Abstract
The effect of pH on the initial-rate kinetic behaviour of BVR-A (biliverdin-IXα reductase) exhibits an alkaline optimum with NADPH as cofactor, but a neutral optimum with NADH as cofactor. This has been described as dual cofactor and dual pH dependent behaviour; however, no mechanism has been described to explain this phenomenon. We present evidence that the apparent peak of activity observed at neutral pH with phosphate buffer and NADH as cofactor is an anion-dependent activation, where inorganic phosphate apparently mimics the role played by the 2′-phosphate of NADPH in stabilizing the interaction between NADH and the enzyme. The enzymes from mouse, rat and human all exhibit this behaviour. This behaviour is not seen with BVR-A from Xenopus tropicalis or the ancient cyanobacterial enzyme from Synechocystis PCC 6803, which, in addition to being refractory to activation by inorganic phosphate, are also differentiated by an acid pH optimum with both nicotinamide nucleotides.
Keywords: anion activation, biliverdin-IXα reductase (BVR-A), NAD, pH optima, Xenopus tropicalis
Abbreviations: BVR-A, biliverdin-IXα reductase; DTT, dithiothreitol; EST, expressed sequence tag; HO-1, haem oxygenase-1; MBP, maltose-binding protein; mBVR-A, murine BVR-A; RT, reverse transcriptase
INTRODUCTION
The haem catabolic pathway is currently receiving attention in a number of fields with the observation that induction of HO-1 (haem oxygenase-1) is cytoprotective [1,2]. Attempts to relate this to production of the immediate product biliverdin [3,4] and/or subsequent reduction to bilirubin [5] or to a redox cycling mechanism between these two linear tetrapyrroles [6] are unresolved; however, there is clear evidence that biliverdin has some unexpected and dramatic effects. Bach and co-workers [7–9] have presented intriguing evidence that administration of biliverdin is cytoprotective for the heart, colon and liver in animal transplantation studies. In a striking report, Bach and co-workers have shown that short-term treatment (3 weeks) with biliverdin is sufficient to induce tolerance in a recipient to the donor heart for 120 days [9]. The introduction of an allogeneic third heart at 120 days was rejected, whereas the introduction of a third syngeneic heart was accepted, clearly indicating tolerance.
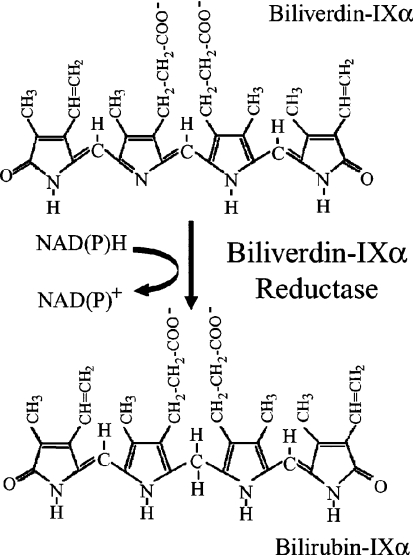
Despite the current interest in the metabolism of linear tetrapyrroles, BVR-A (biliverdin-IXα reductase) has received relatively little attention. The reaction catalysed by BVR-A is illustrated in Scheme 1. BVR-A is a cytosolic monomer (34 kDa), although the human enzyme migrates anomalously on SDS/PAGE with a mobility corresponding to a molecular mass reported variously as 46 kDa [10] or 41 kDa [11]. BVR-A catalyses the transfer of the B-face hydrogen [12] to a variety of verdin acceptors [13]. The structure of the rat enzyme [14] shows that the enzyme contains two domains, an N-terminal domain characteristic of a dinucleotide-binding fold (Rossmann fold) and a C-terminal domain that is predominantly an antiparallel six-stranded β-sheet. A structure for the rat enzyme with NADH bound has also been reported [15]; however, the adenine moiety folds back over the nicotinamide ring and this may, as the authors discuss, be an artefact of crystallization, where Lys209 from a symmetry-related molecule precludes the adenine ring, adopting the open structure predicted to exist in solution [14].
Scheme 1. BVR-A-catalysed reduction of biliverdin-IXα.
BVR-A is a small enzyme that exhibits complicated kinetic behaviour. The enzyme shows pronounced substrate inhibition (which may be modulated by intracellular biliverdin-binding proteins [16]) and also has two distinct pH optima depending on whether NADH or NADPH is the nicotinamide nucleotide. NADH is preferentially used between pH 6 and 7, whereas NADPH is the preferred cofactor between pH 8.5 and 8.7. This behaviour is seen with both native and recombinant human and rat BVR-A [17–20], the ox kidney [21] and the pig spleen enzymes [22]. Interestingly, one notable exception to this dual pH behaviour is the cyanobacterial BVR-A from Synechocystis PCC6803, which displays maximal activity at pH 5.8 when using either NADPH or NADH as the cofactor [23]. During a re-investigation of this phenomenon (extended to include the recombinant enzymes from Xenopus tropicalis and mouse) we have discovered that the enzyme activity with NADH as the cofactor is markedly activated by inorganic phosphate at neutral pH, and present a novel mechanism to explain this observation.
MATERIALS AND METHODS
Details on the purification of the salmon [24], rat [12], human [13] and Synechocystis [23] enzymes have been reported previously. The pBS152V plasmid encoding Synechocystes BVR-A was given kindly by Dr W. Schluchter (Department of Molecular and Cell Biology, University of California, Berkeley, CA, U.S.A.).
The mouse enzyme was cloned by RT (reverse transcriptase)–PCR using mRNA from Balb/c mouse liver. The specific primer used for the RT reaction was 5′-TGAAGCTTCCTCCTCACTGCTTCCGG-3′, and the primers for the PCR reaction were: forward primer, 5′-CCTCTAGAATGAGTACTGAGCCAA-3′, and reverse primer, 5′-CCAAGCTTCCTCCTCACTGCTTCC-3′. The PCR product was digested with XbaI and HindIII to allow directional cloning into the MBP (maltose-binding protein) fusion vector ppMAL. DNA sequencing of the mBVR-A (murine BVR-A) cDNA construct was carried out and compared with four ESTs (expressed sequence tags). The cDNA sequence of EST-1 (accession number AK002231.1) appears to have a stop codon 21 bp after the stop codon of all other cDNA sequences aligned including that of the cloned product. An additional variation occurs in the codon for Leu27 (TTG). In two of these sequences (accession numbers AK002231.1 and AK010847.1) and also the murine ppMAL-mBVR-A construct, the codon was as expected; however, in two other ESTs (accession numbers BC052146.1 and NM_026678.3), codon 27 reads TCG, which codes for a lysine residue at this position. The MBP–mBVR-A fusion protein was produced in Escherichia coli strain BL21 transformed with ppMAL-mBVR-A. After induction with IPTG (isopropyl β-D-thiogalactoside) (0.2 mM for 12 h at 27 °C) the cytosol was loaded on to an amylose affinity column (1.5 cm×10 cm) to bind the MBP–BVR-A fusion protein. This was washed extensively with 50 mM Tris/HCl (pH 7.0), 150 mM NaCl, 1 mM EDTA and 1 mM DTT (dithiothreitol), and the fusion protein was then eluted by 10 mM maltose in 50 mM Tris/HCl (pH 7.0), 150 mM NaCl, 1 mM EDTA and 1 mM DTT. The eluate was tested for BVR-A activity and the peak fractions were pooled. From 1 litre of culture, 20 mg of MBP fusion protein was routinely obtained. The MBP was cleaved using PreScission™ Protease (Amersham Biosciences).
The enzyme from X. tropicalis was cloned from the I.M.A.G.E cDNA clone (IMAGE: 5308696), which was acquired from U.K. HGMP (Human Genome Mapping Project, Hinxton, Cambridge, U.K. [24a]. Using the following primers: forward, 5′-GGATGGATCCATGTTTGGCGCAGTAGT-3′ and reverse, 5′-CCTCGAGCCATGGTTAATGCTCACATTTC-3′, an amplified product of the predicted size (approx. 882 bp) was obtained. The product was then digested with BamHI and XhoI to allow directional cloning into pGEX-KG. The resulting plasmid, pGEX-KG-XeBVR-A, was transformed into E. coli BL21 cells, and the glutathione S-transferase fusion protein was isolated by affinity chromatography on glutathione–agarose (Chromatrin). Site-directed mutagenesis of the rat enzyme was carried out using PCR as described previously [25].
Initial-rate kinetics were conducted by following the production of bilirubin by monitoring the increase in absorbance at 460 nm as described previously [13]. Assays were conducted at 30 °C using a Unicam 8625 UV/Vis spectrophotometer connected to a Kipp and Zonen flat-bed chart recorder. For routine assays the incubation mixture contained 0.1 M sodium phosphate buffer (pH 7.2), 700 μM NADH and 10 μM biliverdin-IXα, and the reaction was started by the addition of enzyme. To determine the stability of BVR-A over the pH range studied, the various enzymes were diluted 10-fold to a concentration of 0.1 mg/ml in 100 mM buffer at each pH value. The diluted mixtures were incubated for 10 min at 30 °C in the buffers indicated, after which time 10 μl aliquots were assayed in 100 mM Tris/HCl (pH 8.0) in a final volume of 2 ml, with a final concentration of 100 μM NADPH and 10 μM biliverdin. Initial rates are described as μmol or nmol produced/min per mg of enzyme. For some experiments, particularly the stopped-flow experiments, the rates are expressed as ΔA/min. The stopped flow experiments were conducted using an Applied Photophysics π*-180 spectrometer and Pro-K global analysis software.
The following buffers (100 mM) were used to determine the effect of pH on the activity of BVR-A: sodium acetate (pH 5.0–6.0), sodium citrate (pH 5.0–6.5), sodium phosphate (pH 6.5–8.0) and Tris/HCl (pH 8.0–9.0). Assays for BVR-A activity were generally conducted in the absence of BSA. The addition of BSA gives an enhanced ΔA460 and maintains linear initial rates for longer; however, it introduces complications when interpreting kinetic results, since the total concentration of biliverdin is not identical with the free concentration, as a significant fraction binds to BSA (or any other biliverdin-binding protein [16,26]). Biliverdin-IXα was prepared by oxidation of bilirubin-IXα [27].
RESULTS AND DISCUSSION
The effects of pH on the stability of various BVR-As
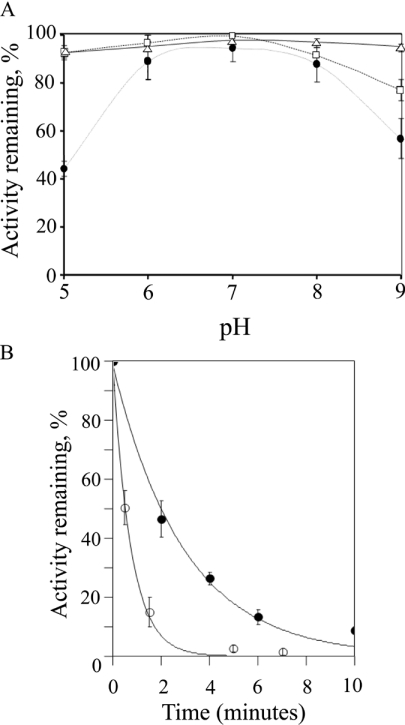
To determine the stability of the rat, murine and human recombinant BVR-A, the enzymes were pre-incubated in various buffers over the pH range 5.0–9.0. The human enzyme (Figure 1A) lost no more than 5–10% of its activity even at the extremes of pH tested (pH 5.0 and pH 9.0). Under the conditions examined, the enzyme was relatively dilute (0.1 mg/ml) and was pre-incubated for 10 min at the various pH values. In kinetic studies the reaction was generally initiated by the addition of enzyme and the initial rate measured from the initial 10–20 s of the reaction. It is unlikely therefore that any significant inactivation of the human enzyme occurred in these initial-rate experiments. The rat enzyme exhibited a similar tolerance to pH, but was marginally less stable at alkaline values, losing 25% of its activity at pH 9.0 after a 10 min pre-incubation (Figure 1A). The mouse enzyme is considerably less stable at both acid and alkaline pH values, and even at pH values between 6 and 8 loses 10–20% activity after a 10 min incubation at 30 °C (Figure 1A). The relative instability of mBVR-A at acid pH values prompted a further investigation to determine the kinetics of inactivation at pH 4.0 and 4.5. After a 10 min pre-incubation at pH 4.5, only 10% of the activity remained (Figure 1B). However, 50% activity remains after 2 min of pre-incubation at pH 4.5 (Figure 1B) and extrapolation suggests that measurements up to 30 s at pH 4.5 will yield 90% of the original activity. It is not possible to extend this level of confidence to pH 4.0, where only 50% activity remains after 30 s of pre-incubation (Figure 1B). The degradation of NADH and NADPH under acid conditions has been reported in numerous studies (for example see [28]). It is clear from the present studies that the pH has a significant effect on the rate of NADPH and NADH degradation. At 30 °C, the half-life of NADPH was reported as just 38 min at pH 5 [28]. As a result stock solutions of NAD(P)H in water were prepared fresh each day and stored on ice.
Figure 1. Effect of pH on the stability of BVR-A and kinetics of inactivation of mBVR-A at acid pH.
(A) The effect of pH on the stability of BVR-A. The enzyme (2 μg) was incubated at the pH indicated for 10 min and then diluted 100-fold into assay buffer (100 mM Tris/HCl, pH 8.0) with 100 μM NADPH and 10 μM biliverdin-IXα. The results are shown for the human (△), rat (□) and mouse (●) enzymes. (B) The kinetics of inactivation of the mBVR-A at acid pH. The enzyme (2 μg) was incubated in 0.1 M sodium acetate at pH 4.0 (○) and pH 4.5 (●) for various times and then diluted 100-fold into assay buffer (100 mM Tris/HCl, pH 8.0) with 100 μM NADPH and 10 μM biliverdin-IXα.
The effect of pH on the activity of BVR-A
It is noteworthy that, of all the BVR-A enzymes characterized to date, the Synechocystis PCC6803 and salmon enzymes are the only forms where activity has been surveyed over the entire pH range studied using both nucleotides [23,29], and these, together with the eel (Anguilla japonica) enzyme [30], support significant activity at acid pH values.
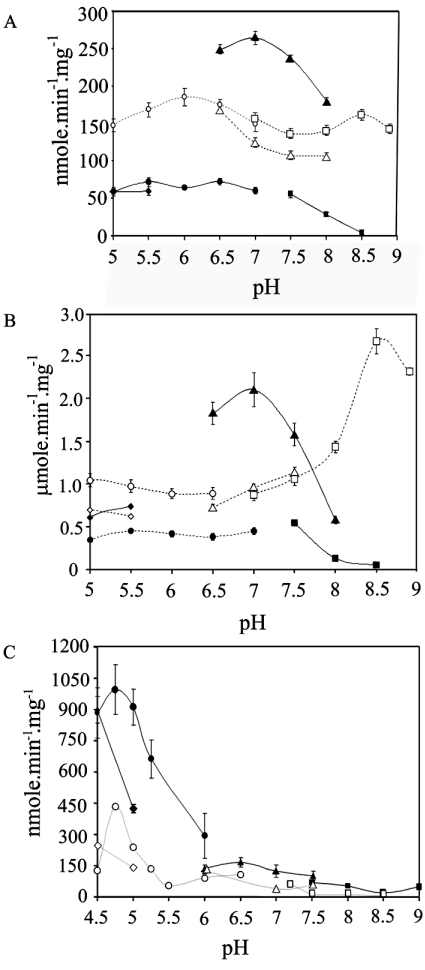
A series of reactions were set up to determine the effects of pH on the activity of human, rat and Xenopus BVR-A, and the results are presented in Figure 2.
Figure 2. Effect of pH on the activity of BVR-A.
The results are shown for human (A), rat (B) and Xenopus (C) enzymes. The enzyme was assayed at the pH indicated in the following buffers: sodium citrate (●), sodium phosphate (▲), sodium acetate (◆) and Tris/HCl (■) using 700 μM NADH as the cofactor and sodium citrate (○), sodium phosphate (△), sodium acetate (◇) and Tris/HCl (□) using 100 μM NADPH as the cofactor. A final concentration of 10 μM biliverdin was used.
During the course of these experiments two interesting observations were made. The first is that the use of phosphate buffer substantially increased the rate for the NADH-dependent reaction over the pH range (pH 6–8) used in these experiments for the human and rat enzymes. The second point worth noting is that the NADPH-dependent activity of the human and rat enzymes is clearly not limited to the pH range of 7.5–9.
The activity of the Xenopus enzyme exhibited a pH optimum of 4.75, which is similar to the behaviour of the Synechocystis enzyme [23]. The Xenopus enzyme is similar to the salmon enzyme [29] in that both are more active with NADH than NADPH.
The effects of various anions on the activity of human BVR-A
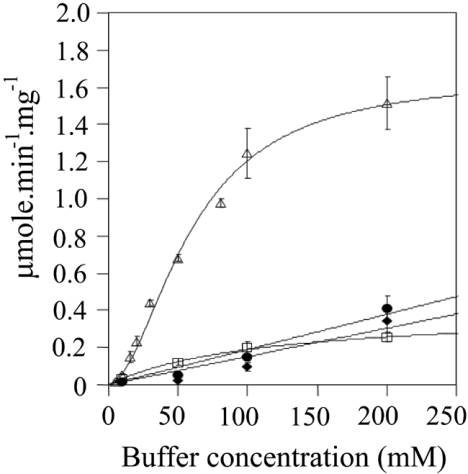
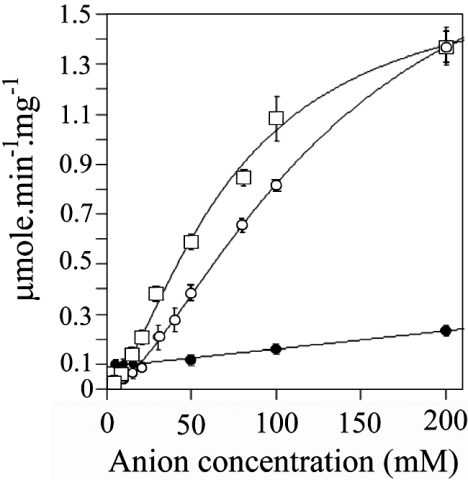
Figure 3 shows the effect of increasing concentrations of phosphate buffer (pH 7) on the activity of human BVR-A with NADH as the cofactor. The data fit well to the Hill equation, as illustrated; however, there are probably effects due to ionic strength in addition to the specific activation by phosphate anion. The activity in 5 mM phosphate is 0.04 μmol/min per mg of protein, and in 200 mM phosphate this increases 37.5-fold to 1.5 μmol/min per mg of protein. To determine whether this effect was specific to phosphate, a range of concentrations of various buffers at pH 7 was tested. Assays were performed with 20 μM biliverdin and 700 μM NADH at 30 °C in various concentrations of sodium phosphate (2, 5, 10, 25, 50, 75, 100 and 200 mM) or sodium carbonate, Tris/HCl or sodium citrate (all at 10, 50, 100 and 200 mM). The reaction was started by the addition of enzyme. The pH of each assay were tested before and after the assay to ensure that pH did not alter over the course of the assay, especially when using salts at the limits of their buffering capacity.
Figure 3. Effect of various buffers on the NADH-dependent activity of human BVR-A.
The activity of the human enzyme was assayed in the presence of 700 μM NADH and 20 μM biliverdin as substrate at 30 °C in the various buffers at the concentrations indicated. The buffers used were: sodium phosphate (△), sodium carbonate (◆), Tris/HCl (●) and sodium citrate (□) at pH 7.0.
The results of the experiment detailed in Figure 3 demonstrate clearly that phosphate is a powerful activator when NADH is used as cofactor. There is a modest effect of ionic strength, as shown by the increase in initial rates seen with increasing concentrations of sodium citrate, Tris/HCl and sodium carbonate up to concentrations of 200 mM. However, the effect of phosphate on enzyme activity is particularly marked. Interestingly, pyrophosphate also displayed an activator effect; indeed the observed potency was greater than that obtained with phosphate (Figure 4). It is also clear from Figure 4 that the marked activation seen with both phosphate and pyrophosphate is specific for NADH. There is only a very modest increase in activity seen with NADPH as the cofactor over the same range. The basis for the activation, however, was unclear from these results. Phosphate may allow substrates to bind more effectively to the enzyme and/or enhance turnover.
Figure 4. Effect of various anions on the NADH- and NADPH-dependent activity of human BVR-A.
The enzyme (5 μg) was assayed using 700 μM NADH as the cofactor and 10 μM biliverdin as the substrate at pH 7.0 in sodium phosphate (○) and sodium pyrophosphate (□). Also shown are the results when human BVR-A was assayed using 100 μM NADPH and 10 μM biliverdin as substrate in sodium phosphate (●), pH 7.0, at the indicated concentrations.
The effects of phosphate on the NADH-dependent activity of human BVR-A
A series of saturation curves were generated with NADH as the variable substrate (20 μM to 3.2 mM) in 10, 50, 100 and 200 mM sodium phosphate (pH 7.0). Biliverdin-IXα at a final concentration of 20 μM was used as substrate and the reaction was performed at 30 °C. Data points were fitted to the Michaelis equation, and the rectangular hyperbolas generated by these fits were used to obtain the apparent Km and Vmax values (Table 1). Increasing phosphate from 10 mM to 200 mM resulted in a dramatic decrease in the apparent Km for NADH (150-fold), with no significant change in the apparent Vmax. Clearly inorganic phosphate promotes NADH binding without enhancing turnover.
Table 1. The apparent Km and Vmax of human BVR-A for NADH with increasing concentrations of sodium phosphate.
Data points in Figure 6 were fitted to the Michaelis equation, and the rectangular hyperbolas generated by these fits were used to obtain the apparent Km and Vmax. The results are shown as means±S.D.
| Phosphate (mM) | Km NADH (app) (mM) | Vmax (app) (μmol/min per mg of protein) |
|---|---|---|
| 10 | 39.20±69 | 2.00±3.77 |
| 50 | 3.05±0.74 | 2.03±0.29 |
| 100 | 0.79±0.18 | 2.00±0.21 |
| 200 | 0.22±0.02 | 2.12±0.07 |
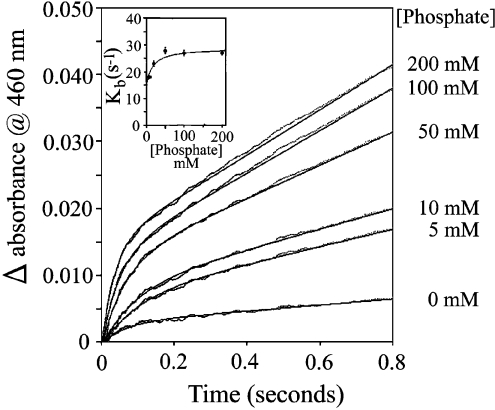
The effect of phosphate on the burst rate constant of human BVR-A
Stopped-flow studies on ox kidney [21] and rat [12] BVR-A show a burst reaction, which has been attributed to the slow release of bilirubin from an enzyme–NADP–bilirubin complex. The effect of phosphate on the pre-steady-state kinetics was determined by monitoring the increase in absorbance at 460 nm at 23 °C. Human BVR-A (1.6 μM) in 100 mM sodium citrate (pH 7.2) was rapidly mixed with 1.4 mM NADH and 14 μM biliverdin-IXα in varying concentrations of sodium phosphate (pH 7.2) (Figure 5). The concentration of phosphate in the substrate syringe was varied to give final concentrations of 5, 10, 20, 50, 100 and 200 mM. The results of these stopped-flow experiments were fitted to a curve of single exponential with slope. The steady-state rate showed that, at 23 °C, there is a pronounced activation by phosphate anion (8-fold compared with the rate in Tris/HCl), and the burst rate constants (kb) obtained from these experiments are shown in the inset to Figure 5. There is a modest effect of phosphate concentration on the burst rate constant, which essentially doubles between 0 and 50 mM phosphate from 0.15 s−1 to 0.28 s−1.
Figure 5. Effect of phosphate on the pre-steady burst of human BVR-A.
The activity of the human enzyme (0.8 μM) was assayed in the presence of 700 μM NADH and 7 μM biliverdin with increasing concentrations of phosphate (0, 5, 10, 50, 100 and 200 mM) in 50 mM Tris/HCl, pH 7.2.
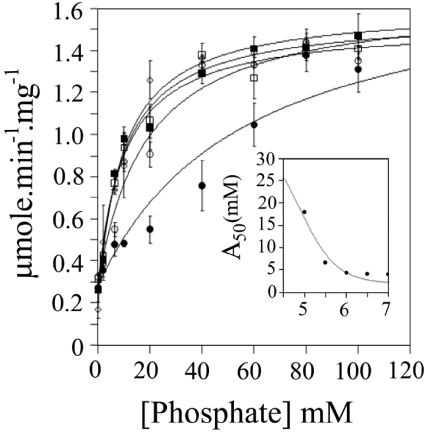
The effect of pH on the activation by phosphate of human BVR-A using NADH as cofactor
The effect of pH on the activity of human BVR-A with increasing concentrations of phosphate was also investigated (Figure 6). The activity was measured in 100 mM sodium citrate over the range pH 5.0–7.0 with 7 μm biliverdin-IXα and 100 μM NADH as substrates. The concentrations of phosphate employed were 5, 10, 15, 20, 40, 60, 80, 100 mM. It is clear that, at pH 5.0 the activation by phosphate anion is less potent than that seen at more neutral pH values. By plotting A50 (the concentration of activator giving 50% activation) against pH, an apparent pK of approx. 4.9 can be identified (see the inset panel for Figure 6). Deprotonation of this residue increases the affinity for phosphate anion 6-fold. A possible candidate for this pK can be tentatively identified as Glu75. The side chain of Glu75 interacts with Arg44 (one of the residues that interacts with inorganic phosphate in the rat structure) and is predicted to do so more strongly as the carboxylate anion. A weaker interaction may occur, as the protonated carboxyl may allow Arg44 to adopt an alternative conformation that partially destabilises the Arg44 interaction with phosphate.
Figure 6. Effect of pH on the phosphate-dependent activation of human BVR-A.
The activity of human BVR-A was measured in 100 mM sodium citrate at pH 5.0 (●), pH 5.5 (○), pH 6.0 (□), pH 6.5 (■) and pH 7.0 (◇) with 7 μm biliverdin-IXα and 700 μM NADH. Phosphate concentration was increased from 0 to 100 mM phosphate at each pH measured. The inset panel shows a plot of A50 against pH fitted to a single pK.
Studies on phosphate activation of rat BVR-A: identification of a putative 2′-phosphate-binding site
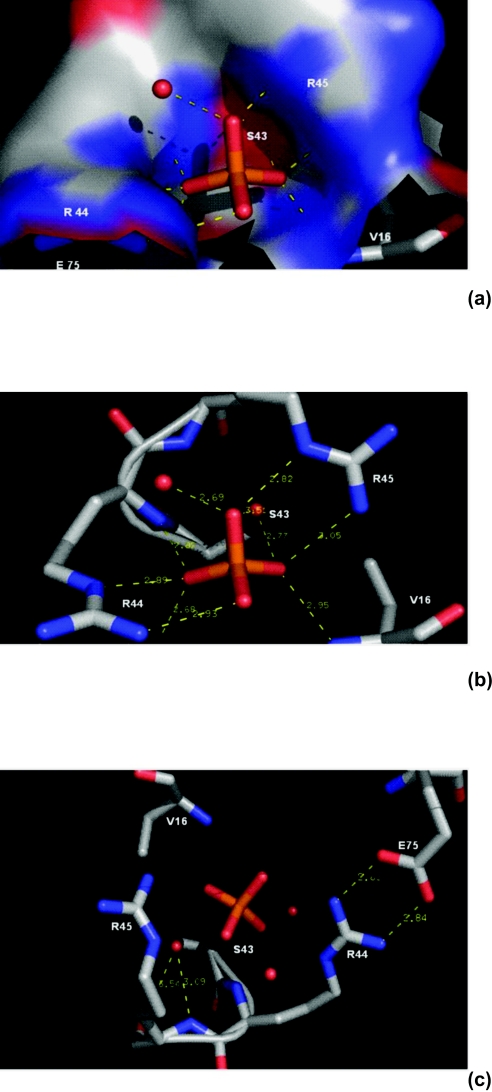
In an effort to understand further the nature of the activation of the NADH-dependent activity of human BVR-A by phosphate and related anions, it was decided to analyse the two BVR-A structures available for the rat enzyme [14,15]. Although not commented on by the authors, rat BVR-A [15] co-crystallized with three inorganic phosphate molecules [15]. Interestingly, one of these phosphates occupies a site which has been proposed as the binding pocket for the 2′-phospho ester of NADPH [14]. In light of this information it was decided to look at the residues involved in binding phosphate in more detail. As shown in Figure 7, Ser43, Arg44 and Arg45 form an intricate lattice of ionic interactions and hydrogen bonds with inorganic phosphate.
Figure 7. Inorganic phosphate pocket of rat BVR-A.
The co-ordinates used for this Figure are taken from the crystal structure of the rat enzyme reported by Whitby et al. [15]. The structure was downloaded from the Protein Data Bank [36], and graphics generated using the pyMol program published by DeLano Scientific [37]. Red dots represent water molecules. A surface diagram of the residues is shown in (a). The phosphate pocket is clearly defined with a large distribution of positive charges contributed by Arg44 and Arg45, which lie along the walls of this pocket (coloured blue) and are seen to co-ordinate with the phosphate anion of the crystal structure, shown in (b) for clarity. (c) The salt bridge formation between Arg44 and Glu75. This interaction may contribute to the stabilization of the loop between β-sheets β2 and β3, and the αD helix in the rat crystal structure. Residues Ser43, Arg44 and Arg45 were targeted for mutation in an attempt to determine their effects on NADH-dependent phosphate activation.
Effect of phosphate concentration on the activity of wild-type and 2′-phosphate-pocket mutants of rat BVR-A
Figure 8 shows the effect of phosphate concentration on the activity of the rat enzyme (wild-type) in Tris/HCl buffer. As with the human enzyme, there is a marked activation by phosphate, with an A50 value of 20 mM. Although all three of the desired mutants were created, only the S43A and the R44A mutants were sufficiently stable to be examined kinetically. The A50 for the S43A mutant increases significantly to 60 mM (Figure 8), indicating that significant binding energy results from the interaction between Ser43 and the phosphate anion. A more dramatic result was seen with the R44A mutant, which is not activated significantly by increasing concentrations of phosphate when NADH is used as the cofactor (Figure 8). Mutation of these two residues in the 2′-phosphate-pocket decreases (S43A) and abolishes (R44A) the ability of inorganic phosphate to activate the NADH-dependent reduction of biliverdin, clearly implicating this as the site involved in the phosphate-activation phenomenon. We have also confirmed an earlier report [14] that the R44A mutation equalizes the apparent Km for NADH and NADPH (results not shown), again implicating this as the binding site for the 2′-phosphoester of NADPH.
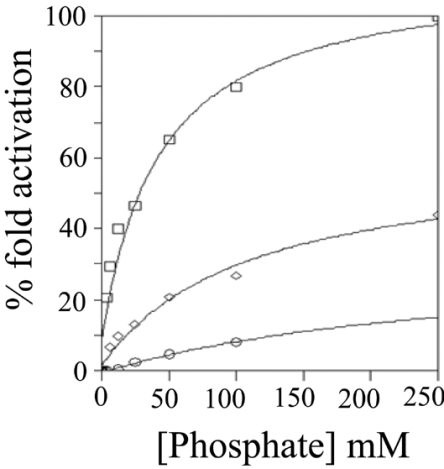
Figure 8. Effect of phosphate on the activity of the wild-type, S43A and R44A mutant rat BVR-A.
The activity of wild-type rat BVR-A (□), BVR-AR44A (○), and BVR-AS43A (◇), was measured in 50 mM Tris/HCl at pH 7.2 with 700 μM NADH and 10 μM biliverdin-IXα as substrate over the phosphate concentration range (0–250 mM).
The conclusions from this study, namely that NAD(P)H binds in an extended conformation, and that inorganic phosphate binds to the 2′-phosphate-pocket for NAD(P)H, have recently been confirmed in the crystal structure of human BVR-A with NADPH, which has been deposited in the Protein Data Bank by the Structural Genomics Consortium with code 2H63. Our model for the mechanism of activation of the NADH-dependent activity of BVR-A by inorganic phosphate therefore proposes that inorganic phosphate mimics the 2′-phosphoester of NADPH. There are clearly questions remaining concerning the nature of this complex, as an additional oxygen atom is present in the enzyme–NADH–Pi complex when compared with the enzyme–NADPH complex. Indeed the ability of pyrophosphate to activate suggests that at least some reorganization of the enzyme structure is necessary to achieve the ternary complexes with anions, and this will be the focus of further studies.
The hypothesis that NADH binding can be augmented by inorganic phosphate mimicking the 2′-phosphate of NADPH is not entirely novel, as a similar conclusion has been reached by Christensen et al. [31] for the Mg-dependent NMDMC (methylenetetrahydrofolate dehydrogenase-cyclohydrolase). These authors point out that NMDMC uses phosphate and magnesium to adapt an NADP site for NAD binding.
The effect of pH and biliverdin-IXα concentrations on the initial rate using NADPH as cofactor
At 2 μM biliverdin-IXα, the human enzyme is active over the entire pH range studied (Figure 9). As the substrate concentration is increased to 5 and 20 μM, the initial rate is significantly suppressed at neutral pH (due to pronounced substrate inhibition), which is far less severe under both acidic and alkaline conditions (Figure 9). This results in a shift in the apparent pH optimum from 8.0 to 8.5, depending on the biliverdin concentration used in the assay. The increased potency of substrate inhibition at neutral pH was also clearly evident in the time course of the reaction. An increase in rate during the time course of the reaction was observed as the substrate concentration decreased below substrate- inhibitory levels. This was particularly apparent at neutral pH values, where substrate inhibition is pronounced, whereas at the acid and alkaline limbs, where substrate inhibition was not apparent more normal P(t) curves were observed (results not shown).
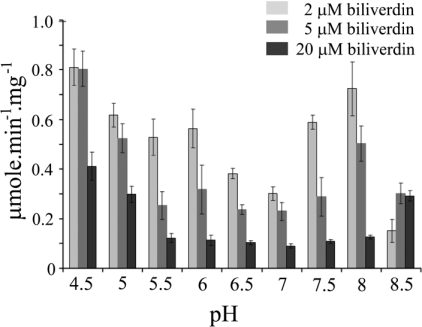
Figure 9. Effect of biliverdin concentration on the pH dependence of human BVR-A activity.
The effect of biliverdin concentration on the pH dependence of human BVR-A activity was investigated. The reaction was monitored at 460 nm at 30 °C, and contained a final concentration of 100 μM NADPH with 2, 5 and 20 μM biliverdin-IXα over the pH range 4.5–8.5.
It is still not clear why NADPH supports a significant rate at pH 8.5 to 9.0, whereas NADH is unable to achieve this. It should be noted that the approximate concentrations of nicotinamide nucleotides in rat liver have been reported as 120 μM for NADPH, 35 μM for NADP+, 70 μM for NADH and 500 μM for NAD+ [32,33]. With the apparent Km for NADPH of biliverdin reductases being in the low μmolar range, typically 1–10 μM, the enzyme is most probably saturated with NADPH in vivo. In contrast, the apparent Km for NADH is generally reported between 300 and 500 μM, and with cytosolic concentrations of NADH generally around 70 μM, this is unlikely to be a physiologically significant substrate. In well-oxygenated perfused rat liver, the cytosolic concentration of inorganic phosphate has been reported as 5.3 mM [34], and therefore is not predicted to impinge on NADH-dependent activity. However, inorganic phosphate in mitochondria can rise to 25 mM, and as this subcellular location has a higher NADH/NAD+ ratio and has also been reported recently to contain BVR-A [35], inorganic phosphate may promote NADH-dependent BVR-A activity in mitochondria.
Acknowledgments
This work was supported by a grant from Science Foundation Ireland.
References
- 1.Foresti R., Green C. J., Motterlini R. Generation of bile pigments by haem oxygenase: a refined cellular strategy in response to stressful insults. Biochem. Soc. Symp. 2004;71:177–192. doi: 10.1042/bss0710177. [DOI] [PubMed] [Google Scholar]
- 2.Ryter S. W., Alam J., Choi A. M. Heme oxygenase-1/carbon monoxide: from basic science to therapeutic applications. Physiol. Rev. 2006;86:583–650. doi: 10.1152/physrev.00011.2005. [DOI] [PubMed] [Google Scholar]
- 3.Fondevila C., Shen X. D., Tsuchiyashi S., Yamashita K., Csizmadia E., Lassman C., Busuttil R. W., Kupiec-Weglinski J. W., Bach F. H. Biliverdin therapy protects rat livers from ischemia and reperfusion injury. Hepatology. 2004;40:1333–1341. doi: 10.1002/hep.20480. [DOI] [PubMed] [Google Scholar]
- 4.Sarady-Andrews J. K., Liu F., Gallo D., Nakao A., Overhaus M., Ollinger R., Choi A. M., Otterbein L. E. Biliverdin administration protects against endotoxin-induced acute lung injury in rats. Am. J. Physiol. Lung Cell Mol. Physiol. 2005;289:L1131–L1137. doi: 10.1152/ajplung.00458.2004. [DOI] [PubMed] [Google Scholar]
- 5.Kapitulnik J. Bilirubin: an endogenous product of heme degradation with both cytotoxic and cytoprotective properties. Mol. Pharmacol. 2004;66:773–779. doi: 10.1124/mol.104.002832. [DOI] [PubMed] [Google Scholar]
- 6.Baranano D. E., Rao M., Ferris C. D., Snyder S. H. Biliverdin reductase: a major physiologic cytoprotectant. Proc. Natl. Acad. Sci. U.S.A. 2002;99:16093–16098. doi: 10.1073/pnas.252626999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nakao A., Otterbein L. E., Overhaus M., Sarady J. K., Tsung A., Kimizuka K., Nalesnik M. A., Kaizu T., Uchiyama T., Liu F., et al. Biliverdin protects the functional integrity of a transplanted syngeneic small bowel. Gastroenterology. 2004;127:595–606. doi: 10.1053/j.gastro.2004.05.059. [DOI] [PubMed] [Google Scholar]
- 8.Nakao A., Neto J. S., Kanno S., Stolz D. B., Kimizuka K., Liu F., Bach F. H., Billiar T. R., Choi A. M., Otterbein L. E., Murase N. Protection against ischemia/reperfusion injury in cardiac and renal transplantation with carbon monoxide, biliverdin and both. Am. J. Transplant. 2005;5:282–291. doi: 10.1111/j.1600-6143.2004.00695.x. [DOI] [PubMed] [Google Scholar]
- 9.Yamashita K., McDaid J., Ollinger R., Tsui T. Y., Berberat P. O., Usheva A., Csizmadia E., Smith R. N., Soares M. P., Bach F. H. Biliverdin, a natural product of heme catabolism, induces tolerance to cardiac allografts. FASEB J. 2004;18:765–767. doi: 10.1096/fj.03-0839fje. [DOI] [PubMed] [Google Scholar]
- 10.Rigney E. M., Phillips O., Mantle T. J. Some physical and immunological properties of ox kidney biliverdin reductase. Biochem. J. 1988;255:431–435. doi: 10.1042/bj2550431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maines M. D., Trakshel G. M. Purification and characterization of human biliverdin reductase. Arch. Biochem. Biophys. 1993;300:320–326. doi: 10.1006/abbi.1993.1044. [DOI] [PubMed] [Google Scholar]
- 12.Ennis O., Maytum R., Mantle T. J. Cloning and over-expression of rat kidney biliverdin IXα reductase as a fusion protein with glutathione S-transferase: stereochemistry of NADH oxidation and evidence that the presence of the glutathione S-transferase domain does not effect BVR-A activity. Biochem. J. 1997;328:33–36. doi: 10.1042/bj3280033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cunningham O., Dunne A., Sabido P., Lightner D., Mantle T. J. Studies on the specificity of the tetrapyrrole substrate for human biliverdin-IXα reductase and biliverdin-IXβ reductase: structure-activity relationships define models for both active sites. J. Biol. Chem. 2000;275:19009–19017. doi: 10.1074/jbc.275.25.19009. [DOI] [PubMed] [Google Scholar]
- 14.Kikuchi A., Park S. Y., Miyatake H., Sun D., Sato M., Yoshida T., Shiro Y. Crystal structure of rat biliverdin reductase. Nat. Struct. Biol. 2001;8:221–225. doi: 10.1038/84955. [DOI] [PubMed] [Google Scholar]
- 15.Whitby F. G., Phillips J. D., Hill C. P., McCoubrey W., Maines M. D. Crystal structure of a biliverdin IXα reductase enzyme-cofactor complex. J. Mol. Biol. 2002;319:1199–1210. doi: 10.1016/S0022-2836(02)00383-2. [DOI] [PubMed] [Google Scholar]
- 16.Phillips O., Mantle T. J., Tuffery A. R., Heyworth C. M., Wilson S. R., Houslay M. D. On the possible role of biliverdin stimulation of cyclic AMP levels as a trigger for liver regeneration in the rat. Biochem. Pharmacol. 1984;33:1963–1967. doi: 10.1016/0006-2952(84)90556-2. [DOI] [PubMed] [Google Scholar]
- 17.Yamaguchi T., Komoda Y., Nakajima H. Biliverdin-IXα reductase and biliverdin-IXβ reductase from human liver: purification and characterization. J. Biol. Chem. 1994;269:24343–24348. [PubMed] [Google Scholar]
- 18.Maines M. D., Polevoda B. V., Huang T. J., McCoubrey W. K., Jr Human biliverdin IXα reductase is a zinc-metalloprotein: characterization of purified and Escherichia coli expressed enzymes. Eur. J. Biochem. 1996;235:372–381. doi: 10.1111/j.1432-1033.1996.00372.x. [DOI] [PubMed] [Google Scholar]
- 19.Kutty R. K., Maines M. D. Purification and characterization of biliverdin reductase from rat liver. J. Biol. Chem. 1981;256:3956–3962. [PubMed] [Google Scholar]
- 20.Fakhrai H., Maines M. D. Expression and characterization of a cDNA for rat kidney biliverdin reductase: evidence suggesting the liver and kidney enzymes are the same transcript product. J. Biol. Chem. 1992;267:4023–4029. [PubMed] [Google Scholar]
- 21.Rigney E., Mantle T. J., Dickinson F. M. The kinetics of ox kidney biliverdin reductase in the pre-steady state: evidence that the dissociation of bilirubin is the rate-determining step. Biochem J. 1989;259:709–713. doi: 10.1042/bj2590709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Noguchi M., Yoshida T., Kikuchi G. Purification and properties of biliverdin reductases from pig spleen and rat liver. J. Biochem. (Tokyo) 1979;86:833–848. doi: 10.1093/oxfordjournals.jbchem.a132615. [DOI] [PubMed] [Google Scholar]
- 23.Schluchter W. M., Glazer A. N. Characterization of cyanobacterial biliverdin reductase: conversion of biliverdin to bilirubin is important for normal phycobiliprotein biosynthesis. J. Biol. Chem. 1997;272:13562–13569. doi: 10.1074/jbc.272.21.13562. [DOI] [PubMed] [Google Scholar]
- 24.Elliott G., Mantle T. J. Purification and properties of salmon liver biliverdin reductase. Biochem. Soc. Trans. 1995;23:389S. doi: 10.1042/bst023389s. [DOI] [PubMed] [Google Scholar]
- 24a.Lennon G. G., Auffray C., Polymeropoulos M., Soares M. B. The I.M.A.G.E. consortium: an integrated molecular analysis of genomes and their expression. Genomics. 1996;33:151–152. doi: 10.1006/geno.1996.0177. [DOI] [PubMed] [Google Scholar]
- 25.Fisher C. L., Pei G. K. Modification of a PCR-based site-directed mutagenesis method. Biotechniques. 1997;23:570–574. doi: 10.2144/97234bm01. [DOI] [PubMed] [Google Scholar]
- 26.Phillips O., Mantle T. J. Some kinetic and physical properties of biliverdin reductase. Biochem. Soc. Trans. 1981;9:275–278. doi: 10.1042/bst0090275. [DOI] [PubMed] [Google Scholar]
- 27.McDonagh A. F. In: The Porphyrins vol. 6. Dolphin D., editor. London: Academic Press; 1979. pp. 453–455. [Google Scholar]
- 28.Wu J. T., Wu L. H., Knight J. A. Stability of NADPH: effect of various factors on the kinetics of degradation. Clin. Chem. 1986;32:314–319. [PubMed] [Google Scholar]
- 29.Elliott G. Ph.D. Thesis. University of Dublin; 1996. [Google Scholar]
- 30.Fang L. S., Lai C. C. Characterisation and purification of biliverdin reductase from the liver of eel, Anguilla japonica. Comp. Biochem. Physiol. B. 1987;88:1151–1155. [Google Scholar]
- 31.Christensen K. E., Mirza I. A., Berghuis A. M., Mackenzie R. E. Magnesium and phosphate ions enable NAD binding to methylenetetrahydrofolate dehydrogenase-methenyltetrahydrofolate cyclohydrolase. J. Biol. Chem. 2005;280:34316–34323. doi: 10.1074/jbc.M505210200. [DOI] [PubMed] [Google Scholar]
- 32.Llorente P., Marco R., Sols A. Regulation of liver pyruvate kinase and the phosphoenolpyruvate crossroads. Eur. J. Biochem. 1970;13:45–54. doi: 10.1111/j.1432-1033.1970.tb00897.x. [DOI] [PubMed] [Google Scholar]
- 33.Marco R., Pestana A., Sebastian J., Sols A. Oxaloacetate metabolic crossroads in liver: enzyme compartmentation and regulation of gluconeogenesis. Mol. Cell. Biochem. 1974;3:53–70. doi: 10.1007/BF01660077. [DOI] [PubMed] [Google Scholar]
- 34.Desmoulin F., Cozzone P. J., Canioni P. Phosphorus-31 nuclearmagnetic-resonance study of phosphorylated metabolites compartmentation, intracellular pH and phosphorylation state during normoxia, hypoxia and ethanol perfusion, in the perfused rat liver. Eur. J. Biochem. 1987;162:151–159. doi: 10.1111/j.1432-1033.1987.tb10555.x. [DOI] [PubMed] [Google Scholar]
- 35.Converso D. P., Taille C., Carreras M. C., Jaitovich A., Poderoso J. J., Boczkowski J. HO-1 is located in liver mitochondria and modulates mitochondrial heme content and metabolism. FASEB J. 2006;20:1236–1238. doi: 10.1096/fj.05-4204fje. [DOI] [PubMed] [Google Scholar]
- 36.Berman H. M., Westbrook J., Feng Z., Gilliland G., Bhat T. N., Weissig H., Shindyalov I. N., Bourne P. E. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.DeLano W. L. The PyMOL Molecular Graphics System. DeLano Scientific: San Carlos; 2002. [Google Scholar]