Abstract
Cn (calcineurin) activity is stabilized by SOD1 (Cu-Zn superoxide dismutase), a phenomenon attributed to protection from superoxide (O2•−). The effects of O2•− on Cn are still controversial. We found that O2•−, generated either in vitro or in vivo did not affect Cn activity. Yet native bovine, recombinant human or rat, and two chimaeras of human SOD1–rat SOD1, all activated Cn, but SOD2 (Mn-superoxide dismutase) did not affect Cn activity. There was also a poor correlation between SOD1 dismutase activity and Cn activation. A chimaera of human N-terminal SOD1 and rat C-terminal SOD1 had little detectable dismutase activity, yet stimulated Cn activity the same as full-length human or rat SOD1. Nevertheless, there was evidence that the active site of SOD1 was involved in Cn activation based on the loss of activation following chelation of Cu from the active site of SOD1. Also, SOD1 engaged in the catalysis of O2•− dismutation was ineffective in activating Cn. SOD1 activation of Cn resulted from a 90-fold decrease in phosphatase Km without a change in Vmax. A possible mechanism for the activation of Cn was identified in our studies as the prevention of Fe and Zn losses from the active site of Cn, suggesting a conformation-dependent SOD1–Cn interaction. In neurons, SOD1 and Cn were co-localized in cytoplasm and membranes, and SOD1 co-immunoprecipitated with Cn from homogenates of brain hippocampus and was present in immunoprecipitates as large multimers. Pre-incubation of pure SOD1 with Cn caused SOD1 multimer formation, an indication of an altered conformational state in SOD1 upon interaction with Cn.
Keywords: brain, calcineurin, hydrogen peroxide, iron, superoxide dismutase, zinc
Abbreviations: ALS, amyotrophic lateral sclerosis; ANS, 8-anilinonaphthalene-l-sulfonic acid; CaM, calmodulin; Cn, calcineurin; co-IP, co-immunoprecipitation; DETC, diethyldithiocarbamate; DTT, dithiothreitol; fALS, familial ALS; GdmCl, guanidinium chloride; 2-ME, 2-mercaptoethanol; MTT, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-2H-tetrazolium bromide; NBT, Nitro Blue Tetrazolium; NTA, nitrilotriacetic acid; PAR, 4-(2-pyridylazo)resorcinol; pNP, p-nitrophenol; pNPP, p-nitrophenylphosphate; PQ, paraquat; sALS, sproradic ALS; SOD1, Cu-Zn superoxide dismutase; hSOD1, human SOD1; rSOD1, rat SOD1; HN-RC, chimaera of N-terminal 76 residues of hSOD1 and C-terminal 78 residues of rSOD1; RN-HC, chimaera of N-terminal 76 residues of rSOD1 and C-terminal 78 residues of hSOD1; SOD2, Mn-superoxide dismutase; WT, wild-type; XO, xanthine oxidase
INTRODUCTION
Cn (calcineurin) is an Fe- and Zn-containing metallophosphatase [1–6]. The active form of the enzyme is a heterodimer of CnA and CnB. Activation of Cn occurs following Ca2+ entry through ion channels [7–11], but Cn activity may also be controlled by the redox state of the metals at the active site [12–16]. The importance of Cn in T-cell activation is well defined [17], as is its role in cardiac muscle structure [18]. In the mammalian brain, Cn is concentrated in regions that are crucial for learning, memory and the control of movement or compulsive behaviours. Changes in Cn levels and activity have been linked to age-related declines in memory formation [19,20], and, in some familial cases of schizophrenia, there is genetic variation in the gene encoding an isoform of CnA [21].
SOD1 (Cu-Zn superoxide dismutase), another enzyme with a dinuclear metal centre, has been shown to activate neuronal Cn [13,14]. The mechanism for the enhancement of Cn activity by WT (wild-type) SOD1 has not been resolved. It was proposed that SOD1 protects Cn from attack by superoxide (O2•−) [13], an attack assumed to occur on the metal centre of CnA. O2•− attack of Cn requires that the metal centre is in the form of Fe2+-Zn2+ [13]. However, the exact state of Fe in Cn in cells is not certain [5,12,22]. Thus the mechanism of activation of Cn by SOD1 remains uncertain. To resolve some of the issues surrounding SOD1 activation of Cn we have undertaken the following experimental approaches: (i) re-examine the effects of O2•− on Cn activity; and (ii) explore possible mechanisms for SOD1 activation of Cn by assessing the role of dismutase activity in Cn activation, structural changes in Cn proteins following interactions with SOD1, effects of differing structures of SOD1 on the activation of Cn, and effects of SOD1–Cn interactions on the Fe-Zn centre of Cn.
One of the most compelling reasons for undertaking a detailed examination of SOD1 interaction with Cn is that a key Ca2+-signalling macromolecule in cells, Cn, may not be fully active in diseases characterized by structural and functional alterations in SOD1, such as ALS (amyotrophic lateral sclerosis). Mutations in SOD1 are linked to the neurodegeneration in fALS (familial ALS). Also, the mutant forms of SOD1 that are Zn-deficient are less efficient in stabilizing or activating Cn [23,24]. The decreased activation of Cn by mutant SOD1 may be a true loss of function of SOD1 in neurons of fALS and sALS (sporadic ALS) patients [25]. However, there is currently no direct demonstration of Cn interaction with SOD1 in the cytoplasm of neurons in vivo. To address this issue, we have examined whether CnA co-localizes with SOD1 in primary neuronal cultures and whether CnA and SOD1 can be co-immunoprecipitated from brain tissue extracts.
EXPERIMENTAL
Materials
XO (xanthine oxidase), trypsin inhibitor, MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-2H-tetrazolium bromide], NBT (Nitro Blue Tetrazolium), bovine kidney and erythrocyte SOD1, Escherichia coli SOD2 (Mn-superoxide dismutase), protease inhibitor cocktail and all cell culture media were from Sigma Chemical Co. Catalase was from Boehringer Mannheim. ANS (8anilinonaphthalene-l-sulfonic acid) was from Fluka. Mouse monoclonal anti-CnA antibody was from Signal Transduction Laboratories. Polyclonal anti-CnB was from Upstate Biotechnology. Rabbit polyclonal anti-SOD1 was from Chemicon. PQ (paraquat) was from ICN Biochemicals. Trisacryl beads and Profound™ mammalian co-immunoprecipitation kit were from Pierce. Purified bovine brain Cn was from Biomol or Sigma–Aldrich. Turbo Pfu polymerase was from Stratagene. pTYB1 vector, chitin column and E. coli ER2566 were from New England Biolabs.
Cn preparations
Brain cytosol preparations were obtained from Sprague–Dawley male rats. All experimental procedures followed those of the Institutional Animal Care and Use Committee of the University of Kansas. Rat brain homogenates and cytosol supernatants were obtained as described in [26]. Protein concentration was estimated by the bicinchoninic acid assay and samples were divided into small aliquots and stored at −70 °C.
Cn was also freshly purified from bovine brain. Cortex homogenate in 0.1 M Tris/HCl, 2 mM EDTA, 10% (v/v) glycerol, pH 7.5, was sonicated three times for 30 s at 0–4 °C and centrifuged at 9000 g for 20 min, and the supernatant was filtered through glass wool. To the filtrate 10 mM 2-ME (2-mercaptoethanol) and 0.1 mM EGTA were added and the solution passed through DEAE-cellulose. Elution was initiated with buffer containing 1 mM imidazole, 1 mM MgSO4, 0.1 mM EGTA, 10 mM 2-ME and 20 mM Tris/HCl, pH 7.0, plus 3% glycerol, followed by elution of Cn with 0.25 M NaCl in the same buffer. After poly(ethylene glycol) precipitation and dialysis, the Cn-enriched fraction was subjected to Blue Sepharose chromatography. Purity was assessed by SDS/PAGE and immunoblotting with anti-CnA antibodies [26].
Recombinant human SOD1 (hSOD1) and rat SOD1 (rSOD1) and dismutase activity measurements
rSOD1 cDNA (pUC13-RCS) and hSOD1 cDNA (pET21b) were used to express and purify the respective proteins and to construct chimaeras of human SOD1–rat SOD1. For the chimaeras, the N- and C-terminal parts of hSOD1 and rSOD1 were amplified by PCR using the primer sets: hSOD1 N-terminus, F1 5′-TTTTCATATGGCCACGAAGGCCGTGTGCGTGCTG-3′ and R1 5′-CTCCAACATGCCTCTCTTCATCCTTTG-3′; hSOD1 C-terminus, F2 5′-CAGAAAACACGGTGGGCCAAAGGATG-3′ and R2 5′-TTTGGTACCCTTGGCAAAGCATTGGGCGATCCCAATTACAC-3′ (underlined sequence is Acc65I restriction site); rSOD1 N-terminus, F3 5′-TTTTCATATGGCGATGAAGGCCGTGTGCGTGCTG-3′ and R3 5′-GTCTCCAACATGCCTCTCTTCATCCGC-3′; rSOD1 C-terminus, F4 5′-TAAGAAACAT-GGCGGTCCAGCGGATG-3′ and R4 5′-TTTGGTACCCTTGGCAAAGCATTGGGCAATCCCAATCACAC-3′. PCR amplification with Turbo Pfu polymerase (50 μl reaction volume containing 2.5 units of polymerase, 15 pmol of each primer, 200 μM dNTP, 10 ng of template and PCR buffer). The amplification programme was 2 min at 95 °C, 32 cycles at 95 °C for 30 s, 60 °C for 30 s and 72 °C for 60 s. Amplified DNA fragments of N-terminal hSOD1 and C-terminal rSOD1 were mixed, digested with EarI, and religated to yield the N-terminal hSOD1–C-terminal rSOD1 cDNA. Identical procedures were used to obtain DNA of N-terminal rSOD1 and C-terminal hSOD1. The ligated products were used as PCR templates to form the two chimaeric DNAs. The primer sets were: hSOD1 F1 5′-TTTTCATATGGCCACGAAGGCCGTGTGCGTGCTG-3′ and rSOD1 R4 5′-TTTGGTACCCTTGGCAAAGCATTGGGCAATCCCAATCACAC-3′ for N-terminal hSOD1–C-terminal rSOD1 chimaera; and rat F3 5′-TTTTCATATGGCGATGAAGGCCGTGTGCGTGCTG-3′ and human R2 5′-TTTGGTACCCTTGGCAAAGCATTGGGCGATCCCAATTACAC-3′ for N-terminal rSOD1–C-terminal hSOD1 chimaera. The DNAs were cut with NdeI and Acc65I and cloned into pTYB1. The cDNAs for intact rSOD1 and hSOD1 were also amplified using forward and reverse primers with NdeI and Acc651.
To express and purify the respective proteins, E. coli strain ER2566 was transformed with pTYB1, induced with 0.4 mM IPTG (isopropyl β-D-thiogalactoside), grown at 20 °C for 15 h, harvested, resuspended in 5 ml of lysis buffer (500 mM NaCl, 20 μg/ml lysozyme, 0.2% Triton X-100 and 20 mM Tris/HCl, pH 8.0), incubated at 20 °C for 1 h, passed ten times through a 25 G needle and centrifuged at 20000 g for 1 h, and the supernatant was loaded on to 2 ml of a chitin matrix. Following washing with 50 ml of lysis buffer without lysozyme and Triton X-100, then with 50 ml of 1 M NaCl and 50 ml of buffer, the intein was activated by buffer containing 40 mM DTT (dithiothreitol). After elution of the first 8 ml, column flow was stopped, the matrix was incubated at 4 °C overnight, and the cleaved proteins were collected in 15 ml of elution buffer.
Purified hSOD1 depleted of Zn as described [23] was used for the measurement of protein aggregation following extraction of Zn from the active site [23]. Formation of protein aggregates by either intact or Zn-depleted SOD1 was estimated as the change in fluorescence of 10 μM ANS bound to 5 μM SOD1 at 23 °C [27].
A different set of recombinant hSOD1 proteins were expressed in BL21 bacterial cells, purified by chromatography through DEAE-Sephacel followed by anion-exchange HPLC and the metal ion content was estimated [23]. SOD activity in native gels was measured by staining with riboflavin/NBT [28] [soaked in 0.2% NBT, 0.28 M TEMED (N,N,N′,N′-tetramethylethylenediamine), 56 μM riboflavin and 50 mM potassium phosphate buffer, pH 7.8, 30 min at 23 °C]. SOD activity in solution was measured as described in [29].
Treatment of Cn with SOD1 and Cn activity assays
Purified SOD1 preparations were pre-incubated with Cn for 25–50 min at 30 °C. Free phosphate in the buffer of SOD1 preparations was removed by filtration through Sephadex G25. The same was done for brain cytosol preparations. Two assays were used to determine Cn activity: hydrolysis of pNPP (p-nitrophenylphosphate) [30] and dephosphorylation of a phosphopeptide substrate, RII phosphopeptide [DLDVPIPGNFDNNVS(-P)VCAE] [31]. All assays were performed in 96-well microplates under aerobic conditions. For the pNPP assay, either 4 μg of rat brain cytosol or 0.5–2.0 μg of purified bovine brain Cn was added to 90 μl of assay buffer [1 mM Ca2+, 0.1 μM CaM (calmodulin), 0.1 M NaCl, 6 mM MgCl2, 0.05 mM MnCl2, 0.1 mg/ml BSA, 0.5 mM DTT and 2.5 mM pNPP in 20 mM Tris/HCl buffer, pH 7.5]. For estimation of background absorbance, 10 μl of buffer was added in place of protein. The reaction mixture was incubated for 30 min at 37 °C, and the absorbance was measured at 405 nm. For the assays using the RII phosphopeptide, the reaction mixture consisted of 7 μl of substrate solution (60 μg RII/ml), 63 μl of buffer (0.2 mM EGTA, 10 mM MgCl2, 0.4 mM CaCl2, 0.1 μM CaM, 0.02% 2-ME, 50 mM imidazole, pH 7.2), and rat brain cytosol or purified Cn (final concentrations 4.7 μg/ml and 0.2 μM respectively). After incubation for 30 min at 37 °C, 50 μl of Malachite Green plus sodium molybdate were added, and, 2 min later, 100 μl of 3.66 M H2SO4. Colour was measured 45 min later (absorbance at 660 nm).
Exposure of Cn preparations to O2•− and H2O2
Different preparations of Cn were pre-incubated for 25 min at 30 °C with O2•− generated by the oxidation of 25 μM (or various concentrations) xanthine by 0.01 units/ml XO. Formation of O2•− was monitored at 570 nm as the reduction of 100 μM NBT [32,33]. Trypsin contaminants of XO preparations were blocked with 5 mg/ml trypsin inhibitor. The trypsin inhibitor had no effect on Cn activity. The effects of H2O2 on Cn were assessed by pre-incubating Cn for 25 min at 30 °C with H2O2, either in the absence or presence of 130 units/ml catalase. To test whether the effect of H2O2 was produced through the oxidation of CaM rather than Cn, brain CaM (0.1 μM) that was fully oxidized through incubation for 10 h with 10 mM H2O2 was used in place of freshly prepared brain CaM.
Neuronal cell cultures and PQ treatment
Primary cultures of cortical neurons were prepared from day-18 embryonic rat brain exactly as described in [34]. Cells were plated in 35-mm-diameter dishes and were maintained either under 5% CO2 and atmospheric O2 (37 °C) or under reduced O2 conditions (CO2/O2/N2, 1:1:18). Cell survival was measured using the MTT reduction assay [35]. Cn activity was measured following exposure of cultures to PQ (0.5, 1.0 or 2.5 mM) added to the medium on day 7. Following incubation for 1 h, the cells were rinsed with saline, harvested and homogenized in the same buffer as brain samples. Following centrifugation at 13400 g for 10 min to remove particulates, Cn activity in cytosol was measured. O2•− formation in cells was measured by following the inactivation of aconitase by O2•− in the cytosol fraction. Aconitase activity was measured as the formation of cis-aconitate (at 240 nm) in buffer containing 20 mM isocitrate, 0.6 mM MnCl2 and 50 mM Tris/HCl buffer, pH 7.4.
Immunohistochemistry of CnA and SOD1 in cortical neurons and co-IP (co-immunoprecipitation) studies
Primary cortical neurons on glass coverslips were fixed and processed for immunohistochemistry as described in [35]. The cells were incubated first with polyclonal anti-SOD1 (1:500 dilution) and monoclonal anti-CnA (1:250 dilution), then rinsed, labelled with Cy5 (indodicarbocyanine)-labelled anti-rabbit and Cy3 (indocarbocyanine)-labelled anti-mouse antibodies and were examined by confocal microscopy.
For the co-IP studies, dissected hippocampi (adult rats) were homogenized in buffer as described in [37] and centrifuged at 500 g for 10 min, and the supernatant was collected and centrifuged at 20000 g for 20 min. The pellet from the last centrifugation was suspended in buffer (150 mM KCl, 1% Triton X-100, 4.5 mM magnesium acetate, 0.5 mM PMSF and 20 mM Mops, pH 7.0) containing a protease inhibitor cocktail tablet. Following incubation at 37 °C for 30 min, the suspension was centrifuged at 1000 g for 60 min and the supernatant was incubated for 16 h at 4 °C with 2.8 μg/μl anti-CnA–agarose beads. Bound proteins were eluted (buffer supplied by manufacturer), and subjected to SDS/PAGE and immunoblot analysis using anti-CnA, anti-CnB and anti-SOD1 (1:1000 dilution). A similar method of co-IP of mixtures of purified Cn and SOD1 was used for determination of complex formation between the two proteins in vitro.
Estimation of metal released from Cn using PAR [4-(2-pyridylazo)resorcinol] chelation
The release of Cn-associated metals was detected by measuring the formation of the metal–PAR2 complex in sodium borate buffer [23]. Freshly purified bovine brain Cn (7 μM) was added to a solution containing either 100 μM PAR in 1.4 ml of 0.1 mM sodium borate (pH 7.8), or in the same solution containing 6 M GdmCl (guanidinium chloride). The increase in absorbance at 500 nm was monitored. When there was no further change in absorbance (after ∼4 min), 0.8 mM NTA (nitrilotriacetic acid) was added, and the decrease in absorbance of the metal–PAR2 complex was recorded. The same procedures were used to determine metal loss from SOD1 or BSA (negative control). The effect of SOD1 on the release of metals from Cn was measured by pre-incubating Cn with 2–15 μM SOD1 for 30 min at 30 °C.
Statistical analyses and curve fitting
Statistical analysis was carried out using Student's t test. Non-linear least squares was used to fit data to Michaelis–Menten, Hill or logistic equations.
RESULTS
O2•− does not affect brain Cn, yet SOD1 activates Cn
The phosphatase activity of brain cytosol was compared with that of purified brain Cn in terms of sensitivity to okadaic acid. Purified Cn preparations had only two proteins, as found by SDS/PAGE: a 61 kDa protein recognized by anti-CnA in immunoblots and a 19 kDa protein recognized by anti-CnB. Okadaic acid at 500 nM nearly completely inhibits the protein phosphatases PP1 and PP2A, but has only a modest effect on Cn. The enzyme activity of purified Cn was inhibited by okadaic acid by 22% and that in brain cytosol by 18% (results not shown). Equivalent inhibition of phosphatase activities by okadaic acid in the two preparations was an indication that neither PP1 nor PP2A contributed to the phosphatase activity being measured in brain cytosol.
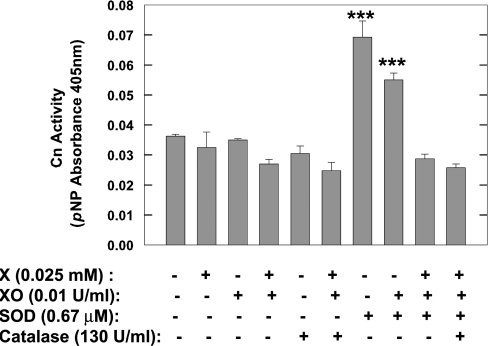
Pre-incubation of brain cytosol with xanthine alone, XO alone or the O2•−-generating system of xanthine plus XO, had no effect on Cn activity (Figure 1). These experiments were repeated using purified bovine brain Cn and, once again, pre-incubation of purified Cn with xanthine plus XO had no effect on the enzyme (results not shown). The inclusion of catalase in the pre-incubation mixture also did not alter Cn activity, irrespective of the presence or absence of the O2•−-generating system (Figure 1). Pre-incubation of brain cytosol with increasing amounts of O2•− generated by varying the concentration of xanthine (0.1–0.5 mM) while maintaining XO constant (0.025 unit/ml) also did not affect Cn activity (results not shown).
Figure 1. Effects of O2•−, SOD1 and catalase on Cn activity in brain cytosol.
Rat brain cytosol was exposed to O2•−-generating system [25 μM xanthine (X) and 0.01 unit/ml XO], 0.67 μM SOD1, or both, in the absence or presence of 130 units/ml catalase. Cn activity was measured following pre-incubation with the various agents for 25 min at 30 °C. Control samples pre-incubated with buffer only. Results are means±S.E.M. Cn specific activities from duplicate determinations from three experiments. ***P<0.001.
Although O2•− generated in vitro did not have an effect on Cn activity, it is possible that O2•− in neurons might inactivate Cn. PQ leads to the formation of O2•− in cells [39] and was used in these studies to generate excess O2•− in primary neurons. Neuronal cultures maintained under atmospheric O2 encounter higher levels of O2 than neurons in the cell body. For this reason, we grew and treated with PQ two types of neuronal cultures: neurons maintained in 5% CO2 and atmospheric O2, and those in a reduced O2 atmosphere of CO2/O2/N2 (1:1:18). Cortical neurons under reduced O2 atmosphere had the same survival at 7, 14 and 21 days in culture as neurons maintained under atmospheric O2 (R. Braceras and E, Michaelis, unpublished work). Exposure of neurons (18–24 h) to concentrations of PQ as low as 40 μM produced substantial cell death (>30%). Therefore we restricted the period of exposure to PQ to 1 h. Loss of cytoplasmic aconitase activity was used to detect intraneuronal generation of O2•−. Incubation of neurons maintained in atmospheric O2 with 250 μM PQ for 1 h, suppressed aconitase by 46% as compared with control neurons; similar incubation with 2.5 mM PQ inhibited aconitase by 80%. Yet incubation of neuronal cultures maintained under atmospheric O2 for 1 h with 2.5 mM PQ, had no effect on Cn activity (control: 6.6±0.3 nmol of Pi/mg of protein; and 2.5 mM PQ: 6.2±0.5 nmol of Pi/mg of protein). Very similar results were obtained from cortical cultures maintained under reduced O2 conditions (control: 6.5±0.4 nmol of Pi/mg of protein; and 2.5 mM PQ: 6.4±0.4 nmol of Pi/mg of protein). Thus a reduced O2 atmosphere had no effect on basal Cn activity, nor did it alter the lack of effect of PQ-generated O2•− on neuronal Cn.
Even though O2•− in the environment of Cn had no effect on enzyme activity, the pre-incubation of rat brain cytosol with SOD1 still led to a marked increase in Cn activity (Figure 1). Chelation of Cu in SOD1 partially inactivates SOD1 [40] and alters its conformation [41,42]. We tested whether chelation of Cu in SOD1 by DETC (diethyldithiocarbamate) before incubation with brain cytosol altered the SOD1-induced activation of Cn. Pre-reaction of bovine erythrocyte SOD1 with 0.2 mM DETC diminished SOD1 activation of Cn, but did not eliminate it (Cn activity: control cytosol, 7.5±1.0 nmol of Pi/min per mg; cytosol+0.2 mM DETC, 6.6±0.9 nmol of Pi/min per mg; cytosol+SOD1, 12.7±1.4 nmol of Pi/min per mg; cytosol+DETC-treated SOD1, 9.3±0.9 nmol of Pi/min per mg). DETC in the absence of SOD1 (cytosol+0.2 mM DETC) had no significant effect on Cn activity.
Engaging the active site of SOD1 in the process of catalysis, i.e. when SOD1 was involved in dismutation of O2•−, led to loss of SOD1 efficacy in activating Cn (Figure 1). The loss of Cn activation by SOD1 might have been the result of formation of H2O2 from O2•− and subsequent inactivation of Cn by H2O2. If this were the case, then catalase should protect Cn from the effects of H2O2. However, Cn activity remained at basal levels regardless of the presence or absence of catalase in the medium (Figure 1). The maximum concentration of H2O2 that might have been generated if all O2•− formed by the xanthine+XO reaction were converted into H2O2 by SOD1 would have been 0.025 mM. But this concentration had no effect on either basal or SOD1-stimulated Cn activity (ratio of activities in the presence and absence of H2O2 was 1.16; n=3). Thus a likely explanation for the loss of the stimulatory effect of SOD1 on Cn might be that the conformation of SOD1 during catalysis disrupted Cn–SOD1 interactions.
Others have reported inhibition of Cn by high concentrations of H2O2 [43]. We too observed significant inhibition of cytosol Cn by H2O2 when [H2O2] was greater than 100 μM (estimated Ki=1 mM). The inhibition by H2O2 was detectable in the presence of DTT or 2-ME, as reported previously [43]. The addition of catalase into the pre-incubation medium prevented the inhibition of Cn by H2O2 (results not shown).
Activation of Cn by SOD1 was dependent on SOD1 concentration
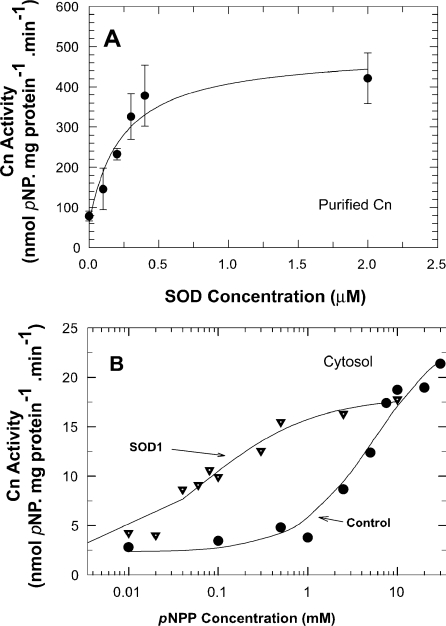
Pre-incubation of rat brain cytosol with various concentrations of bovine SOD1 enhanced the activity of Cn in a concentration-dependent manner (e.g. 5.3 μM SOD1 increased Cn activity nearly 4-fold; P<0.005). Bovine SOD1 similarly increased the activity of purified brain Cn in a concentration-dependent manner (Figure 2A). The estimated Kact for SOD1 activation of Cn was 0.23 μM. At that concentration, the molar ratio of SOD1 to Cn (0.2 μM in the assay) was 1.1. Cn activity reached a plateau when the SOD1 concentration was 0.4 μM or higher, i.e. at a molar ratio of SOD1 to Cn of 2.
Figure 2. Kinetics of activation of Cn by SOD1.
(A) Concentration-dependent effects of SOD1 on purified Cn. Cn (0.2 μM) was pre-incubated with various concentrations of SOD1 and activity was measured. Results are means±S.E.M. for three determinations from two experiments. Data were fitted to the equation V=V0+Vmax·[SOD1]/(Kact+[SOD1]). Basal Cn activity: 78.3 nmol/min per mg of protein. (B) SOD1 effects on substrate activation of Cn. Cn in rat brain cytosol was pre-incubated with or without 2.7 μM SOD1 for 25 min at 30 °C, and Cn activity was measured using increasing concentrations of pNPP. Results are from two determinations from two experiments. Curves are non-linear least-squares fitting of data to the equation V=Vmax·[pNPP]h/(Km+[pNPP]h).
Activation of brain Cn was specific to SOD1 and was due to a decrease in Km
The activation by SOD1 of Cn in brain cytosol and purified preparations was observed when either pNPP or the RII phosphopeptide were used as substrates. Also, the activation by SOD1 was observed in the presence or absence of Mn2+, a bivalent cation that activates Cn. Thus neither the type of substrate used nor the presence of Mn2+ affected the activation of Cn by SOD1.
To determine whether the activation of cytosol Cn by SOD1 was due to contaminating small-molecule enhancers, such as ascorbate [13], bovine kidney SOD1 was boiled before it was applied to the Cn reaction mixture. Boiled SOD1 did not increase Cn activity, whereas native SOD1 did [control Cn=0.029±0.0004, Cn+SOD1=0.052±0.002 (P<0.05) and Cn+boiled SOD1=0.025±0.001 pNP (p-nitrophenol) absorbance units; n=3]. These results indicated that SOD1 activation of Cn was due to the SOD1 protein, not a thermostable contaminant.
Pre-incubation times of 20 s to 20 min with SOD1 led to approximately equal levels of activation of bovine brain Cn when the ratio of SOD1 to Cn was 2:1 (control Cn activity=40.2±0.7, Cn pre-incubated with SOD1 for 20 s=63.3±4.1, Cn pre-incubated with SOD1 for 1 min=56.2±2.2, Cn pre-incubated with SOD1 for 5 min=59.3±4.8, and Cn pre-incubated with SOD1 for 20 min=53.3±0.4 nmol/min per mg of protein; n=3). Thus, kinetically, the activation of Cn by SOD1 was a rapid process.
The effect of SOD1 on Cn activity in rat brain cytosol resulted in a nearly 90-fold decrease in the Km for pNPP (from 5.5 to 0.06 mM) without a significant change in Vmax (control=22.9; SOD1-pre-treated=18.1 nmol of Pi/min per mg of protein) (Figure 2B). Pre-incubation of the cytosol with SOD1 induced small negative co-operativity in Cn. The Hill coefficient (h) for the two enzymatic reactions was control=1.0 and SOD1-pre-treated=0.7.
The primary and tertiary structure of bovine SOD1 differs from that of hSOD1 [42]. hSOD1 and rSOD1 have two additional residues (Glu24 and Ser25 for hSOD1, Ala24 and Ser25 for rSOD1) compared with bovine SOD1. Pre-incubation at molar ratios of bovine SOD1 to recombinant human Cn equal to 2:1, yielded a 74±17% increase above Cn activity in the absence of SOD1 (n=5). For hSOD1 and rSOD1, the activation of Cn was 47±24 and 53±15% increase above control (n=3) respectively. Thus, despite small differences in structure, the activation of Cn by all three forms was not significantly different.
Pre-incubation of Cn with SOD1 prevents loss of Zn from Cn
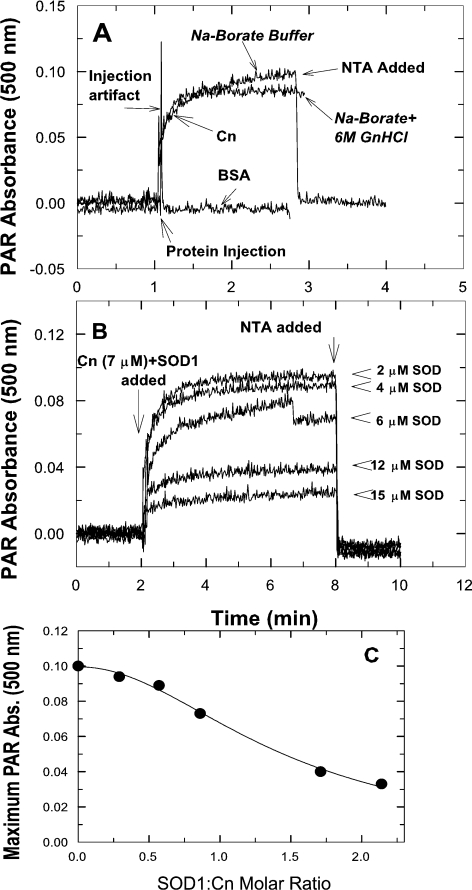
Activation of Cn by SOD1 might reflect a change in the metal content and thus the conformation of Cn. We probed the ease with which freshly purified bovine brain Cn might lose Fe or Zn from the metal centre by monitoring the formation of metal–PAR complexes [23]. As reported previously [23], SOD1 in PAR/borate buffer did not lose either Cu or Zn from its active site, but lost both metals upon exposure to 6 M GdmCl (results not shown). Unlike SOD1, Cn released as much of the protein-associated metal ions in the absence of GdmCl as it did in the presence of GdmCl (Figure 3A). All, or almost all, chromogenic PAR complexes formed following incubation of Cn (2–4 min) in the PAR/borate buffer system, either in the presence or the absence of GdmCl, could subsequently be chelated by the addition of NTA (Figure 3A). NTA would be expected to chelate both Fe and Zn (log complexation constants: Fe2+=8.8, Fe3+=16.3, Zn=10.7 [44]). To assess how much of the PAR complex detected might have been due to the formation of Fe2+–PAR or Fe3+–PAR, 8 M desferrioxamine mesylate, an iron chelator, was introduced to abstract the Fe in such complexes. Addition of desferrioxamine after a stable absorbance measurement was attained (2 min after the introduction of Cn) diminished the absorbance maximum at 500 nm by 16–19% (results not shown). These results indicated that the PAR complexes formed after the introduction of Cn were mostly Zn–PAR complexes. Introduction of BSA, a negative control, to the PAR/borate solution produced no changes in absorbance (Figure 3A).
Figure 3. Loss of metal ions from Cn in the absence or presence of GdmCl (A) and protection of Cn from metal loss by SOD1 (B and C).
(A) Chelation of metals associated with Cn by PAR was determined in either sodium borate buffer or the same buffer with 6 M GdmCl (GnHCl). Spectrophotometric signals from BSA were a negative control. Addition of NTA after 2 min of incubation of Cn plus PAR brought the measured absorbance to zero. (B and C) Interaction of SOD1 with Cn (30 min pre-incubation) prevented the loss of metals from Cn. SOD1 ranging from 0.29 to 2.1 molar ratios to Cn was used. All metals bound to PAR were chelated by NTA (B). Relationship of SOD1/Cn ratio to retention of metals by Cn (C). Data were fitted to a logistic function. Abs., absorbance.
Pre-incubation of Cn with SOD1 prevented metal losses from Cn. This effect of SOD1 on Cn was dependent on the ratio of SOD1 to Cn (Figure 3B). At a molar ratio of 2:1, most of the metal loss from Cn was suppressed. The maximum absorbance at 500 nm achieved by incubating Cn in the presence of various molar ratios of SOD1 to Cn led to an estimate of the ratio of SOD1 to Cn at half-maximal suppression of the signal. This ratio was 0.99 (Figure 3C), i.e. almost identical with the ratio needed for half-maximal activation of Cn by SOD1.
SOD1 dismutase activity was not necessary for activation of Cn
Pre-incubation of either brain cytosol or purified brain Cn with bacterial SOD2 did not show any change in Cn activity (results not shown). Extending the pre-incubation period of Cn with SOD2 up to 120 min did not alter the results obtained, suggesting that the lack of an effect on Cn was not the result of slow kinetics of activation by SOD2. Therefore, although SOD2 was fully active as a dismutase, it did not activate Cn.
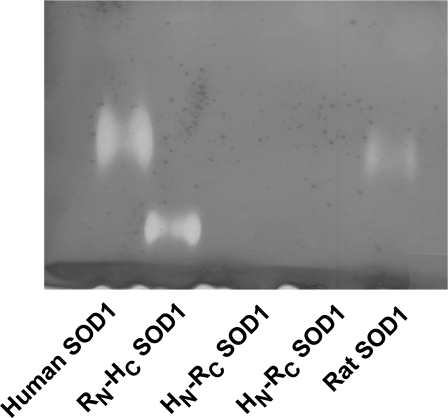
We also discovered a chimaeric form of SOD1 that lacked dismutase activity, yet was fully capable of activating Cn. Bovine SOD1, hSOD1 and rSOD1 differ most from each other in the primary structure of the N-terminal half of the protein. We created two chimaeras of SOD1 that contained either the N-terminal 76 residues of hSOD1 and C-terminal 78 residues of rSOD1 (HN-RC), or the reverse structures (RN-HC), and measured their dismutase activity and their capacity to increase Cn activity. For measurements of Cn activation, all assays were performed using a 2:1 ratio of SOD1 to Cn. Both chimaeras of SOD1 activated Cn and this activation was nearly identical with that obtained with either hSOD1 or rSOD1 alone (control Cn=202.5±22.5, Cn+hSOD1=298.5±18.5, Cn+rSOD1=310.0±10.0, Cn+HN-RC=318.0±28.0, and Cn+RC-HN=291±41 pmol). However, of the four recombinant species of SOD1, the HN-RC chimaera exhibited little or no dismutase activity in solution (results not shown) or in-gel assays (Figure 4). The reason for the loss of SOD1 activity in HN-RC is not known, but there appeared to be no relationship between activation of Cn and dismutase activity.
Figure 4. Dismutase activity of rat, HN-RC and RN-HC SOD1 chimaeras.
Dismutase activity was determined by gel assays. Note the lack of activity for the HN-RC SOD1 (two different preparations of HN-RC tested).
SOD1 and Cn proteins interact in vivo and in vitro
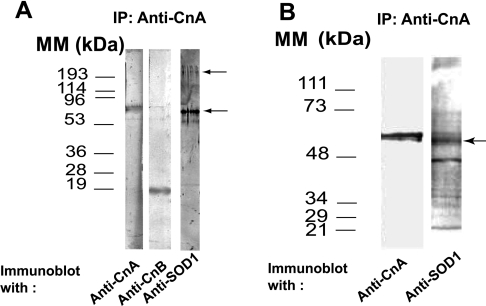
As shown in Figure 5(A), immunoprecipitate from hippocampus extracts with anti-CnA led to the co-immunoprecipitation not only of CnB, but also of a protein recognized by anti-SOD1 antibodies. However, the protein band reacting most strongly with anti-SOD1 had an estimated size of 60 kDa. There was only weak labelling of a band with the expected size of SOD1 monomers (∼20 kDa). The anti-SOD1 also labelled weakly a band of ∼250 kDa (Figure 5A). The ∼60 kDa protein band might have been a stable aggregated form of SOD1 or a different protein recognized by the anti-SOD1 antibodies.
Figure 5. Interaction of SOD1 with Cn in vivo and in vitro.
(A) Co-IP of Cn and SOD1 from brain extracts. Monoclonal anti-CnA antibodies were used in IP (immunoprecipitation), and immunoprecipitates were subjected to SDS/PAGE (15% gels). Immunoblots were developed with antibodies against CnA, CnB or SOD1. Molecular-mass standards (left) and the ∼60 and 250 kDa bands reacting with anti-SOD1 (right) are marked. (B) Results of IP of CnA from a mixture of SOD1 and Cn. SDS/PAGE was performed on 9% polyacrylamide gels. Antibodies used, molecular-mass standards (left) and bands labelled by anti-SOD1 (right) are indicated.
To resolve this issue, pure Cn was pre-incubated with pure bovine kidney SOD1 at a 2:1 molar ratio for 25 min at 30 °C, and Cn was immunoprecipitated using monoclonal anti-CnA. Both CnA- and SOD1-reactive bands were in the precipitates (Figure 5B). The major protein recognized by the anti-SOD1 antibody had an estimated size of 58–60 kDa. The anti-SOD1 antibodies also recognized, albeit more weakly, additional protein bands, including one of 250 kDa, i.e. the ∼60 and 250 kDa bands were SOD1 aggregates. When the ratio of Cn to bovine SOD1 in the incubation medium was increased, the amount of high-molecular-mass aggregates of SOD1 also increased (results not shown). There was no loss of SOD1 activity at any Cn/SOD1 ratios used (SOD1 activity, no Cn=1.47±0.08, 0.5 Cn/SOD1 ratio=1.48±0.06, 1.0 Cn/SOD1 ratio=1.54±0.08, and 2.0 Cn/SOD1 ratio=1.48±0.09 μmol/min per mg of protein, NBT reduced; n=3). Finally, pre-incubation of purified hSOD1 with purified Cn and subsequent electrophoretic separation yielded very similar results to those seen with bovine SOD1 (results not shown).
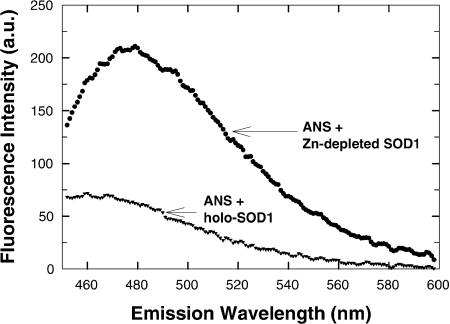
Since several mutant forms of hSOD1 are Zn-deficient [23], exhibit a high tendency toward aggregate formation [27] and decreased efficiency in activating Cn [24], we generated Zn-depleted but Cu-repleted WT hSOD1 [23,27] and tested them for their effect on Cn. Depletion of Zn from SOD1 caused very substantial conformational changes in recombinant WT hSOD1 as determined by changes in ANS binding and fluorescence (Figure 6). These measures fit previous observations [45]. In addition to conformational changes, the loss of Zn from hSOD1 caused nearly complete loss of SOD1-induced activation of Cn (Cn activity in presence of Zn-depleted SOD1: 105.0±6.9% control Cn activity; n=3).
Figure 6. Fluorescence emission spectra of ANS bound to intact hSOD1 (holo-SOD1) and Zn-depleted hSOD1.
Either holo- or Zn-depleted SOD1 (5 μM) in 10 mM Tris/acetate, pH 7.0, was incubated with 10 μM ANS (20 min at 23 °C). Excitation wavelength was 372 nm. Background fluorescence (absence of protein) was subtracted from spectra. a.u., arbitrary units.
Cn and SOD1 co-localize in the cytoplasm and plasma membrane of neurons
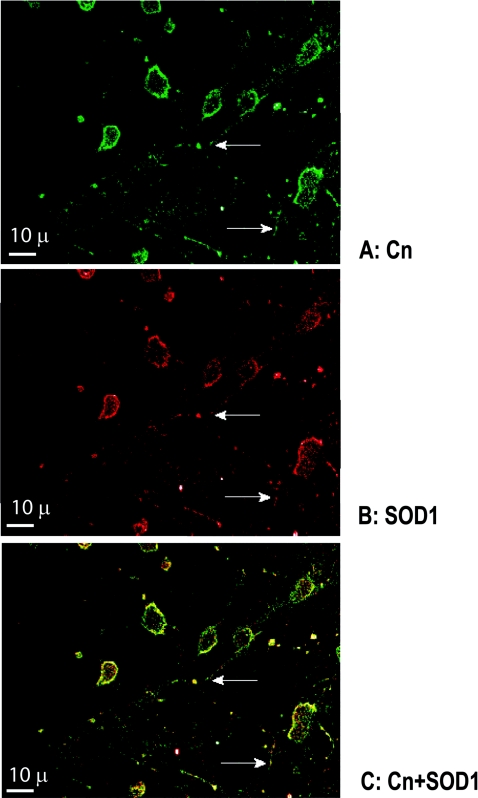
CnA and SOD1 were closely located in many regions of neuronal cytoplasm and plasma membranes, in both the cell bodies and neurites of cortical neurons in culture (Figure 7). SOD1 was somewhat more abundant and distributed more widely than CnA in the cytoplasm. Both CnA and SOD1 were present in neuritic processes, primarily in punctate-like accumulations (Figure 7, arrows). Pre-staining of neurons and glial cells (glia were less than 10% of all cells) with calcein dye, revealed very low to almost absent glial staining by either anti-CnA or anti-SOD1 antibodies (results not shown). It has been shown previously that glia do not have immunoreactive sites for SOD1 [46].
Figure 7. Localization of Cn (A) and SOD1 (B), and co-localization of the two enzymes (C) in primary neurons.
Images in (A) and (B) were obtained as described in the Experimental section and were superimposed to form the image shown in (C). Arrows show neuritic processes. Scale bar, 10 μm.
DISCUSSION
The present studies have identified several new aspects of the interaction between SOD1 and Cn. SOD1 enhancement of Cn activity was not due to protection of Cn from O2•−-induced inactivation. Inactivation of Cn by O2•− was not observed either in in vitro assays or in primary neurons. SOD1 activation of Cn was a property of bovine SOD1, hSOD1 and rSOD1, but not of SOD2. Dismutase activity of SOD1 did not correlate with efficacy of activation of Cn. Chimaeras of human SOD1–rat SOD1 with little dismutase activity stimulated Cn activity as much as those with high dismutase activity. Activation of Cn by SOD1 was the result of a 90-fold decrease in Km for the substrate and was dependent on the structure of SOD1. Activation of Cn was negatively affected by chelation of SOD1 Cu, the removal of Zn, or the process of engaging SOD1 in the catalysis of O2•− dismutation. SOD1 and Cn did interact in the intracellular environment of neurons and such interaction apparently induced a stable multimeric conformation in SOD1. Finally, the binding of SOD1 to Cn prevented the loss of metals by Cn following exposure of this enzyme to a metal-chelating system, indicative of altered Cn conformation in the presence of SOD1.
Although the activation of Cn by SOD1 was not due to protection of Cn from O2•− attack, the introduction of O2•− into the reaction mixture at the same time as SOD1 eliminated the effect of SOD1 on Cn. This was not due to H2O2 formation as the presence of catalase did not prevent the loss of the SOD1 effect on Cn. We propose that the reaction cycle of O2•− with Cu might alter the conformation of SOD1 and disrupt its interaction with Cn. A loss of Cn activation by SOD1 was observed when Cu in SOD1 was chelated by DETC. DETC-treated SOD1 loses its enzymatic activity, but we showed that dismutase activity did not correlate with activation of Cn. Thus we favour the idea that the removal of either Zn or Cu from the active site of SOD1 alters the conformation of SOD1 and prevents productive interactions with Cn.
Increasing concentrations of SOD1 brought about increased retention of metals by Cn and the molar ratios of SOD1 to Cn required for such enhanced retention of metals were nearly identical with those required for activation of Cn by SOD1. These observations were suggestive of a direct relationship between Cn activity and metal release from the protein and are concordant with previous observations of Cn metal content and activity [47]. The 90-fold decrease in the Km of Cn catalytic activity following pre-incubation with SOD1 might be the result of retention of the dinuclear metal centre following interaction of Cn with SOD1 and a resultant increased affinity of Cn for its substrate.
The study of the interactions between SOD1 and Cn is of physiological relevance because both proteins are involved in signal transduction in neurons and because SOD1 mutations lead to the neurodegeneration seen in fALS. The observed co-localization of Cn and SOD1 in cortical neurons in culture provided support for in vivo regulation of Cn activity by SOD1. Furthermore, the co-immunoprecipitation of SOD1 with Cn from cell extracts of brain hippocampus was indicative of strong interactions between these two proteins within the cell environment.
SOD1 is stable in terms of Cu content, and Cu depletion is not a likely cause of changes in SOD1 activity or structure in the mutant forms of SOD1 in fALS [48,49]. On the other hand, mutant SOD1 in fALS is frequently Zn-deficient and such Zn depletion makes the SOD1 protein likely to undergo aggregation under oxidative conditions [27]. We found that depletion of Zn from hSOD1 led to changes in protein conformation and to the loss of SOD1-induced activation of Cn. We hypothesize that the persistent loss of Cn activity in lymphocytes from ALS patients [25] may be in part due to altered conformations of SOD1, possibly those caused by Zn depletion. But, since not all forms of mutant SOD1 in fALS are Zn-deficient [23], and since fALS and sALS cases have lower Cn activity than controls [25], the loss of Cn activity in sALS patients may be due to some other molecular event. A common feature in cells of fALS and sALS patients is the presence of higher oxidative conditions than in cells from normal individuals [25]. Higher levels of reactive oxygen species may cause abnormal aggregation of SOD1, particularly in motor neurons of the spinal cord [50]. Abnormal aggregation may impede SOD1–Cn interactions and diminish activation of Cn by SOD1. Formation of SOD1 aggregates as a result of interactions with Cn may actually protect SOD1 dimers from oxidative modification.
Acknowledgments
We thank Dr David Nichols, Microscopy and Analytical Imaging Laboratory, University of Kansas, for assistance with confocal microscopy; Dr T. V. O'Halloran, Department of Chemistry, Northwestern University, Evanston, IL, U.S.A., and Dr Y.-S. Ho, Institute of Chemical Toxicology, Wayne State University, Detroit, MI, U.S.A., for their contribution of the human and rat cDNA for SOD1; Dr L. J. Hayward, Department of Neurology, University of Massachusetts Medical School, Worcester, MA, U.S.A., for supplying some of the recombinant hSOD1; Dr M. Richter, Department of Molecular Biosciences, University of Kansas, and Dr M.L. Michaelis, Department of Pharmacology and Toxicology, University of Kansas, for scientific advice. The support of the Higuchi Biosciences Center, University of Kansas, is acknowledged. This work was funded by grants from NIA (National Institute of Aging), AG12993, and NICHD (National Institute of Child Health and Human Development), HD02528.
References
- 1.King M. M., Huang C. Y. The calmodulin-dependent activation and deactivation of the phosphoprotein phosphatase, calcineurin, and the effect of nucleotides, pyrophosphate, and divalent metal ions: identification of calcineurin as a Zn and Fe metalloenzyme. J. Biol. Chem. 1984;259:8847–8856. [PubMed] [Google Scholar]
- 2.Klee C. B., Draetta G. F., Hubbard M. J. Calcineurin. Adv. Enzymol. Relat. Areas Mol. Biol. 1988;61:149–200. doi: 10.1002/9780470123072.ch4. [DOI] [PubMed] [Google Scholar]
- 3.Kincaid R. L., Higuchi S., Tamura J., Giri P., Martensen T. M. Structural isoforms of the catalytic subunit of calmodulin-dependent phosphoprotein phosphatase. Adv. Protein Phosphatases. 1991;6:73–99. [Google Scholar]
- 4.Klee C. B., Ren H., Wang X. Regulation of the calmodulin-stimulated protein phosphatase, calcineurin. J. Biol. Chem. 1998;273:13367–13370. doi: 10.1074/jbc.273.22.13367. [DOI] [PubMed] [Google Scholar]
- 5.Rusnak F., Mertz P., Reiter T. A., Yu L. Regulation of the protein phosphatase calcineurin by redox: implications for catalysis and signal transduction. In: Ferreira G. C., Franco R., Moura J. J. G., editors. Inorganic Biochemistry and Regulatory Mechanisms of Iron Metabolism. Weinheim: Wiley-VCH; 1999. pp. 35–50. [Google Scholar]
- 6.Yu L., Haddy A., Rusnak F. Evidence that calcineurin accommodates an active site binuclear metal center. J. Am. Chem. Soc. 1995;117:10147–10148. [Google Scholar]
- 7.Mansuy I. M., Mayford M., Jacob B., Kandel E. R., Bach M. E. Restricted and regulated overexpression reveals calcineurin as a key component in the transition from short-term to long-term memory. Cell. 1998;92:39–49. doi: 10.1016/s0092-8674(00)80897-1. [DOI] [PubMed] [Google Scholar]
- 8.Hodgkiss J., Kelly J. Only ‘de novo’ long-term depression (LTD) in the rat hippocampus in vitro is blocked by the same low concentration of FK506 that blocks LTD in the visual cortex. Brain Res. 1995;705:241–246. doi: 10.1016/0006-8993(95)01168-4. [DOI] [PubMed] [Google Scholar]
- 9.Torii N., Kamishita T., Otsu Y., Tsumoto T. An inhibitor for calcineurin, FK506, blocks induction of long-term depression in rat visual cortex. Neurosci. Lett. 1995;185:1–4. doi: 10.1016/0304-3940(94)11210-a. [DOI] [PubMed] [Google Scholar]
- 10.Yasuda H., Higashi H., Kudo Y., Inoue T., Hata Y., Mikoshiba K., Tsumoto T. Imaging of calcineurin activated by long-term depression-inducing synaptic inputs in living neurons of rat visual cortex. Eur. J. Neurosci. 2003;17:287–297. doi: 10.1046/j.1460-9568.2003.02449.x. [DOI] [PubMed] [Google Scholar]
- 11.Zhuo M., Zhang W., Son H., Mansuy I., Sobel R. A., Seidman J., Kandel E. A selective role of calcineurin Aa in synaptic depotentiation in hippocampus. Proc. Natl. Acad. Sci. U.S.A. 1999;96:4650–4655. doi: 10.1073/pnas.96.8.4650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rusnak F., Reiter T. Sensing electrons: protein phosphatase redox regulation. Trends Biochem. Sci. 2000;25:527–529. doi: 10.1016/s0968-0004(00)01659-5. [DOI] [PubMed] [Google Scholar]
- 13.Wang X., Culotta V. C., Klee C. B. Superoxide dismutase protects calcineurin from inactivation. Nature. 1996;383:434–437. doi: 10.1038/383434a0. [DOI] [PubMed] [Google Scholar]
- 14.Ferri A., Gabbianelli R., Casciati A., Paolucci E., Rotilio G., Carri M. T. Calcineurin activity is regulated both by redox compounds and by mutant familial amyotrophic lateral sclerosis-superoxide dismutase. J. Neurochem. 2000;75:606–613. doi: 10.1046/j.1471-4159.2000.0750606.x. [DOI] [PubMed] [Google Scholar]
- 15.Beiqing L., Chen M., Whisler R. L. Sublethal levels of oxidative stress stimulate transcriptional activation of c-jun and suppress IL-2 promoter activation in Jurkat T cells. J. Immunol. 1996;157:160–169. [PubMed] [Google Scholar]
- 16.Bogumil R., Namgaladze D., Schaarschmidt D., Schmachtel T., Hellstern S., Mutzel R., Ullrich V. Inactivation of calcineurin by hydrogen peroxide and phenylarsine oxide: evidence for a dithiol-disulfide equilibrium and implications for redox regulation. Eur. J. Biochem. 2000;267:1407–1415. doi: 10.1046/j.1432-1327.2000.01133.x. [DOI] [PubMed] [Google Scholar]
- 17.Jain J., Loh C., Rao A. Transcriptional regulation of the IL-2 gene. Curr. Opin. Immunol. 1995;7:333–342. doi: 10.1016/0952-7915(95)80107-3. [DOI] [PubMed] [Google Scholar]
- 18.Molkentin J. D., Lu J. R., Antos C. L., Markham B., Richardson J., Robbins J., Grant S. R., Olson E. N. A calcineurin-dependent transcriptional pathway for cardiac hypertrophy. Cell. 1998;93:215–228. doi: 10.1016/s0092-8674(00)81573-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Foster T. C., Norris C. M. Age-associated changes in Ca2+-dependent processes: relation to hippocampal synaptic plasticity. Hippocampus. 1997;7:602–612. doi: 10.1002/(SICI)1098-1063(1997)7:6<602::AID-HIPO3>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 20.Foster T. C., Sharrow K. M., Masse J. R., Norris C. M., Kumar A. Calcineurin links Ca2+ dysregulation with brain aging. J. Neurosci. 2001;21:4066–4073. doi: 10.1523/JNEUROSCI.21-11-04066.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gerber D. J., Hall D., Miyakawa T., Demars S., Gogos J. A., Karayiorgou M., Tonegawa S. Evidence for association of schizophrenia with genetic variation in the 8p21.3 gene, PPP3CC, encoding the calcineurin γ subunit. Proc. Natl. Acad. Sci. U.S.A. 2003;100:8993–8998. doi: 10.1073/pnas.1432927100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reiter T. A., Rusnak F. Electrochemical studies of the mono-Fe, Fe-Zn, and Fe-Fe metalloisoforms of bacteriophage lambda protein phosphatase. Biochemistry. 2004;43:782–790. doi: 10.1021/bi0356956. [DOI] [PubMed] [Google Scholar]
- 23.Crow J. P., Sampson J. B., Zhuang Y., Thompson J. A., Beckman J. S. Decreased zinc affinity of amyotrophic lateral sclerosis-associated superoxide dismutase mutants leads to enhanced catalysis of tyrosine nitration by peroxynitrite. J. Neurochem. 1997;69:1936–1944. doi: 10.1046/j.1471-4159.1997.69051936.x. [DOI] [PubMed] [Google Scholar]
- 24.Volkel H., Scholz M., Link J., Selzle M., Werner P., Tunnemann R., Jung G., Ludolph A. C., Reuter A. Superoxide dismutase mutations of familial amyotrophic lateral sclerosis and the oxidative inactivation of calcineurin. FEBS Lett. 2001;503:201–205. doi: 10.1016/s0014-5793(01)02730-2. [DOI] [PubMed] [Google Scholar]
- 25.Ferri A., Nencini M., Battistini S., Giannini F., Siciliano G., Casali C., Damiano M. G., Ceroni M., Chio A., Rotilio G., Carri M. T. Activity of protein phosphatase calcineurin is decreased in sporadic and familial amyotrophic lateral sclerosis patients. J. Neurochem. 2004;90:1237–1242. doi: 10.1111/j.1471-4159.2004.02588.x. [DOI] [PubMed] [Google Scholar]
- 26.Agbas A., Zaidi A., Michaelis E. K. Decreased activity and increased aggregation of brain calcineurin during aging. Brain Res. 2005;1059:59–71. doi: 10.1016/j.brainres.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 27.Rakhit R., Cunningham P., Furtos-Matei A., Dahan S., Qi X. F., Crow J. P., Cashman N. R., Kondejewski L. H., Chakrabartty A. Oxidation-induced misfolding and aggregation of superoxide dismutase and its implications for amyotrophic lateral sclerosis. J. Biol. Chem. 2002;277:47551–47556. doi: 10.1074/jbc.M207356200. [DOI] [PubMed] [Google Scholar]
- 28.Beauchamp C., Fridovich I. Superoxide dismutase: improved assays and an assay applicable to acrylamide gels. Anal. Biochem. 1971;44:276–287. doi: 10.1016/0003-2697(71)90370-8. [DOI] [PubMed] [Google Scholar]
- 29.Beauchamp C. O., Fridovich I. Isozymes of superoxide dismutase from wheat germ. Biochim. Biophys. Acta. 1973;317:50–64. doi: 10.1016/0005-2795(73)90198-0. [DOI] [PubMed] [Google Scholar]
- 30.Bessey O. A., Lowry O. H., Brock M. J. A method for the rapid determination of alkaline phosphatase with five cubic millimeters of serum. J. Biol. Chem. 1946;164:321. [PubMed] [Google Scholar]
- 31.Penny C. L. A simple micro-assay for inorganic phosphate. Anal. Biochem. 1976;75:201–210. doi: 10.1016/0003-2697(76)90071-3. [DOI] [PubMed] [Google Scholar]
- 32.Roerig D. L., Mascaro L., Jr, Aust S. D. Microsomal electron transport: tetrazolium reduction by rat liver microsomal NADPH-cytochrome c reductase. Arch. Biochem. Biophys. 1972;153:475–479. doi: 10.1016/0003-9861(72)90365-7. [DOI] [PubMed] [Google Scholar]
- 33.Winterbourn C. C., Hawkins R. E., Brian M., Carrell R. W. The estimation of red cell superoxide dismutase activity. J. Lab. Clin. Med. 1975;85:337–341. [PubMed] [Google Scholar]
- 34.Chen X., Moore-Nichols D., Nguyen H., Michaelis E. K. Calcium influx through NMDA receptors, chronic receptor inhibition by ethanol and 2-amino-5phosponopentanoic acid, and receptor protein expression. J. Neurochem. 1999;72:1969–1980. doi: 10.1046/j.1471-4159.1999.0721969.x. [DOI] [PubMed] [Google Scholar]
- 35.Xia Y., Ragan R. E., Seah E. E., Michaelis M. L., Michaelis E. K. Developmental expression of N-methyl-D-aspartate (NMDA)-induced neurotoxicity, NMDA receptor function, and the NMDAR1 and glutamate-binding protein subunits in cerebellar granule cells in primary cultures. Neurochem. Res. 1995;20:617–629. doi: 10.1007/BF01694545. [DOI] [PubMed] [Google Scholar]
- 36. Reference deleted.
- 37.Ling S., Sheng J. Z., Braun J. E., Braun A. P. Syntaxin 1A co-associates with native rat brain and cloned large conductance, calcium-activated potassium channels in situ. J. Physiol. 2003;553:65–81. doi: 10.1113/jphysiol.2003.051631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Reference deleted.
- 39.Bus J. S., Gibson J. E. Paraquat: model for oxidant-initiated toxicity. Environ. Health Perspect. 1984;55:37–46. doi: 10.1289/ehp.845537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Heikkila R. E., Cabbat F. S., Cohen G. Inactivation of superoxide dismutase by several thiocarbamic acid derivatives. Experientia. 1978;34:1553–1554. doi: 10.1007/BF02034668. [DOI] [PubMed] [Google Scholar]
- 41.Turner B. J., Lopes E. C., Cheema S. S. Inducible superoxide dismutase 1 aggregation in transgenic amyotrophic lateral sclerosis mouse fibroblasts. J. Cell. Biochem. 2004;91:1074–1084. doi: 10.1002/jcb.10782. [DOI] [PubMed] [Google Scholar]
- 42.Strange R. W., Antonyuk S., Hough M. A., Doucette P. A., Rodriguez J. A., Hart P. J., Hayward L. J., Valentine J. S., Hasnain S. S. The structure of holo and metal-deficient wild-type human Cu, Zn superoxide dismutase and its relevance to familial amyotrophic lateral sclerosis. J. Mol. Biol. 2003;328:877–891. doi: 10.1016/s0022-2836(03)00355-3. [DOI] [PubMed] [Google Scholar]
- 43.Ferri A., Gabbianelli R., Casciati A., Celsi F., Rotilio G., Carri M. T. Oxidative inactivation of calcineurin by Cu,Zn superoxide dismutase G93A, a mutant typical of familial amyotrophic lateral sclerosis. J. Neurochem. 2001;79:531–538. doi: 10.1046/j.1471-4159.2001.00558.x. [DOI] [PubMed] [Google Scholar]
- 44.Anderegg G. Critical survey of stability constants of NTA complexes. Pure Appl. Chem. 1982;54:2693–2758. [Google Scholar]
- 45.Banci L., Bertini I., D'Amelio N., Gaggelli E., Libralesso E., Matecko I., Turano P., Valentine J. S. Fully metallated S134N Cu,Zn-superoxide dismutase displays abnormal mobility and intermolecular contacts in solution. J. Biol. Chem. 2005;280:35815–35821. doi: 10.1074/jbc.M506637200. [DOI] [PubMed] [Google Scholar]
- 46.Pei J. J., Grundke-Iqbal I., Iqbal K., Bogdanovic N., Winblad B., Cowburn R. F. Elevated protein levels of protein phosphatases PP-2A and PP-2B in astrocytes of Alzheimer's disease temporal cortex. J. Neural Transm. 1997;104:1329–1338. doi: 10.1007/BF01294734. [DOI] [PubMed] [Google Scholar]
- 47.Wang X., Randall C. R., True A. E., Que L., Jr X-ray absorption spectroscopic studies of the FeZn derivative of uteroferrin. Biochemistry. 1996;35:13946–13954. doi: 10.1021/bi961436n. [DOI] [PubMed] [Google Scholar]
- 48.Subramaniam J. R., Lyons W. E., Liu J., Bartnikas T. B., Rothstein J., Price D. L., Cleveland D. W., Gitlin J. D., Wong P. C. Mutant SOD1 causes motor neuron disease independent of copper chaperone-mediated copper loading. Nat. Neurosci. 2002;5:301–307. doi: 10.1038/nn823. [DOI] [PubMed] [Google Scholar]
- 49.Malinowski D. P., Fridovich I. Subunit association and side-chain reactivities of bovine erythrocyte superoxide dismutase in denaturing solvents. Biochemistry. 1979;18:5055–5060. doi: 10.1021/bi00590a005. [DOI] [PubMed] [Google Scholar]
- 50.Borchelt D. R., Wong P. C., Becher M. W., Pardo C. A., Lee M. K., Xu Z. S., Thinakaran G., Jenkins N. A., Copeland N. G., Sisodia S. S., et al. Axonal transport of mutant superoxide dismutase 1 and focal axonal abnormalities in the proximal axons of transgenic mice. Neurobiol. Dis. 1998;5:27–35. doi: 10.1006/nbdi.1998.0178. [DOI] [PubMed] [Google Scholar]