Abstract
In a recent study, we described that UV-C irradiation resulted in redox-dependent activation and relocalization of A-SMase (acid sphingomyelinase) to the external surface of raft membrane microdomains, hydrolysis of SM (sphingomyelin) associated with the plasma membrane outer leaflet, ceramide generation and apoptosis. In the present study, we have investigated the influence of PKCζ (protein kinase Cζ), an atypical form of PKC on this pathway. This study shows that PKCζ overexpression resulted in the abrogation of UV-C-induced A-SMase translocation and activation into the raft microdomains, lack of ceramide generation and apoptosis inhibition. Moreover, PKCζ overexpression resulted in a decrease in UV-C-induced ROS (reactive oxygen species) production, which correlated with increased gene expression level of various antioxidant enzymes, including TRx (thioredoxin), TR (thioredoxin reductase) 1, TR2 and peroxiredoxin 1/TPx2 (thioredoxin peroxidase 2). Importantly, enforced TPx2 gene expression inhibited UV-C-induced A-SMase translocation. Finally, PKCζ inhibition led to a significant reduction in TPx2 protein expression. Altogether, these results suggest that PKCζ interferes with the UV-activated sphingolipid signalling pathway by regulating the TRx system. These findings may have important consequences for UV-induced carcinogenesis and resistance to phototherapy.
Keywords: acid sphingomyelinase, antioxidant defence, protein kinase Cζ (PKCζ), raft, thioredoxin peroxidase, UV-C
Abbreviations: A-SMase, acid sphingomyelinase; Ct, threshold cycle; DAPI, 4′,6-diamidino-2-phenylindole; DEPC, diethyl pyrocarbonate; FCS, fetal calf serum; MBS, Mes-buffered saline; PAG, proliferation-associated gene; PKC, protein kinase C; ROS, reactive oxygen species; RT, reverse transcription; SM, sphingomyelin; TPx2, thioredoxin peroxidase 2; TR, thioredoxin reductase; TRx, thioredoxin
INTRODUCTION
Mammalian cells respond to UV irradiation by activating a complex signalling network which involves both serine/threonine and tyrosine kinase stimulation, resulting in activation of transcription factors which, in turn, regulate a large variety of genes involved in cell growth and apoptosis. This pathway is under the control of two main types of mediators: ROS (reactive oxygen species) and lipid messengers, including diacylglycerol, arachidonic acid, phosphatidic acid and ceramide [1–3]. Previous studies have extensively documented the role of the SM (sphingomyelin)–ceramide pathway in apoptosis induced by both UV-B and UV-C [4]. In this signalling cascade, UV irradiation stimulates an acid sphingomyelinase (A-SMase), SM hydrolysis and the generation of ceramide, which, in turn, induces apoptosis through a JNK (c-Jun N-terminal kinase)-dependent pathway [5]. However, the mechanism by which A-SMase operates remained unclear until a recent study in which we described that UV-C irradiation resulted in a multistep process consisting of redox-dependent activation and relocalization of a Zn2+-independent A-SMase to the external surface of raft membrane microdomains, hydrolysis of plasma membrane outer leaflet-associated SM and accumulation of ceramide at the cell surface [6]. These results have been confirmed by another group using a different cellular model [7]. From these studies, one can speculate that any mechanism that could inhibit either A-SMase activation or relocalization might seriously interfere with UV-induced SM–ceramide pathway activation and apoptosis.
In this context, we have speculated that PKCζ (protein kinase Cζ) could be one of these regulators. Three lines of evidence support this hypothesis. First, we have described previously that this enzyme is a potent inhibitor of apoptosis induced by genotoxic agents [8,9]. Secondly, our group has reported that enforced expression of PKCζ confers a significant protection against UV-C irradiation [10]. Thirdly, we have reported that PKCζ contributes to the detoxication of ROS [8], the latter being an important contributor to UV-C-mediated A-SMase activation and membrane externalization [6].
In the present study, we show that PKCζ does interfere with UV-induced A-SMase relocalization and activation by limiting UV-induced ROS production.
EXPERIMENTAL
Materials
Silica gel 60 TLC plates were obtained from Merck. All other drugs and reagents were purchased from Sigma Chemical Co., Alexis Biochemicals, Matreya Biochemicals, Carlo Erba or Prolabo. Peptide PKCζ inhibitory pseudosubstrate was purchased from Millegen.
Cell culture
The human myeloblastic cell line U937 was obtained from the A.T.C.C. (Manassas, VA, U.S.A.). U937-ζJ cells were obtained from separated co-transfections with a full-length rat PKCζ cDNA construct subcloned into the pSV2M(2)6 vector and neomycin-resistant plasmid. U937-neo cells were obtained by transfection with the empty vector. All of these cell lines were kindly provided by Dr D. K. Ways (Lilly Corporate Center, Indianapolis, IN, U.S.A.) [11]. These cells displayed a 3-fold increase of both PKCζ expression and activity, as we described in two previous studies [12,13]. U937-TPx2 cells were obtained by stable transfection with TPx2 (thioredoxin peroxidase 2)/PAG (proliferation-associated gene) cDNA subcloned into the pEGFP-N1 vector. The plasmid encoding for PAG was kindly provided by Dr Goubin (Institut Curie, Paris, France). U937-TPx2 neo cells were obtained by transfection with the empty vector. Clone selection was carried out using geneticin.
All cells were cultured in RPMI 1640 medium supplemented with 10% FCS (fetal calf serum) at 37 °C under 5% CO2. Culture medium was supplemented with 2 mM L-glutamine, 200 units/ml penicillin and 100 μg/ml streptomycin (all from Eurobio).
Cell irradiation
Cells were irradiated with UV-C light (254 nm) in PBS during 30 s corresponding to 30 J/m2, at a concentration of 106 cells/ml.
DAPI (4′,6-diamidino-2-phenylindole) nuclear staining assay
Changes in cellular nuclear chromatin were evaluated by fluorescence microscopy (Diaplan, Leica) by DAPI staining using the following method. Briefly, cells were fixed in 4% (w/v) paraformaldehyde in PBS, pH 7.4, for 15 min. After washing in PBS, cells were dried and stained by 0.5 μg/ml DAPI.
Metabolic cell labelling and quantification of ceramide
Total cellular ceramide was performed by labelling 5×106 cells to isotopic equilibrium with 1 μCi/ml of [9, 10-3H]palmitic acid (53.0 Ci/mmol) (Amersham Biosciences) for 48 h in complete medium as described previously [14]. Cells were then washed and resuspended in serum-free medium for kinetic experiments. Lipids were extracted and resolved by TLC developed in chloroform/methanol/ethanoic (acetic) acid/methanoic (formic) acid/water (65:30:10:4:2, by vol.) up to two-thirds of the plate and then in chloroform/methanol/ethanoic acid (94:5:5, by vol.). Ceramide was scraped and quantified by liquid-scintillation counting. Lipid standards were used to identify the various metabolic products.
Isolation of membrane raft microdomains
Raft microdomains were isolated from cells as described previously [15]. For each isolation, 108 cells were washed twice with PBS. Cells were pelleted by centrifugation at 194 g for 5 min, resuspended in 1 ml of ice-cold MBS (Mes-buffered saline) (150 mM NaCl and 25 mM Mes, pH 6.5), containing 1% (w/v) Triton X-100. After 30 min on ice, cells were homogenized further by ten strokes of a Dounce homogenizer on ice. Then, 1.5 ml of ice-cold MBS was added and 2 ml of this suspension was mixed with 2 ml of 80% (w/v) sucrose in MBS. This mixture was subsequently loaded on to a linear gradient consisting of 8 ml of 5–40% (w/v) sucrose in MBS. All solutions contained the following protease inhibitors: 100 μM PMSF, 1 mM EDTA and 1 μM each of aprotinin, leupeptin and pepstatin A. Gradients were centrifuged in a Beckman SW 41 swinging-rotor at 39000 rev./min for 20 h at 4 °C. Twelve fractions of 1 ml each were collected (from top to bottom), vortex-mixed and stored at −80 °C. The protein content of both fractions and the total initial cell suspension were measured using BSA as standard ([16], but see [16a]). GM1 was used as a marker of rafts [17,18].
A-SMase activity
A-SMase activity was evaluated as we described previously [6,19]. Briefly, whole cells were irradiated for different times with 30 J/m2 of UV-C. For A-SMase activity, the substrate solution consisted of [methylcholine-14C]SM (105 d.p.m./assay) (NEN) and 0.1% (w/v) Triton X-100 in 200 mM Tris/HCl buffer (pH 5) containing 10 mM DTT (dithiothreitol) and 10 mM MgCl2. After 2 h of incubation at 37 °C, reactions were terminated by adding 300 μl of water and 2.5 ml of chloroform/methanol (2:1, v/v). Phases were separated by centrifugation at 1000 g for 5 min, and the amount of released radioactive phosphocholine was determined by subjecting 700 μl of the upper phase to scintillation counting.
The amount of radiolabelled substrate hydrolysed during an assay never exceeded 10% of the total amount of substrate added. For calculation of the specific radioactivities in total cell homogenates, values were corrected for protein content, reaction time and specific radioactivity of the substrate.
A-SMase activity was assayed in 250 μl of each raft fraction, and reactions were started by adding 250 μl of substrate solution as described previously [19].
A-SMase translocation
A-SMase translocation was visualized by two different methods, FACS and confocal microscopy, as we described previously [6,19].
FACS analysis
Cells were irradiated or not with 30 J/m2 of UV-C, fixed for 10 min in 4% (w/v) paraformaldehyde in PBS. Cells were then washed and incubated further for 45 min with rabbit polyclonal anti-A-SMase antibody (Santa Cruz Biotechnology) at 2 μg/ml. Cells were then washed in PBS containing 1% FCS, and stained for 45 min with FITC-labelled goat anti-rabbit (Jackson ImmunoResearch Laboratories). After a final wash in PBS, cells were analysed using a FACSCalibur flow cytometer (Becton-Dickinson).
Confocal microscopy
Cells were irradiated with 30 J/m2 of UV-C, fixed for 10 min in 4% (w/v) paraformaldehyde in PBS and washed with PBS containing 3% BSA (w/v) and 1 mM Hepes (PBS-BSA). Cells were then incubated for 45 min with a rabbit polyclonal anti-A-SMase antibody at 2 μg/ml. Cells were then washed in PBS-BSA and stained for 45 min with 200 ng/ml Cy5 (indodicarbocyanine)-labelled goat anti-rabbit antibody (Jackson ImmunoResearch Laboratories). After a final wash in PBS, cells were mounted on glass coverslips with Dako mounting medium. Control staining was performed without primary antibody. Slides were examined with a Carl Zeiss LSM confocal microscope using a Plan-Apochromat 63× oil-immersion objective [1.4 NA (numerical aperture)]. An argon laser at 488 nm was used to excite FITC (emission wavelength 515–540 nm).
Western blotting
Cells (106) were washed in PBS before addition of Laemmli sample buffer (2% SDS, 10% glycerol, 5% 2-mercaptoethanol, 60 mM Tris/HCl, pH 6.8, and 0.001% Bromophenol Blue). Samples were sonicated for 15–20 s and boiled for 5 min at 95 °C. The samples were resolved by SDS/PAGE (10% gels), transferred on to nitrocellulose membrane (Hybond-C, Amersham Biosciences), blocked with 10% non-fat dried milk powder in Tris-buffered saline/0.1% Tween 20 at 4 °C for 2 h and incubated overnight at 4 °C with a rabbit anti-TPx2 (PAG) antibody (Santa Cruz Biotechnology), and for 30 min with horseradish-peroxidase-conjugated anti-rabbit antibody. Bound proteins were detected using the ECL® (enhanced chemiluminescence) detection system (Amersham Biosciences).
Determination of ROS
Production of ROS was detected using a C2938 fluorescent probe (Molecular Probes). Briefly, exponentially growing cells were labelled with 0.5 μM C2938 for 1 h and then irradiated with 30 J/m2 of UV-C light. The cells were washed in PBS, and cell fluorescence was determined using flow cytometry on a FACScan cytometer (BD Biosciences).
Total RNA extraction
Total RNA was isolated by TRIzol® Reagent (Invitrogen) according to the manufacturer's protocol and resuspended in DEPC (diethyl pyrocarbonate)-treated water. RNA quantification, purity and integrity were determined by spectrophotometry (A260, A280 and A320 measurements) and 1.5% agarose gel electrophoresis.
RT (reverse transcription)–PCR
The RT–PCR multiplex reaction was carried out with Superscript™ kit one-step RT–PCR with Platinum® Taq (Invitrogen) according to the manufacturer's recommendations, on 0.2 μg of total RNA with the following cycles: reverse transcription for 30 min at 50 °C, then amplification at 94 °C for 5 min, 30 cycles of 94 °C for 1 min, 60 °C for 1 min and 72 °C for 1 min, and a final elongation at 72 °C for 10 min. PCR amplification was carried out using the following primer pairs: TPx2 fwd (5′-TTTGGTATCAGACCCGAAGC-3′) and TPx2 rev (5′-TCCCCATGTTTTCAGTGAA-3′), TR1 fwd (5′-TGTGGACTGACCAAAAAGCA-3′) and TR1 rev (5′-CTGCCAAATGTCAGCTTCAA-3′), TR2 fwd (5′-GACCAGCAAATGTCCTCCAT-3′) and TR2 rev (5′-ATCGCTATGGGTGTCAGCTC-3′), and β-actin fwd (5′-ACACTGTGCCCATCTACGAGG-3′) and β-actin rev (5′-AGGGGCCGGACTCGTCATACT-3′). Primer concentrations were 1 μM for TPx2, TR (thioredoxin reductase) 1 or TR2 and 0.1 μM for β-actin. Distilled DEPC-treated water was used as a negative control. RT–PCR products were analysed after electrophoresis in a 2% agarose gel stained with 0.5 μg/ml ethidium bromide, and visualized under UV light. Results are expressed as mRNA level fold increase with U937-neo cells as 100%.
Real-time quantitative PCR
Total cellular RNA was extracted with TRIzol®. The cDNA was synthesized using random hexamers and oligo(dT) from 4 μg of mRNA and performed with the SuperScript™ First-Strand Synthesis System for RT–PCR (Invitrogen Life Technologies). Primers were TR1 rev (5′-TTGCAGTCTTGGCAACAGCATC-3′) and TR1 fwd (5′-CAGCTGGACAGCACAATTGGAA-3′), TR2 rev (5′-CCAACGTTTTTCCAGGGGATTC-3′) and TR2 fwd (5′-CAGCCGATCACATCATCATTGC-3′), TPx2 rev (5′-GCACACTTCCCCATGTTTGTCA-3′) and TPx2 fwd (5′-CATCTCGTTCAGGGGCCTTTTT-3′), and TRx rev (5′-GATCATTTTGCAAGGCCCACAC-3′) and TRx fwd (5′-TGGTGAAGCAGATCGAGAGCAA-3′). Real-time PCR was performed using an iCycler thermal cycler (Applied Biosystems 7000 Real-Time PCR system) according to the manufacturer's instructions. Reactions were performed with 0.3 μM primers. Nucleotides, Taq DNA polymerase and buffer were included in SYBR Green JumpStart™ Taq ReadyMix™ for quantitative PCR. cDNA amplification consisted of one cycle at 95 °C for 90 s, followed by 40 cycles of denaturation at 95 °C for 15 s and annealing and extension at 60 °C for 1 min. The threshold cycle (Ct) values were determined by iCycler software (ABI Prism 7000), and the quantification data were analysed following the ΔΔCt method using S14 as reference. We have checked that PCR efficiency (E) of the amplification was similar regardless of which primers were used, and we calculated the relative amount (RA): RA=(1+E)−ΔΔCt.
Statistics
Student's t test was performed to evaluate the statistical significance.
RESULTS
Effect of PKCζ on UV-C-induced apoptosis
The influence of PKCζ overexpression on UV-C-induced apoptosis was evaluated in U937-neo and PKCζ-overexpressing U937-ζJ cells. DAPI nuclear staining revealed that, 12 h after 30 J/m2 of irradiation, UV-C induced 55±11% of apoptotic cells in U937-neo, whereas only 16±10% of apoptotic cells were observed in U937-ζJ cells.
Effect of PKCζ on A-SMase redistribution
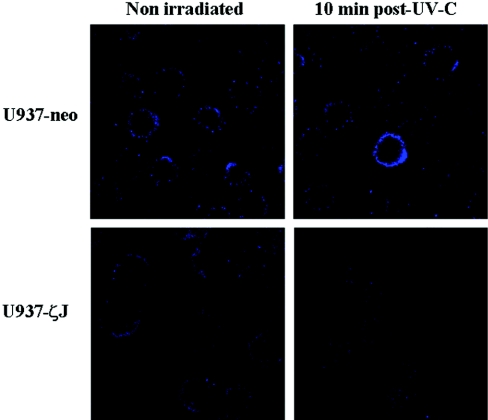
In our previous study, we showed that UV-C induced A-SMase translocation from the cytosol to the cell surface and enzyme activation [6]. On the basis of these results, we first evaluated the effect of PKCζ on A-SMase translocation. Flow cytometry (results not shown) and confocal microscopy (Figure 1) analysis performed on non-permeabilized U937-neo and U937-ζJ cells with an anti-A-SMase antibody revealed that UV-C induced A-SMase accumulation at the external surface in U937-neo cells, but not in U937-ζJ cells, suggesting that the enzyme did interfere with A-SMase translocation. Moreover, PKCζ overexpression had no impact on A-SMase expression as revealed by Western blot analysis (results not shown).
Figure 1. Effect of PKCζ on UV-C-induced A-SMase localization.
U937-neo and U937-ζJ cells were irradiated or not with 30 J/m2 of UV-C. A-SMase translocation was observed at 10 min post-UV-C by confocal microscopy on non-permeabilized cells using a FITC-conjugated rabbit anti-A-SMase antibody. Results are representative of three independent experiments.
Effect of PKCζ on A-SMase stimulation in intact cells
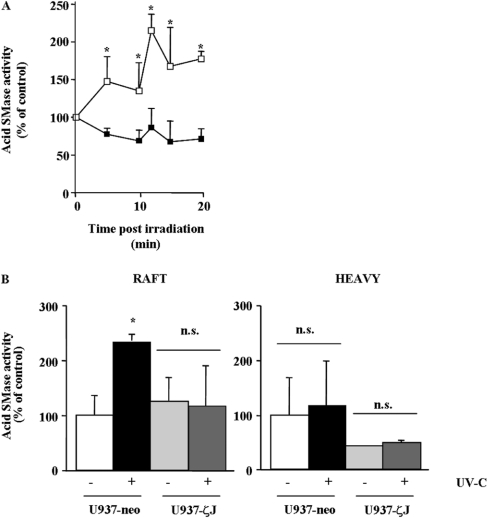
We therefore hypothesized that PKCζ could interfere with A-SMase stimulation. In untreated cells, basal A-SMase activity was similar in U937-ζJ cells compared with U937-neo cells (40.8±3.3 and 40±2.2 pmol/h per mg of protein respectively). Moreover, as expected from our previous study, UV-C induced a significant A-SMase stimulation in U937-neo cells, which was detected as soon as 5 min after irradiation and peaked at 10–15 min. However, no A-SMase stimulation was detectable in U937-ζJ cells following irradiation up to 20 min (Figure 2A).
Figure 2. Effect of PKCζ on UV-C-induced A-SMase stimulation on whole cells and on raft microdomains.
(A) U937-neo (□) and U937-ζJ (■) cells were irradiated or not with 30 J/m2 of UV-C. Total cellular A-SMase activity was measured at various times as described in the Experimental section. Results are means+S.D. of triplicate determinations of a representative experiment (one of three independent experiments). *P<0.05. (B) U937-neo and U937-ζJ cells were irradiated with 30 J/m2 of UV-C, lysed in cold Triton X-100 and fractionated on a sucrose density gradient. Aliquots were collected and analysed for A-SMase activity. Peak A-SMase activities (10–15 min) are expressed compared against the amount of proteins in each fraction, rafts (4–6) and heavy (7–10). Results are means+S.D. of triplicate determinations of a representative experiment (one of three independent experiments). *P<0.05; n.s., not significant.
Then, we evaluated the influence of PKCζ in UV-induced stimulation in a raft-associated fraction of A-SMase. Cells were irradiated or not with 30 J/m2 of UV-C, then at 12 min postUV-C irradiation (corresponding to the peak of A-SMase stimulation in U937-neo), cells were lysed in cold Triton X-100, separated on a sucrose density gradient, and A-SMase was measured on pooled fractions. U937 rafts were previously characterized by the Triton-insoluble material (fractions 4–6), the high SM content and the raft marker ganglioside GM1 (results not shown). As shown in Figure 2(B), UV-C induced an increase in A-SMase activity exclusively in raft fractions in U937-neo, but not in U937-ζJ cells.
These results suggest that PKCζ interferes with the SM–ceramide pathway by inhibiting UV-induced A-SMase translocation and consequently its stimulation into raft microdomains.
Effect of PKCζ on ceramide production
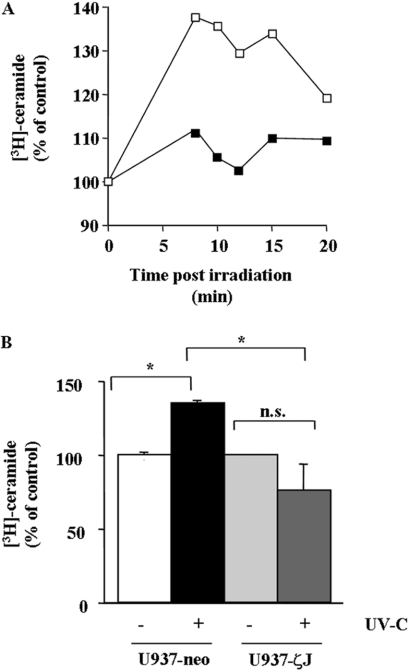
We next investigated the role of PKCζ in the UV-C-induced SM–ceramide pathway. For this reason, U937-neo and U937-ζJ cells were pre-labelled with [9, 10-3H]palmitic acid to equilibrium for 48 h and then irradiated in PBS at 30 J/m2 of UV-C. Ceramide was quantified as described in the Experimental section. As shown in Figure 3, UV-C induced ceramide accumulation in U937-neo, but not in U937-ζJ cells.
Figure 3. Effect of PKCζ on UV-C-induced ceramide generation.
U937-neo (□) and U937-ζJ (■) cells were pre-labelled with [9, 10-3H]palmitic acid to equilibrium for 48 h and then irradiated or not with 30 J/m2 of UV-C. (A) Ceramide was extracted at various times post-irradiation as described in the Experimental section. Results are representative of three independent experiments. (B) Results were obtained at 12 min post-UV-C, which represents the peak of ceramide generation, and are the means+S.D. for three independent experiments. *P<0.05; n.s., not significant.
Effect of PKCζ on UV-C-induced ROS generation
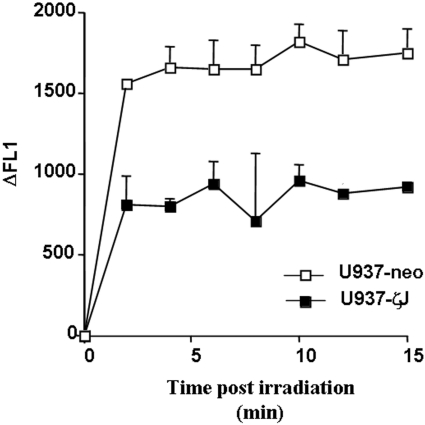
As we have recently described, UV-C-induced A-SMase translocation and its activation are mediated by ROS production, generated 5 min after irradiation [6]. In this context, we determined whether PKCζ could interfere with this UV-C-induced ROS production. As shown in Figure 4, overexpression of PKCζ resulted in the partial, although significant, inhibition of ROS accumulation in UV-treated cells.
Figure 4. Effect of PKCζ overexpression on UV-C-induced ROS production.
U937-neo (□) and U937-ζJ (■) cells were labelled with C2938, a fluorescent probe, and irradiated with 30 J/m2 of UV-C light, and ROS production was then analysed at different time points by flow cytometry. Results correspond to the difference of fluorescence (ΔFL1) between treated and untreated cells and are means+S.D. of triplicate determinations of a representative experiment.
Altogether, these results suggest that PKCζ acts upstream of A-SMase stimulation by interfering with UV-mediated ROS production.
Effect of PKCζ on H2O2 detoxication system
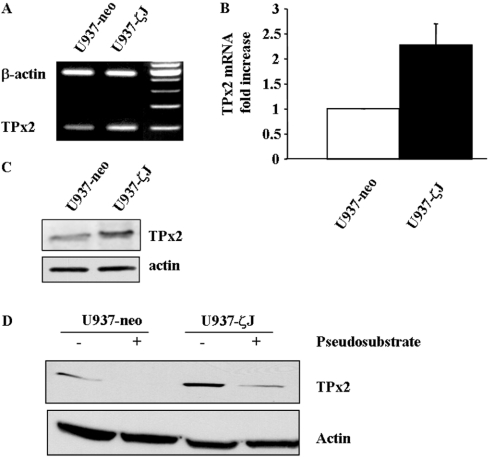
We have reported previously that PKCζ overexpression resulted in an increased in H2O2 detoxication and that U937-ζJ cells were resistant to a high level of exogenous H2O2 [8]. H2O2 may be detoxified by a variety of enzymes, including catalase, glutathione peroxidase and the thioredoxin system. Therefore we hypothesized that PKCζ may influence the expression level of these enzymes. No difference on either catalase or glutathione peroxidase mRNA level was observed in U937-ζJ compared with U937-neo cells (results not shown). However, as measured by RT–PCR (Figure 5A), real-time quantitative PCR (Figure 5B) and Western blotting (Figure 5C), PKCζ-overexpressing cells displayed much higher mRNA and protein levels of TPx2/PAG, the prime candidate for H2O2 detoxication (for review, see [20]). TRx (thioredoxin) and TRs mRNA were also increased in PKCζ-overexpressing cells (results not shown). Moreover, treatment of both U937-neo and U937-ζJ cells with PKCζ pseudosubstrate resulted in an impressive reduction of TPx2 protein expression level (Figure 5D).
Figure 5. Effect of PKCζ overexpression on TPx2/PAG.
The mRNA level of TPx2 was analysed by RT–PCR (A) or by real-time quantitative PCR (B) in U937-neo and U937-ζJ cells as described in the Experimental section. Results are representative of (A) or are the means+S.D. for (B) three independent experiments. (C) TPx2 protein levels were visualized by Western blotting using specific anti-PAG antibody in U937-neo and U937-ζJ cells. (D) U937-neo and U937-ζJ cells were treated for 1 h with PKCζ pseudosubstrate at 50 μM. TPx2 expression was evaluated by Western blotting using anti-PAG antibody. Actin was used as a control of protein expression.
Effect of TPx2 overexpression on A-SMase delocalization and ceramide formation
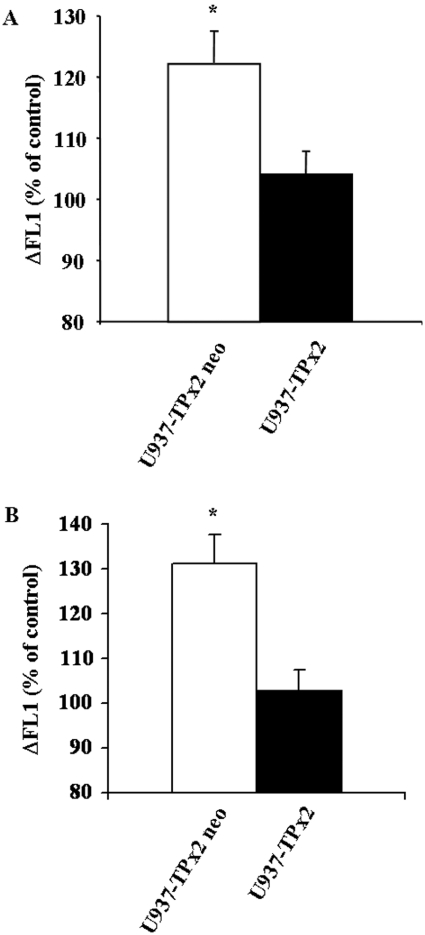
To confirm further the role of TPx2 in the regulation of UV-induced SM–ceramide pathway stimulation, U937 cells were stably transfected with a plasmid encoding for TPx2/PAG. TPx2 overexpression resulted in the inhibition of both A-SMase membrane translocation (Figure 6A) and ceramide generation at the cell surface (Figure 6B) upon UV-C irradiation, thus mimicking the effect of PKCζ. The A-SMase expression level was unaffected by TPx2 overexpression (results not shown).
Figure 6. Effect of TPx2/PAG overexpression on A-SMase delocalization and ceramide generation.
U937-TPx2 neo and U937-TPx2 cells were irradiated at 30 J/m2 of UV-C. A-SMase delocalization (A) or ceramide generation (B) was determined in non-permeabilized cells as described in the Experimental section by FACS analysis 10 min post-UV-C. Results correspond to the difference of fluorescence (ΔFL1) between treated and untreated cells and are means+S.D. for three independent experiments. *P<0.01. Results are representative of all clones tested.
DISCUSSION
In the present study, we describe for the first time that the atypical PKCζ inhibits UV-C-induced apoptosis. This observation may have important implications, since this enzyme is regulated by a large variety of signalling pathways, including tyrosine kinase receptors, Ras oncogenic variants and phospholipase C and D, through various messengers such as ceramide, diacylglycerol and phosphatidic acid [21–24]. Therefore it is conceivable that each of these pathways are regulatory candidates for UV-induced apoptosis through PKCζ. On the basis of previous studies, it appears that classic and novel PKC isoenzymes are also implicated in the negative regulation of UV-induced apoptosis [25,26]. Altogether, these observations suggest a general protective function for PKC family enzymes in the context of UV irradiation. This may have significant consequences not only for malignant transformation through survival of cells with damaged DNA, but also for the resistance to phototherapy in the treatment of malignant or inflammatory cutaneous diseases.
Several studies carried out in our laboratory have described that PKCζ may exert a protective function following other genotoxic stress, including treatment with topoisomerase II inhibitors [9,27], nucleoside analogues [8], cisplatin and alkylating agents [10]. These findings suggest a global function of this enzyme in cellular protection against various stress conditions. In the latter studies, we presented evidence that PKCζ acts by regulating the level of DNA damage. As UV irradiation is concerned, we have reported previously that PKCζ significantly enhances nucleotide excision repair capacity [10]. The present study shows that PKCζ may also act by regulating post-damage apoptotic pathways such as the SM cycle. These observations suggest that PKCζ, and perhaps other PKC isoenzymes, may exert their cellular protective function by regulating both the level of damage and the post-damage response.
We have shown that PKCζ interferes with UV-mediated ROS production, which is an important contributor for A-SMase stimulation [6]. In the present study, we propose that the antioxidant effect of PKCζ is related to its capacity to regulate the expression of different antioxidant enzymes and, particularly, the enzymes of the TRx system. Indeed, we show here for the first time that PKCζ regulates the expression of TPx2, which, in turn, acts as a potent inhibitor of A-SMase translocation upon UV-C irradiation. However, it is possible that other antioxidant systems are involved, although it appears that catalase and the glutathione-related enzymes are unlikely to be involved in the PKCζ antioxidant effect (see also [8]). The mechanism by which PKCζ regulates TPx2 remains to be elucidated. Interestingly, a previous study described phorbol ester-dependent activation of TPx2 via PKC [28], suggesting that classic or novel PKC isoenzymes may also regulate TPx2. Combined with the present study, these findings suggest that PKC in general could be a critical regulator of the TRx system. In a previous study, we described that PKCζ overexpression did not confer protection against cell-permeant ceramide [8]. Based on these findings and the present study, we can therefore assume that PKCζ exerts its protective effect at an early step of UV-C signalling by interfering with ceramide production.
To conclude, our study shows that PKCζ is a potent regulator of UV-induced apoptosis. We propose that this regulatory mechanism operates through the inhibition of the UV-induced ROS production and subsequent inhibition of A-SMase activation and translocation to cell surface into raft microdomains. These results indicate that PKCζ is an important player in the UV response.
Acknowledgments
This work was supported by la Ligue Nationale Contre le Cancer and les Comités Départementaux du Gers, de l'Aveyron et de la Haute-Garonne (J.-P. J.). A. C. is the recipient of a grant from la Fondation pour la Recherche Médicale. We acknowledge Dr Gérard Goubin (Laboratoire d’Oncogenèse, FRE 2584 CNRS, Institut Curie, Paris, France) for providing PAG cDNA and Catherine Trichard (INSERM U563, CPTP, Toulouse, France) for technical assistance.
References
- 1.Gilchrest B. A., Park H. Y., Eller M. S., Yaar M. Mechanisms of ultraviolet light-induced pigmentation. Photochem. Photobiol. 1996;63:1–10. doi: 10.1111/j.1751-1097.1996.tb02988.x. [DOI] [PubMed] [Google Scholar]
- 2.Komatsu M., Takahashi T., Abe T., Takahashi I., Ida H., Takada G. Evidence for the association of ultraviolet-C and H2O2-induced apoptosis with acid sphingomyelinase activation. Biochim. Biophys. Acta. 2001;1533:47–54. doi: 10.1016/s1388-1981(01)00139-1. [DOI] [PubMed] [Google Scholar]
- 3.Schieven G. L., Kirihara J. M., Gilliland L. K., Uckun F. M., Ledbetter J. A. Ultraviolet radiation rapidly induces tyrosine phosphorylation and calcium signaling in lymphocytes. Mol. Biol. Cell. 1993;4:523–530. doi: 10.1091/mbc.4.5.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chatterjee M., Wu S. Cell line dependent involvement of ceramide in ultraviolet light-induced apoptosis. Mol. Carcinog. 2001;30:47–55. doi: 10.1023/a:1011083818452. [DOI] [PubMed] [Google Scholar]
- 5.Verheij M., Bose R., Lin X. H., Yao B., Jarvis W. D., Grant S., Birrer M. J., Szabo E., Zon L. I., Kyriakis J. M., et al. Requirement for ceramide-initiated SAPK/JNK signalling in stress-induced apoptosis. Nature. 1996;380:75–79. doi: 10.1038/380075a0. [DOI] [PubMed] [Google Scholar]
- 6.Charruyer A., Grazide S., Bezombes C., Muller S., Laurent G., Jaffrezou J. P. UV-C light induces raft-associated acid sphingomyelinase and JNK activation and translocation independently on a nuclear signal. J. Biol. Chem. 2005;280:19196–19204. doi: 10.1074/jbc.M412867200. [DOI] [PubMed] [Google Scholar]
- 7.Rotolo J. A., Zhang J., Donepudi M., Lee H., Fuks Z., Kolesnick R. Caspase-dependent and -independent activation of acid sphingomyelinase signaling. J. Biol. Chem. 2005;280:26425–26434. doi: 10.1074/jbc.M414569200. [DOI] [PubMed] [Google Scholar]
- 8.Bezombes C., de Thonel A., Apostolou A., Louat T., Jaffrezou J. P., Laurent G., Quillet-Mary A. Overexpression of protein kinase Cζ confers protection against antileukemic drugs by inhibiting the redox-dependent sphingomyelinase activation. Mol. Pharmacol. 2002;62:1446–1455. doi: 10.1124/mol.62.6.1446. [DOI] [PubMed] [Google Scholar]
- 9.Mansat V., Laurent G., Levade T., Bettaieb A., Jaffrezou J. P. The protein kinase C activators phorbol esters and phosphatidylserine inhibit neutral sphingomyelinase activation, ceramide generation, and apoptosis triggered by daunorubicin. Cancer Res. 1997;57:5300–5304. [PubMed] [Google Scholar]
- 10.Louat T., Canitrot Y., Jousseaume S., Baudouin C., Canal P., Laurent G., Lautier D. Atypical protein kinase C stimulates nucleotide excision repair activity. FEBS Lett. 2004;574:121–125. doi: 10.1016/j.febslet.2004.08.024. [DOI] [PubMed] [Google Scholar]
- 11.Ways D. K., Posekany K., deVente J., Garris T., Chen J., Hooker J., Qin W., Cook P., Fletcher D., Parker P. Overexpression of protein kinase Cζ stimulates leukemic cell differentiation. Cell Growth Differ. 1994;5:1195–1203. [PubMed] [Google Scholar]
- 12.Mansat-De Mas V., de Thonel A., Gaulin V., Demur C., Laurent G., Quillet-Mary A. Protein kinase Cζ overexpression induces erythroid phenotype in the monocytic leukaemia cell line U937. Br. J. Haematol. 2002;118:646–653. doi: 10.1046/j.1365-2141.2002.03625.x. [DOI] [PubMed] [Google Scholar]
- 13.Plo I., Hernandez H., Kohlhagen G., Lautier D., Pommier Y., Laurent G. Overexpression of the atypical protein kinase Cζ reduces topoisomerase II catalytic activity, cleavable complexes formation, and drug-induced cytotoxicity in monocytic U937 leukemia cells. J. Biol. Chem. 2002;277:31407–31415. doi: 10.1074/jbc.M204654200. [DOI] [PubMed] [Google Scholar]
- 14.Jaffrezou J. P., Levade T., Bettaieb A., Andrieu N., Bezombes C., Maestre N., Vermeersch S., Rousse A., Laurent G. Daunorubicin-induced apoptosis: triggering of ceramide generation through sphingomyelin hydrolysis. EMBO J. 1996;15:2417–2424. [PMC free article] [PubMed] [Google Scholar]
- 15.Lisanti M. P., Tang Z., Scherer P. E., Kubler E., Koleske A. J., Sargiacomo M. Caveolae, transmembrane signalling and cellular transformation. Mol. Membr. Biol. 1995;12:121–124. doi: 10.3109/09687689509038506. [DOI] [PubMed] [Google Scholar]
- 16.Smith P. K., Krohn R. I., Hermanson G. T., Mallia A. K., Gartner F. H., Provenzano M. D., Fujimoto E. K., Goeke N. M., Olson B. J., Klenk D. C. Measurement of protein using bicinchoninic acid. Anal. Biochem. 1985;150:76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- 16a.Erratum. Anal. Biochem. 1987;163:279. [Google Scholar]
- 17.Solomon S., Masilamani M., Rajendran L., Bastmeyer M., Stuermer C. A., Illges H. The lipid raft microdomain-associated protein reggie-1/flotillin-2 is expressed in human B cells and localized at the plasma membrane and centrosome in PBMCs. Immunobiology. 2002;205:108–119. doi: 10.1078/0171-2985-00114. [DOI] [PubMed] [Google Scholar]
- 18.Nichols B. J. GM1-containing lipid rafts are depleted within clathrin-coated pits. Curr. Biol. 2003;13:686–690. doi: 10.1016/s0960-9822(03)00209-4. [DOI] [PubMed] [Google Scholar]
- 19.Bezombes C., Grazide S., Garret C., Fabre C., Quillet-Mary A., Muller S., Jaffrezou J. P., Laurent G. Rituximab antiproliferative effect in B-lymphoma cells is associated with acid-sphingomyelinase activation in raft microdomains. Blood. 2004;104:1166–1173. doi: 10.1182/blood-2004-01-0277. [DOI] [PubMed] [Google Scholar]
- 20.Rhee S. G., Chae H. Z., Kim K. Peroxiredoxins: a historical overview and speculative preview of novel mechanisms and emerging concepts in cell signaling. Free Radical Biol. Med. 2005;38:1543–52. doi: 10.1016/j.freeradbiomed.2005.02.026. [DOI] [PubMed] [Google Scholar]
- 21.Berra E., Diaz-Meco M. T., Dominguez I., Municio M. M., Sanz L., Lozano J., Chapkin R. S., Moscat J. Protein kinase Cζ isoform is critical for mitogenic signal transduction. Cell. 1993;74:555–563. doi: 10.1016/0092-8674(93)80056-k. [DOI] [PubMed] [Google Scholar]
- 22.Muller G., Ayoub M., Storz P., Rennecke J., Fabbro D., Pfizenmaier K. PKCζ is a molecular switch in signal transduction of TNF-α, bifunctionally regulated by ceramide and arachidonic acid. EMBO J. 1995;14:1961–1969. doi: 10.1002/j.1460-2075.1995.tb07188.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakanishi H., Brewer K. A., Exton J. H. Activation of the ζ isozyme of protein kinase C by phosphatidylinositol 3,4,5-trisphosphate. J. Biol. Chem. 1993;268:13–16. [PubMed] [Google Scholar]
- 24.Nakanishi M., Adami G. R., Robetorye R. S., Noda A., Venable S. F., Dimitrov D., Pereira-Smith O. M., Smith J. R. Exit from G0 and entry into the cell cycle of cells expressing p21Sdi1 antisense RNA. Proc. Natl. Acad. Sci. U.S.A. 1995;92:4352–4356. doi: 10.1073/pnas.92.10.4352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaneko Y. S., Ikeda K., Nakanishi M. Phorbol ester inhibits DNA damage-induced apoptosis in U937 cells through activation of protein kinase C. Life Sci. 1999;65:2251–2258. doi: 10.1016/s0024-3205(99)00490-7. [DOI] [PubMed] [Google Scholar]
- 26.Hussaini I. M., Carpenter J. E., Redpath G. T., Sando J. J., Shaffrey M. E., Vandenberg S. R. Protein kinase C-η regulates resistance to UV- and γ-irradiation-induced apoptosis in glioblastoma cells by preventing caspase-9 activation. Neuro-oncology. 2002;4:9–21. doi: 10.1093/neuonc/4.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Plo I., Hernandez H., Kohlhagen G., Lautier D., Pommier Y., Laurent G. Overexpression of the atypical protein kinase Cζ reduces topoisomerase II catalytic activity, cleavable complexes formation, and drug-induced cytotoxicity in monocytic U937 leukemia cells. J. Biol. Chem. 2002;277:31407–31415. doi: 10.1074/jbc.M204654200. [DOI] [PubMed] [Google Scholar]
- 28.Hess A., Wijayanti N., Neuschafer-Rube A. P., Katz N., Kietzmann T., Immenschuh S. Phorbol ester-dependent activation of peroxiredoxin I gene expression via a protein kinase C, Ras, p38 mitogen-activated protein kinase signaling pathway. J. Biol. Chem. 2003;278:45419–45434. doi: 10.1074/jbc.M307871200. [DOI] [PubMed] [Google Scholar]