K1 cells were grown in Ham's F12 medium (Invitrogen) with the addition of 25 mM Hepes and 10% (v/v) FCS and the human trachea epithelial cell line BEAS 2B was cultured in RPMI 1640 medium (Cambrex BioScience, Verviers, Belgium) supplemented with 10% (v/v) FCS. All media contained 100 units/ml penicillin and 100 μg/ml streptomycin, and cells were grown at 37 °C in a 5% CO2 atmosphere.
WST assay
BHK cells were plated into a 96-well plate. The next day virosomes were added in concentrations of 50–200 ng of DNA per well. A further 24 h later the virosomes were removed and 100 μl fresh medium and 10 μl WST reagent (Roche Molecular Biochemicals, Mannheim, Germany) were added. After 30 min the molar absorption at 440 nm was measured using a Powerwave X microtitre plate reader (Bio-Tek, Winooski, U.S.A.).
Transfections
Cells (BHK: 5×104; MDCK: 4×104; CHO-K1: 5×104; BEAS 2B: 10×104) were seeded in 24 well-plates containing 1 ml of culture medium. After 24 h, virosomes containing the pCMV/EGFP plasmid were added to the cells at the indicated amounts. To examine whether virosome-mediated transfection was pH-dependent, NH4Cl (to a final concentration of 20 mM) was added to the BHK-21 cells 20 min prior to transfection. To reveal the role of HA in transfection, experiments were performed with DNA–virosomes that had been inactivated by low pH treatment prior to transfection [33]. For control experiments, BHK cells were transfected with FuGENE® 6 Transfection Reagent (Roche Diagnostics, Indianapolis, IN, U.S.A.) according to the manufacturer's instructions. After 48 h, cells were analysed on a Zeiss Axiovert microscope equipped with epifluorescence. Subsequently, cells were washed, trypsinized and analysed for EGFP expression by flow cytometry (Epics Elite, Coulter, Miami, U.S.A.).
RESULTS
Production and physical characterization of virosomes with encapsulated plasmid DNA
As indicated above, we have developed previously a virosomal delivery device for pDNA in which the DNA is attached to the surface of pre-formed virosomes [16]. In order to improve the protection of the DNA, in the present study we set out to encapsulate pDNA in the lumen of virosomes by reconstituting the virosomal membranes in the presence of cationic lipid and pDNA. Since detergent removal under vigorous shaking, as performed previously, is not compatible with DNA encapsulation (J. de Jonge, unpublished work) we developed a new procedure allowing membrane reconstitution by dialysis. For this purpose we used for solubilization of the viral membrane DCPC, a short-chain phospholipid with detergent-like properties and a high critical micelle concentration suitable for removal by dialysis [29].
Influenza virus was treated with 100 mM DCPC, resulting in solubilization of the viral membrane lipids and glycoproteins HA and NA (neuraminidase), as shown previously [29]. After sedimentation of the nucleocapsid by ultracentrifugation, the viral membrane components were mixed with pDNA and either DODAC or DOTAP at a concentration of 35 mol% relative to the viral phospholipids. As shown below, this amount of cationic lipid results in optimal cell transfection. Reconstitution of the virus membrane was achieved by removal of the DCPC by means of extensive dialysis. The resulting preparation was further purified on a discontinuous sucrose gradient.
When recovered from the gradient the virosome preparation contained approx. 40% of the initial viral membrane proteins, approx. 50% of the initial viral lipids and approx. 55% of the initial amount of pDNA, irrespective of the cationic lipid used. As for the viral lipid and protein, these recoveries are similar to those obtained with virosomes without encapsulated DNA, which were generated according to the same DCPC dialysis procedure [29].
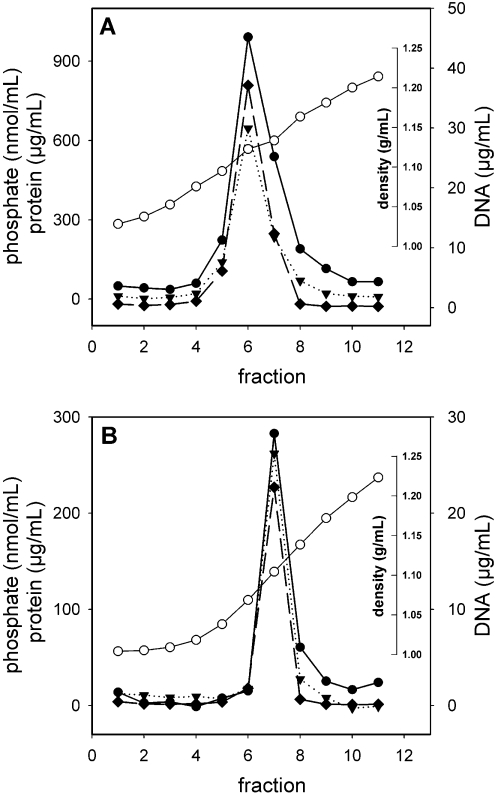
Equilibrium density-gradient centrifugation was performed to further characterize the properties of the purified DODAC or DOTAP virosome preparations. Phospholipid, protein and pDNA co-migrated in the same peak fraction(s), with a density different from the densities of either of the individual components (Figure 1). This co-migration indicates that the three components are associated physically. The densities of the DODAC and DOTAP virosomes were very similar (1.12 g/ml and 1.11 g/ml respectively). The recovery of phospholipid, protein and pDNA from a single fraction and the absence of these components from the other fractions of the gradient indicate that the preparation was highly homogenous and devoid of non-incorporated material.
Figure 1. Equilibrium density-gradient analysis of DNA virosomes.
DODAC virosomes (A) and DOTAP virosomes (B) with encapsulated pDNA were centrifuged to equilibrium on a 10–60% linear sucrose density-gradient. Fractions were analysed for protein (filled circles), phosphate (filled triangles) and DNA (filled diamonds). The density of the fractions (open circles) was determined by measuring the refractive index.
Electron microscopy
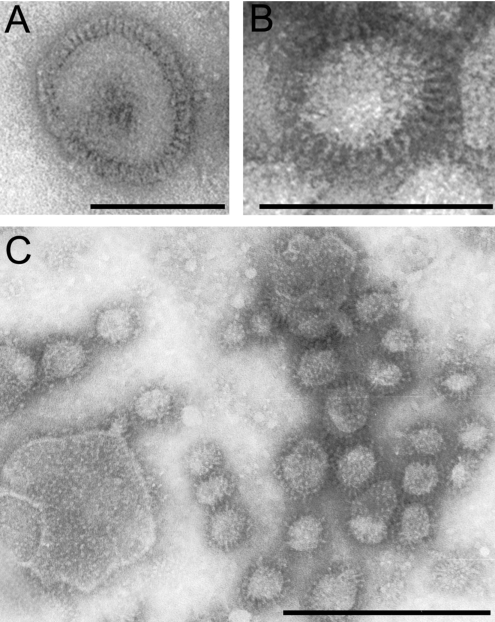
The nature of the complexes detected in the linear sucrose density-gradient were examined further by electron microscopy. The preparation consisted mainly of vesicles with spikes protruding from their membranes (Figure 2; DODAC DNA–virosomes). The morphology of the DNA–virosomes was different from that of native influenza virus. The spike density appeared lower than that on the virus membrane (Figures 2A and 2B). Moreover, the average diameter of the DNA–virosomes (21 nm, calculated from 60 virosomes; Figure 2C) was approximately one fifth of that of native influenza virus (100 nm; [34]), with the exception of some sporadic larger structures (>100 nm; Figure 2C). The standard deviation of the diameter of the major population was approx. 8 nm, indicating that the DNA–virosomes were fairly homogenous in size. Electron microscopical analysis of DOTAP DNA–virosomes revealed similar properties (results not shown).
Figure 2. Analysis of virus and DNA virosomes by negative stain transmission electron microscopy.
(A) Influenza virus. (B) DODAC virosomes with encapsulated pDNA. (C) Overview of DODAC virosomes with encapsulated pDNA. Bars in (A) and (C) represent 100 nm and the bar in (B) represents 50 nm.
Fusogenic properties
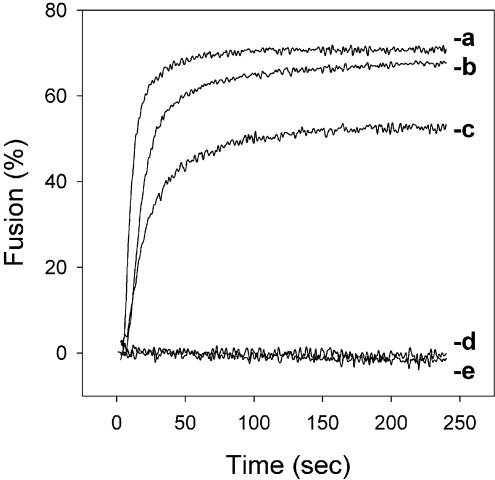
For virosome-mediated transport of pDNA across the endosomal membrane, preservation of the fusogenic properties of the reconstituted virosomal HA is essential. Therefore we examined the membrane fusion activity of the DNA–virosomes using a pyrPC-based fusion assay. DNA–virosomes fused with the same kinetics and to approximately the same extent as empty virosomes prepared without cationic lipids (Figure 3). Moreover, fusion was strictly pH-dependent, occurring only upon acidification of the medium to pH 5.5, and not at neutral pH. To investigate whether fusion of DNA virosomes is mediated by the viral HA, DNA–virosomes were exposed to low pH in the absence of target membranes prior to the fusion assay. This treatment induces a premature conformational change of the HA, which in the absence of target membranes, results in irreversible inactivation of the membrane fusion capacity of HA [35]. In Figure 3, DNA–virosomes that underwent pH-inactivation indeed had completely lost their membrane fusion activity. In conclusion, these results show that virosomes containing pDNA are fusion-active and retain the functional characteristics of the viral HA.
Figure 3. Fusion characteristics of DNA–virosomes.
Fusion with erythrocyte ghosts of empty virosomes prepared without cationic lipid (a) and DOTAP virosomes (b) or DODAC virosomes (c) with encapsulated pDNA was induced by acidification of the buffer to pH 5.5. Alternatively, fusion of DOTAP virosomes with encapsulated pDNA was measured at pH 7.4 (d), or at pH 5.5, after inactivation of HA by pre-exposure of the virosomes to low pH (e). Results of a representative experiment are shown.
Protection of virosomal DNA from nuclease degradation
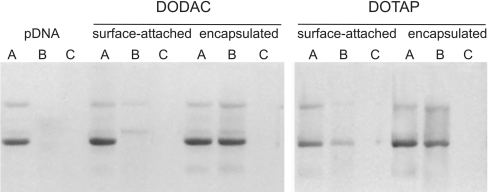
As we aimed at an improved protection of pDNA from nucleases by encapsulation of the pDNA in the virosomal lumen, we next compared the sensitivity to nuclease degradation of the virosomal pDNA in our current preparation with that of pDNA attached to the surface of virosomes, which has been used previously [16]. Samples from both DNA–virosome preparations, as well as naked pDNA, were treated with benzonase in the presence or absence of detergent (Figure 4, lanes B and C). Naked pDNA was completely degraded under either condition. DNA attached to the surface of virosomes prepared with either cationic lipid was also highly sensitive to nuclease treatment (Figure 4, compare lanes A and B under ‘surface-attached pDNA’). In contrast, pDNA in virosomes prepared according to the present protocol, with either DODAC or DOTAP, was almost completely protected from nuclease degradation (Figure 4, compare lanes A and B under ‘encapsulated pDNA’) indicating that the DNA is highly condensed and probably enclosed within the interior of the virosomes. Only in the presence of detergent, resulting in solubilization of the virosomes, the plasmid was accessible for the nuclease and was completely degraded (Figure 4, lane C under ‘encapsulated pDNA’).
Figure 4. Protection of pDNA from nuclease degradation.
Naked pDNA, virosomes with pDNA attached to the surface or virosomes with encapsulated pDNA were treated with detergent alone (total DNA) (A), benzonase (protection) (B), or detergent and benzonase (complete digestion) (C). Virosomes were prepared either with DODAC or DOTAP as indicated. Samples were analysed on a 1% (w/v) agarose gel stained with ethidium bromide. The results shown are one representative of three experiments.
Transfection efficiency of DNA–virosomes
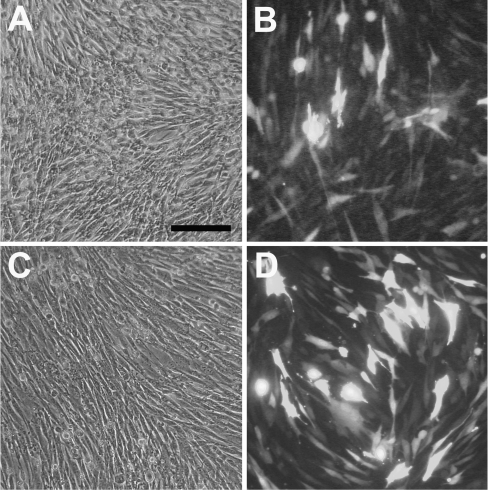
To study the ability of DNA–virosomes to deliver pDNA to the cytosol of target cells, transfection experiments were performed. To this end, BHK-21 cells were exposed to DODAC or DOTAP virosomes containing an EGFP reporter plasmid. Analysis of cell viability using the WST assay revealed 70–80% surviving cells after 24 h of exposure to DNA–virosomes. Moreover, BHK-21 cells incubated with DNA–virosomes for 48 h appeared to be in good condition, as observed by light microscopy (Figures 5A and 5C). This shows that the DNA–virosomes do not induce severe toxic effects. The transfected cells expressed large amounts of EGFP, as observed by fluorescence microscopy (Figures 5B and 5D), indicating that the virosomes had successfully delivered the encapsulated EGFP reporter plasmid to the cytosol.
Figure 5. Microscopy analyses of transfection of BHK-21 cells.
Cells (5×104) were incubated with DODAC virosomes (A) and (B), or DOTAP virosomes (C) and (D) containing the CMV/EGFP plasmid (1 μg DNA). After 48 h the cells were examined by phase contrast (A) and (C), and fluorescence microscopy (B) and (D). Results of a typical experiment are shown.
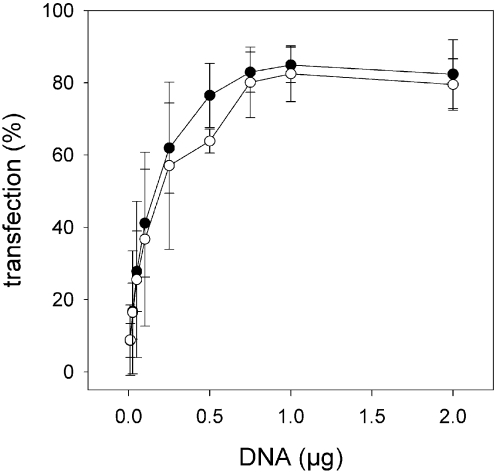
To determine optimal transfection conditions, BHK-21 cells (5×104) were exposed to increasing concentrations of virosome-encapsulated EGFP plasmid. Flow cytometric analysis revealed that the percentage of transfected cells increased in a dose-dependent manner up to 0.75–1 μg pDNA (Figure 6). At this amount of pDNA approx. 80% of the cells were transfected. There were no apparent differences in transfection efficiency between virosomes prepared with DODAC or DOTAP. Dose-dependence was also observed for the average fluorescence per transfected cell, which increased linearly with increasing amounts of virosomes up to 1 μg pDNA. At 2 μg pDNA only a slight further increase in average fluorescence was observed (results not shown).
Figure 6. Flow cytometric characterization of the transfection efficiency of DNA–virosomes.
BHK-21 (5×104) cells were incubated with increasing amounts of pCMV/EGFP encapsulated in DODAC (●) or DOTAP (○) virosomes. After 48 h, the percentage of transfected cells was determined by flow cytometry. The results are the means±S.D. for three independent experiments.
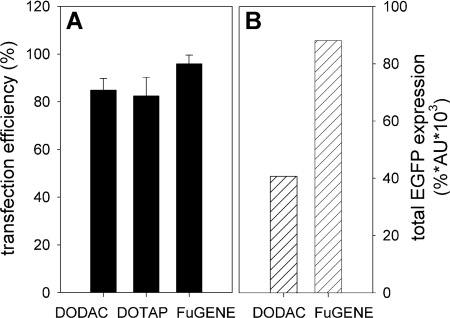
To put the transfection efficiency of the DNA–virosomes into perspective, a comparative study was performed with FuGENE®, a commercially available reagent for in vitro transfection. BHK-21 cells were incubated for 48 h with either 1 μg CMV/EGFP plasmid complexed with FuGENE® or encapsulated in virosomes prepared with 35% DODAC or DOTAP. Flow cytometric analysis revealed that approx. 95% of the BHK-21 cells expressed EGFP when the transfection was performed with FuGENE® and approx. 80–85% of the cells expressed EGFP when incubated with DNA–virosomes (Figure 7A). The level of expressed EGFP was approximately twice as high for transfections performed with FuGENE® compared with transfections performed with DNA–virosomes (Figure 7B). This shows that transfection levels obtained with DNA–virosomes are of the same order as those obtained with one of the most successful in vitro transfection reagents available.
Figure 7. Comparison of transfection by DNA–virosomes with transfection by FuGENE® 6.
BHK-21 (5×104) cells were incubated for 48 h with 1 μg pCMV/EGFP plasmid either complexed to FuGENE® 6 or encapsulated in virosomes prepared with 35% DODAC or DOTAP. (A) Percentage of transfection as determined by flow cytometry. The results are the means±S.D. for three independent experiments. (B) Total EGFP fluorescence as determined by multiplying the percentage of transfected cells (%) with the average fluorescence of the EGFP-positive cells (AU).
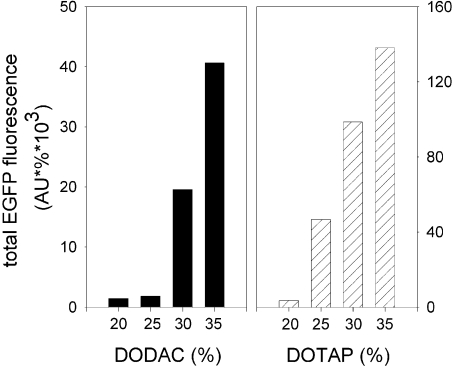
In order to investigate the influence of virosome composition on the transfection efficiency of DNA–virosomes, virosomes were prepared with a range of percentages of cationic lipid (20–35 mol %) and a fixed amount of pDNA. BHK-21 cells (5×104) were exposed to the various virosome preparations containing 1 μg of CMV/EGFP plasmid. Analysis of the cells by flow cytometry revealed that EGFP expression increased with increasing percentages of DODAC or DOTAP (Figure 8). For virosomes prepared with 35% cationic lipid transfection efficiencies were highly reproducible and transfection resulted in consistently high levels of protein expression. In contrast, virosomes prepared with less than 35% cationic lipid showed somewhat variable transfection efficiencies. Since increasing the amount of cationic lipid further was expected to result in cytotoxicity, these conditions were not examined.
Figure 8. Effect of virosome composition on transfection efficiency of DNA–virosomes.
Virosomes were prepared with the indicated percentages of cationic lipid (relative to total lipid) and a fixed amount of pDNA. BHK-21 cells (5×104) were exposed to either DODAC (closed bars) or DOTAP (hatched bars) virosomes containing 1 μg of the CMV/EGFP plasmid. After 48 h incubation, the cells were analysed by flow cytometry. Total EGFP fluorescence was determined by multiplying the percentage of transfected cells (%) with the average fluorescence of the EGFP-positive cells (AU). Data for virosomes prepared with the different cationic lipids were obtained from separate experiments and total EGFP fluorescence is therefore not comparable.
The major advantage of encapsulating pDNA into the lumen of the virosomes, as opposed to binding of pDNA to the virosome surface, is the improved protection of the DNA against nucleases. We therefore tested virosomes before and after benzonase treatment for their transfection properties. For virosomes with encapsulated pDNA, transfection efficiency was not affected by benzonase treatment. Using 500 ng of virosome-encapsulated DNA, the transfection efficiency observed without benzonase treatment was 57%, whereas after benzonase treatment it was 62%. In contrast, transfection efficiency of virosomes with surface-bound pDNA was reduced by 40–50%, depending on the pDNA/virosome ratio.
Mechanism of virosome-mediated transfection
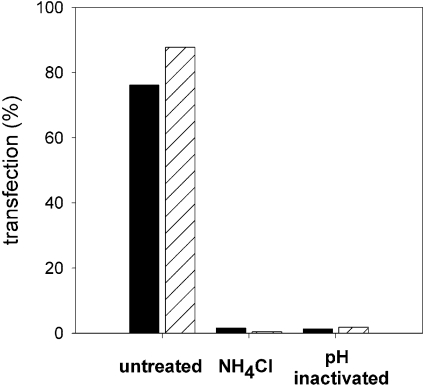
In Figure 3, it is demonstrated that fusion of DNA-virosomes with target membranes is pH-dependent and mediated by the viral HA. To evaluate whether the fusogenic properties of DNA–virosomes are indeed responsible for transfection of cultured cells, two parameters were investigated. First, BHK-21 cells were exposed to virosomes with encapsulated EGFP plasmid in the continuous presence of NH4Cl, which is known to inhibit endosomal acidification [36]. Flow cytometric analysis showed that transfection was blocked completely under these conditions (Figure 9). This indicates that transfection of cells by DNA–virosomes was pH-dependent and that encapsulated pDNA was delivered via the endosomal pathway. Secondly, the fusion activity of the virosomal HA was inhibited by exposure of the virosomes to low pH prior to transfection [33]. BHK-21 cells incubated with these fusion-inactivated DNA–virosomes showed no sign of transfection (Figure 9), demonstrating that transfection was strictly dependent on the membrane fusion activity of HA. Thus, virosomes indeed deliver the encapsulated pDNA to the target cell cytosol by HA-mediated fusion with the endosomal membrane.
Figure 9. Transfection characteristics of DNA–virosomes.
BHK-21 cells (5×104) were incubated with untreated DODAC (closed bars) or DOTAP (hatched bars) virosomes containing the CMV/EGFP plasmid (750 ng). The BHK-21 cells were grown in standard culture medium or in medium supplemented with NH4Cl, which is known to inhibit endosomal acidification, as indicated. Alternatively, DNA–virosomes were fusion-inactivated by pre-exposure to low pH prior to transfection. After 48 h the transfection percentages were determined by flow cytometry. The results shown are derived from a typical experiment.
Transfection of various cell lines
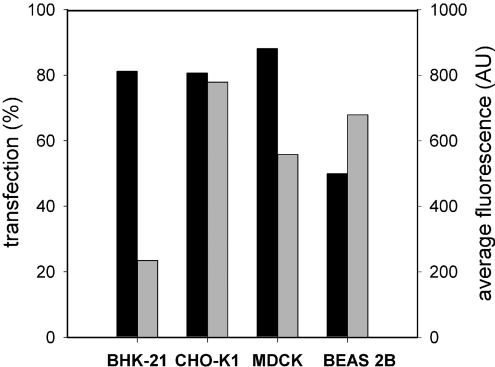
The cellular receptors for the influenza virus HA are sialic acid residues, which are present on the majority of cells. Accordingly, DNA–virosomes should be able to transfect many different cell lines. To investigate whether this holds true, BHK-21, CHO-K1, MDCK and BEAS 2B cells were exposed to virosomes with encapsulated EGFP plasmid. Flow cytometric analysis showed that transfection efficiencies of more than 80%, similar to those observed for BHK-21 cells were obtained for CHO-K1 and MDCK cells after 48 h (Figure 10). The percentage of transfected BEAS 2B cells was lower, but still 50% of the cells expressed EGFP. The average EGFP fluorescence per cell was up to three times higher for the CHO-K1, MDCK and BEAS 2B cell lines as compared with BHK-21 cells. Hence, virosomes are efficient transport devices for the delivery of encapsulated pDNA to a variety of cell lines.
Figure 10. Transfection of various cell lines mediated by DNA–virosomes.
BHK-21 (5×104), CHO-K1 (5×104), MDCK (4×104) and BEAS 2B (1×105) cells were incubated with DOTAP virosomes containing the CMV/EGFP plasmid (750 ng). After 48 h the transfection percentages (closed bars) and average fluorescence per cell (grey bars) were determined by flow cytometry.
DISCUSSION
In the present study, we show that, using a dialysis procedure for virosome reconstitution, pDNA can be highly condensed and probably encapsulated in the lumen of influenza virosomes, such that it is effectively protected from degradation by nucleases. Encapsulation required the simultaneous presence of pDNA and cationic lipid during reconstitution of the virus membrane. Reconstitution resulted in small membrane vesicles consisting of pDNA, phospholipid and protein and displaying prominent spikes on their surface. These DNA–virosomes retained the receptor-binding and membrane fusion activity of the native influenza virus. The DNA–virosomes efficiently transfected various cell lines in an HA-dependent manner. The transfection efficiency of the DNA–virosomes was of the same order of magnitude as that of FuGENE®, one of the most effective and popular reagents for in vitro transfection.
Extracellular nucleases form a potential problem for the delivery of DNA to cells in vivo. The presence of these nucleases was demonstrated by Barry and colleagues [37], who found that all mouse tissues and blood contain varying levels of calcium-dependent and acidic nucleases. Indeed, rapid degradation of naked DNA has been observed in different serum samples, lung fluids, tumours and after intravenous injection into mice [38–41]. Even successful expression of foreign genes upon intramuscular or subcutaneous injection of naked DNA is accompanied by degradation of 99% of the injected DNA by extracellular nucleases within 90 min [37]. These observations imply that the efficiency of gene delivery may be improved considerably through proper protection of the DNA.
With our previously reported method to prepare DNA–virosomes, it did not appear possible to improve the protection of the surface-attached pDNA by increasing the charge ratio and thus DNA condensation without a negative effect on the transfection efficiency [16]. With the new procedure presented here, high charge ratios and thus high condensation were found to be very well compatible with high transfection levels. In DNA–virosomes prepared according to the new protocol, pDNA is strongly condensed and efficiently protected, presumably due to encapsulation in the virosomal lumen, which shields the pDNA from nuclease activity. A similar procedure using dialysis of a mixture of detergent-dissolved DODAC, DOPE, PEG [poly(ethylene glycol)] and pDNA has been applied earlier to encapsulate pDNA in SPLPs (stabilized plasmid–lipid particles; [42,43]). Cryo-electron microscopic analysis revealed that the DNA is encapsulated in these SPLPs [43]. Although these results cannot be extrapolated directly to the current system, they do show that it is feasible to encapsulate DNA in lipid vesicles using a dialysis procedure.
Reconstitution of the viral influenza membrane enables the exploitation of the receptor-binding and membrane fusion properties of the HA spike protein for the delivery of pDNA to the cellular cytoplasm. Indeed, virosomes efficiently delivered encapsulated pDNA to a range of target cells. Approx. 80% of these cells expressed the EGFP protein after incubation with virosomes containing an EGFP reporter plasmid (Figures 5, 6 and 10). Transfection of cells with DNA–virosomes is mediated by the low pH-dependent fusogenic properties of the virosomal HA as indicated by the fact that inactivation of the virosomal HA or prevention of endosome acidification abolished transfection completely (Figure 9).
Encapsulation of pDNA in virosomes was achieved by condensing the pDNA using either one of two different cationic lipids, DODAC or DOTAP. These cationic lipids are distinct only in their polar headgroups. The headgroup of DODAC is coupled directly to two aliphatic chains, whereas the headgroup of DOTAP is separated from the anchor by a propyl chain. Lower accessibility of the polar headgroup, due to steric hindrance, has been described to influence the condensation efficiency of cationic lipids [44,45]. When we assessed the condensation of the encapsulated pDNA in the final virosome preparation, using the intercalating dye PicoGreen, the extent of condensation for the DOTAP virosomes was higher than for the DODAC virosomes (results not shown). This is probably related to the lower accessibility of the headgroup of DODAC. However, with respect to the other physical characteristics, fusion properties and transfection efficiency, we observed no differences between virosomes prepared with the two different cationic lipids. This implies that the procedure of DNA–virosome preparation tolerates minor modifications in the structure of the cationic lipid. This adds to the flexibility of the procedure, which is of potential advantage, since the choice of cationic lipids rapidly grows and future cationic lipids may be more efficient in DNA condensation and induce less toxic effects, and yet be compatible with incorporation in DNA-virosomes.
Potential drawbacks of virosomes for the purpose of in vivo DNA delivery, which are also associated with viral gene delivery systems, are related to their immunogenicity. This might not be much of a problem if the virosomes are directly administered to immuno-priviledged sites, such as the eye or the brain, or to sites where antibody induction occurs only in the presence of a strong adjuvant, such as the lungs [46]. However, administration of virosomes to other sites will induce HA-specific and NA-specific antibodies, which could interfere with subsequent attempts to deliver DNA with the same virosomes. Moreover, pre-existing antibodies induced by prior influenza infection or vaccination may impede application of DNA–virosomes for gene delivery purposes. Yet, there are several possibilities to circumvent these problems. Virosomes may be prepared from a strain of influenza virus which is not recognized by pre-existing antibodies. Alternatively, the virosomal surface can be coated with poly(ethylene glycol), a procedure which shields the HA molecules without necessarily impairing fusion activity after uptake into endosomes [47,48].
In other cases, for example if DNA–virosomes are to be used for DNA vaccination purposes, the presence of antibodies might in fact be beneficial, since opsonization of the virosomes with these antibodies will target them to cells with Fc-receptors, such as dendritic cells [49]. Fusion of influenza virus with the endosomal membrane is not impaired by antibody binding, as suggested by the phenomenon of antibody-mediated-enhanced uptake of influenza virus [46,50]. It is therefore likely that virosomes maintain their delivery properties in the presence of HA-specific antibodies, at least to cells bearing Fc-receptors.
Efficient in vivo gene delivery requires the availability of transport systems that circumvent several obstacles encountered by the gene construct on the way from administration to final expression. Virosomes, by combining specific features of viral and non-viral gene delivery systems, exhibit a number of attractive characteristics in this respect. First, as shown in the present study, the potential problem of degradation of the gene of interest by extracellular nucleases can be resolved by encapsulation of the DNA construct in the lumen of virosomes. This represents a considerable improvement over current procedures involving naked DNA or DNA bound to the surface of virosomes [16], and is likely to facilitate future in vivo application of DNA–virosomes. Secondly, properly reconstituted virosomes preserve the receptor-binding and membrane fusion properties of the virus they are derived from. Thus, virosomes, generated from influenza virus, target cells with sialic acid residues and trigger virosome uptake by endocytosis upon receptor-binding. Re-targeting of influenza-derived virosomes to specific cells may be accomplished by the incorporation in the virosomal membrane of PEG-derivatized lipids coupled to Fab fragments directed against a receptor on the desired target cell [48]. Thirdly, once taken up by endocytosis, virosomes overcome the physical barrier of the endosomal membrane by virtue of the membrane fusion activity of the viral envelope glycoproteins embedded in the virosomal membrane. In the case of influenza-derived virosomes, HA mediates active delivery of the gene of interest to the cellular cytosol. In conclusion, virosomes prepared in the presence of pDNA represent an efficient carrier system for gene delivery to cultured cells, and may be highly suitable for defined in vivo applications.
Acknowledgments
This work was supported by The Netherlands Organization for Scientific Research (NWO), under the auspices of the Chemical Foundation (CW) and the Technology Foundation (STW, Grant 790.33.571). We thank Solvay Pharmaceuticals (Weesp, The Netherlands) for generous supplies of influenza virus. Dr Klaas Sjollema, Department of Eukaryotic Microbiology, University of Groningen is acknowledged for excellent technical help with the electron microscopy experiments.
References
- 1.Kay M. A., Glorioso J. C., Naldini L. Viral vectors for gene therapy: the art of turning infectious agents into vehicles of therapeutics. Nat. Med. 2001;7:33–40. doi: 10.1038/83324. [DOI] [PubMed] [Google Scholar]
- 2.Thomas C. E., Ehrhardt A., Kay M. A. Progress and problems with the use of viral vectors for gene therapy. Nat. Rev. Genet. 2003;4:346–358. doi: 10.1038/nrg1066. [DOI] [PubMed] [Google Scholar]
- 3.Hacein-Bey-Abina S., von Kalle C., Schmidt M., McCormack M. P., Wulffraat N., Leboulch P., Lim A., Osborne C. S., Pawliuk R., Morillon E., et al. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302:415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- 4.Morral N., O'Neal W. K., Rice K., Leland M. M., Piedra P. A., Aguilar-Cordova E., Carey K. D., Beaudet A. L., Langston C. Lethal toxicity, severe endothelial injury, and a threshold effect with high doses of an adenoviral vector in baboons. Hum. Gene Ther. 2002;13:143–154. doi: 10.1089/10430340152712692. [DOI] [PubMed] [Google Scholar]
- 5.Kafri T., Morgan D., Krahl T., Sarvetnick N., Sherman L., Verma I. Cellular immune response to adenoviral vector infected cells does not require de novo viral gene expression: implications for gene therapy. Proc. Natl. Acad. Sci. U.S.A. 1998;95:11377–11382. doi: 10.1073/pnas.95.19.11377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schmidt-Wolf G. D., Schmidt-Wolf I. G. Non-viral and hybrid vectors in human gene therapy: an update. Trends Mol. Med. 2003;9:67–72. doi: 10.1016/s1471-4914(03)00005-4. [DOI] [PubMed] [Google Scholar]
- 7.Kumar V. V., Singh R. S., Chaudhuri A. Cationic transfection lipids in gene therapy: successes, set-backs, challenges and promises. Curr. Med. Chem. 2003;10:1297–1306. doi: 10.2174/0929867033457458. [DOI] [PubMed] [Google Scholar]
- 8.Dass C. R. Lipoplex-mediated delivery of nucleic acids: factors affecting in vivo transfection. J. Mol. Med. 2004;82:579–591. doi: 10.1007/s00109-004-0558-8. [DOI] [PubMed] [Google Scholar]
- 9.Pedroso de Lima M. C., Neves S., Filipe A., Düzgünes N., Simoes S. Cationic liposomes for gene delivery: from biophysics to biological applications. Curr. Med. Chem. 2003;10:1221–1231. doi: 10.2174/0929867033457430. [DOI] [PubMed] [Google Scholar]
- 10.Miller A. D. The problem with cationic liposome/micelle-based non-viral vector systems for gene therapy. Curr. Med. Chem. 2003;10:1195–1211. doi: 10.2174/0929867033457485. [DOI] [PubMed] [Google Scholar]
- 11.Gao X., Huang L. A novel cationic liposome reagent for efficient transfection of mammalian cells. Biochem. Biophys. Res. Commun. 1991;179:280–285. doi: 10.1016/0006-291x(91)91366-k. [DOI] [PubMed] [Google Scholar]
- 12.Porteous D. J., Dorin J. R., McLachlan G., Davidson-Smith H., Davidson H., Stevenson B. J., Carothers A. D., Wallace W. A., Moralee S., Hoenes C., et al. Evidence for safety and efficacy of DOTAP cationic liposome mediated CFTR gene transfer to the nasal epithelium of patients with cystic fibrosis. Gene Ther. 1997;4:210–218. doi: 10.1038/sj.gt.3300390. [DOI] [PubMed] [Google Scholar]
- 13.Noone P. G., Hohneker K. W., Zhou Z., Johnson L. G., Foy C., Gipson C., Jones K., Noah T. L., Leigh M. W., Schwartzbach C., et al. Safety and biological efficacy of a lipid–CFTR complex for gene transfer in the nasal epithelium of adult patients with cystic fibrosis. Mol. Ther. 2000;1:105–114. doi: 10.1006/mthe.1999.0009. [DOI] [PubMed] [Google Scholar]
- 14.Earp L. J., Delos S. E., Park H. E., White J. M. The many mechanisms of viral membrane fusion proteins. Curr. Top. Microbiol. Immunol. 2004;285:25–66. doi: 10.1007/3-540-26764-6_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jardetzky T. S., Lamb R. A. Virology: a class act. Nature. 2004;427:307–308. doi: 10.1038/427307a. [DOI] [PubMed] [Google Scholar]
- 16.Schoen P., Chonn A., Cullis P. R., Wilschut J., Scherrer P. Gene transfer mediated by fusion protein hemagglutinin reconstituted in cationic lipid vesicles. Gene Ther. 1999;6:823–832. doi: 10.1038/sj.gt.3300919. [DOI] [PubMed] [Google Scholar]
- 17.de Jonge J., Holtrop M., Wilschut J., Huckriede A. Reconstituted influenza virus envelopes as an efficient carrier system for cellular delivery of small-interfering RNAs. Gene Ther. 2006;13:400–411. doi: 10.1038/sj.gt.3302673. [DOI] [PubMed] [Google Scholar]
- 18.Correale P., Cusi M. G., Sabatino M., Micheli L., Pozzessere D., Nencini C., Valensin P. E., Petrioli R., Giorgi G., Zurbriggen R., et al. Tumour-associated antigen (TAA)-specific cytotoxic T cell (CTL) response in vitro and in a mouse model, induced by TAA-plasmids delivered by influenza virosomes. Eur. J. Cancer. 2001;37:2097–2103. doi: 10.1016/s0959-8049(01)00241-6. [DOI] [PubMed] [Google Scholar]
- 19.Daemen T., de Mare A., Bungener L., de Jonge J., Huckriede A., Wilschut J. Virosomes for antigen and DNA delivery. Adv. Drug Deliv. Rev. 2005;57:451–463. doi: 10.1016/j.addr.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 20.Ramani K., Hassan Q., Venkaiah B., Hasnain S. E., Sarkar D. P. Site-specific gene delivery in vivo through engineered Sendai viral envelopes. Proc. Natl. Acad. Sci. U.S.A. 1998;95:11886–11890. doi: 10.1073/pnas.95.20.11886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ponimaskin E., Bareesel K. K., Markgraf K., Reszka R., Lehmann K., Gelderblom H. R., Gawaz M., Schmidt M. F. Sendai virosomes revisited: reconstitution with exogenous lipids leads to potent vehicles for gene transfer. Virology. 2000;269:391–403. doi: 10.1006/viro.2000.0233. [DOI] [PubMed] [Google Scholar]
- 22.Shoji J., Tanihara Y., Uchiyama T., Kawai A. Preparation of virosomes coated with the vesicular stomatitis virus glycoprotein as efficient gene transfer vehicles for animal cells. Microbiol. Immunol. 2004;48:163–174. doi: 10.1111/j.1348-0421.2004.tb03502.x. [DOI] [PubMed] [Google Scholar]
- 23.Uchida T., Kim J., Yamaizumi M., Miyake Y., Okada Y. Reconstitution of lipid vesicles associated with HVJ (Sendai virus) spikes: purification and some properties of vesicles containing nontoxic fragment A of diphtheria toxin. J. Cell Biol. 1979;80:10–20. doi: 10.1083/jcb.80.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stegmann T., Morselt H. W. M., Booy F. P., Van Breemen J. F. L., Scherphof G., Wilschut J. Functional reconstitution of influenza virus envelopes. EMBO J. 1987;6:2651–2659. doi: 10.1002/j.1460-2075.1987.tb02556.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paternostre M., Viard M., Meyer O., Ghanam M., Ollivon M., Blumenthal R. Solubilization and reconstitution of vesicular stomatitis virus envelope using octylglucoside. Biophys. J. 1997;72:1683–1694. doi: 10.1016/S0006-3495(97)78814-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dzau V. J., Mann M. J., Morishita R., Kaneda Y. Fusigenic viral liposome for gene therapy in cardiovascular diseases. Proc. Natl. Acad. Sci. U.S.A. 1996;93:11421–11425. doi: 10.1073/pnas.93.21.11421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peterson G. L. A simplification of the protein assay method of Lowry et al. which is more generally applicable. Anal. Biochem. 1977;83:346–356. doi: 10.1016/0003-2697(77)90043-4. [DOI] [PubMed] [Google Scholar]
- 28.Böttcher C. J. F., van Gent C. M., Pries C. A rapid and sensitive sub-micro phosphorus determination. Anal. Chim. Acta. 1961;24:203–204. [Google Scholar]
- 29.de Jonge J., Schoen P., ter Veer W., Stegmann T., Wilschut J., Huckriede A. Use of a dialyzable short-chain phospholipid for efficient solubilization and reconstitution of influenza virus envelopes. Biochim. Biophys. Acta. 2006;1758:527–536. doi: 10.1016/j.bbamem.2006.03.011. [DOI] [PubMed] [Google Scholar]
- 30.Ferrari M. E., Nguyen C. M., Zelphati O., Tsai Y., Felgner P. L. Analytical methods for the characterization of cationic lipid–nucleic acid complexes. Hum. Gene Ther. 1998;9:341–351. doi: 10.1089/hum.1998.9.3-341. [DOI] [PubMed] [Google Scholar]
- 31.Galla H. J., Hartmann W. Excimer-forming lipids in membrane research. Chem. Phys. Lipids. 1980;27:199–219. doi: 10.1016/0009-3084(80)90036-5. [DOI] [PubMed] [Google Scholar]
- 32.Stegmann T., Schoen P., Bron R., Wey J., Bartoldus I., Ortiz A., Nieva J. L., Wilschut J. Evaluation of viral membrane-fusion assays: comparison of the octadecylrhodamine dequenching assay with the pyrene excimer assay. Biochemistry. 1993;32:11330–11337. doi: 10.1021/bi00093a009. [DOI] [PubMed] [Google Scholar]
- 33.Bungener L., Serre K., Bijl L., Leserman L., Wilschut J., Daemen T., Machy P. Virosome-mediated delivery of protein antigens to dendritic cells. Vaccine. 2002;20:2287–2295. doi: 10.1016/s0264-410x(02)00103-2. [DOI] [PubMed] [Google Scholar]
- 34.Lamb R. A., Krug R. M. Orthomyxoviridae: the viruses and their replication. In: Knipe D. M., Howley P. M., editors. Fields Virology. Philadelphia: Lippincott Williams & Wilkins; 2001. pp. 1487–1531. [Google Scholar]
- 35.Puri A., Booy F. P., Doms R. W., White J. M., Blumenthal R. Conformational changes and fusion activity of influenza virus hemagglutinin of the H2 and H3 subtypes: effects of acid pretreatment. J. Virol. 1990;64:3824–3832. doi: 10.1128/jvi.64.8.3824-3832.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ohkuma S., Poole B. Fluorescence probe measurement of the intralysosomal pH in living cells and the perturbation of pH by various agents. Proc. Natl. Acad. Sci. U.S.A. 1978;75:3327–3331. doi: 10.1073/pnas.75.7.3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barry M. E., Pinto-Gonzalez D., Orson F. M., McKenzie G. J., Petry G. R., Barry M. A. Role of endogenous endonucleases and tissue site in transfection and CpG-mediated immune activation after naked DNA injection. Hum. Gene Ther. 1999;10:2461–2480. doi: 10.1089/10430349950016816. [DOI] [PubMed] [Google Scholar]
- 38.Houk B. E., Hochhaus G., Hughes J. A. Kinetic modeling of plasmid DNA degradation in rat plasma. AAPS. PharmSci. 1999;1:1–6. doi: 10.1208/ps010309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kawabata K., Takakura Y., Hashida M. The fate of plasmid DNA after intravenous injection in mice: involvement of scavenger receptors in its hepatic uptake. Pharm. Res. 1995;12:825–830. doi: 10.1023/a:1016248701505. [DOI] [PubMed] [Google Scholar]
- 40.Glasspool-Malone J., Steenland P. R., McDonald R. J., Sanchez R. A., Watts T. L., Zabner J., Malone R. W. DNA transfection of macaque and murine respiratory tissue is greatly enhanced by use of a nuclease inhibitor. J. Gene Med. 2002;4:323–322. doi: 10.1002/jgm.259. [DOI] [PubMed] [Google Scholar]
- 41.Walther W., Stein U., Siegel R., Fichtner I., Schlag P. M. Use of the nuclease inhibitor aurintricarboxylic acid (ATA) for improved non-viral intratumoral in vivo gene transfer by jet-injection. J. Gene Med. 2005;7:477–485. doi: 10.1002/jgm.690. [DOI] [PubMed] [Google Scholar]
- 42.Wheeler J. J., Palmer L., Ossanlou M., MacLachlan I., Graham R. W., Zhang Y. P., Hope M. J., Scherrer P., Cullis P. R. Stabilized plasmid–lipid particles: construction and characterization. Gene Ther. 1999;6:271–281. doi: 10.1038/sj.gt.3300821. [DOI] [PubMed] [Google Scholar]
- 43.Saravolac E. G., Ludkovski O., Skirrow R., Ossanlou M., Zhang Y. P., Giesbrecht C., Thompson J., Thomas S., Stark H., Cullis P. R., Scherrer P. Encapsulation of plasmid DNA in stabilized plasmid–lipid particles composed of different cationic lipid concentration for optimal transfection activity. J. Drug Target. 2000;7:423–437. doi: 10.3109/10611860009102217. [DOI] [PubMed] [Google Scholar]
- 44.Floch V., Loisel S., Guenin E., Herve A. C., Clement J. C., Yaouanc J. J., des Abbayes H., Ferec C. Cation substitution in cationic phosphonolipids: a new concept to improve transfection activity and decrease cellular toxicity. J. Med. Chem. 2000;43:4617–4628. doi: 10.1021/jm000006z. [DOI] [PubMed] [Google Scholar]
- 45.Ilies M. A., Seitz W. A., Balaban A. T. Cationic lipids in gene delivery: principles, vector design and therapeutical applications. Curr. Pharm. Des. 2002;8:2441–2473. doi: 10.2174/1381612023392748. [DOI] [PubMed] [Google Scholar]
- 46.Tamura M., Webster R. G., Ennis F. A. Subtype cross-reactive, infection-enhancing antibody responses to influenza A viruses. J. Virol. 1994;68:3499–3504. doi: 10.1128/jvi.68.6.3499-3504.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chams V., Bonnafous P., Stegmann T. Influenza hemagglutinin mediated fusion of membranes containing poly(ethylene-glycol) grafted lipids: new insights into the fusion mechanism. FEBS Lett. 1999;448:28–32. doi: 10.1016/s0014-5793(99)00333-6. [DOI] [PubMed] [Google Scholar]
- 48.Mastrobattista E., Schoen P., Wilschut J., Crommelin D. J., Storm G. Targeting influenza virosomes to ovarian carcinoma cells. FEBS Lett. 2001;509:71–76. doi: 10.1016/s0014-5793(01)03112-x. [DOI] [PubMed] [Google Scholar]
- 49.Cusi M. G., Terrosi C., Savellini G. G., Di Genova G., Zurbriggen R., Correale P. Efficient delivery of DNA to dendritic cells mediated by influenza virosomes. Vaccine. 2004;22:735–739. doi: 10.1016/j.vaccine.2003.08.024. [DOI] [PubMed] [Google Scholar]
- 50.Ochiai H., Kurokawa M., Matsui S., Yamamoto T., Kuroki Y., Kishimoto C., Shiraki K. Infection enhancement of influenza A NWS virus in primary murine macrophages by anti-hemagglutinin monoclonal antibody. J. Med. Virol. 1992;36:217–221. doi: 10.1002/jmv.1890360312. [DOI] [PubMed] [Google Scholar]