Abstract
Sequential activation of caspases is critical for the execution of apoptosis. Recent evidence suggests caspase 2 is a significant upstream caspase capable of initiating mitochondrial events, such as the release of cytochrome c. In particular, in vitro studies using recombinant proteins have shown that cleaved caspase 2 can induce mitochondrial outer membrane permeabilization directly or by cleaving the BH3-only protein BID (BH3 interacting domain death agonist). However, whether interchain cleavage or activation of procaspase 2 occurs prior to Apaf-1-mediated procaspase 9 activation under more natural conditions remains unresolved. In the present study, we show that Apaf-1-deficient Jurkat T-lymphocytes and mouse embryonic fibroblasts were highly resistant to DNA-damage-induced apoptosis and failed to cleave or activate any apoptotic procaspase, including caspase 2. Significantly, drug-induced cytochrome c release and loss of mitochondrial membrane potential were inhibited in cells lacking Apaf-1. By comparison, procaspase proteolysis and apoptosis were only delayed slightly in Apaf-1-deficient Jurkat cells upon treatment with anti-Fas antibody. Our data support a model in which Apaf-1 is necessary for the cleavage or activation of all procaspases and the promotion of mitochondrial apoptotic events induced by genotoxic drugs.
Keywords: caspase, cytochrome c, DNA damage, Fas, Jurkat cell, mitochondrial outer membrane permeabilization (MOMP)
Abbreviations: ΔΨ, mitochondrial membrane potential; AMC, 7-amino-4-methylcoumarin; AIF, apoptosis-inducing factor; Apaf, apoptosis protease-activating factor; BID, BH3 interacting domain death agonist; CARD, caspase recruitment domain; DIABLO, direct IAP (inhibitor of apoptosis protein)-binding protein with low pI; DTT, dithiothreitol; ETOP, etoposide; MEF, mouse embryonic fibroblast; MITOX, mitoxantrone; MOMP, mitochondrial outer membrane permeabilization; PI, propidium iodide; PIDD, p53-induced protein with a death domain; PS, phosphatidylserine; pSUP, pSUPER.neo; RAIDD, RIP (receptor-interacting protein)-associated ICH-1/CED (Ice and ced-3 homologue-1/cell-death determining-3)-3-homologous protein with a death domain; RNAi, RNA interference; siRNA, small interfering RNA; Smac, second mitochondria-derived activator of caspase; topo2, topoisomerase II
INTRODUCTION
Genotoxic chemotherapeutic drugs, including topo2 (topoisomerase II) inhibitors, trigger a series of intracellular events characteristic of apoptosis. Normally, topo2 alters the supercoiling of DNA by forming double-stranded cleavage complexes through which an intact helix can pass [1,2]. Topo2 inhibitors, e.g. ETOP (etoposide) and MITOX (mitoxantrone), do not inhibit entirely the activity of topo2. Instead, they selectively exploit the catalytic activity of this enzyme by increasing the number and duration of DNA cleavage complexes, resulting ultimately in permanent double-stranded breaks that are lethal to the cell [2,3].
Within the current paradigm of chemotherapy-induced apoptosis, mitochondria are key participants. In particular cytochrome c is released, which together with Apaf-1 (apoptosis protease-activating factor 1) and dATP lead to the recruitment and activation of initiator procaspase 9 within the apoptosome complex [4]. Subsequently, caspase 9 activates effector procaspases 3 and 7, which, in turn, cleave various protein substrates leading to morphological and biochemical features of apoptosis. Although there is little doubt about the importance of caspase 9 during DNA-damage-induced apoptosis, whether activation of procaspase 9 within the apoptosome complex represents the apical caspase event has been questioned recently. In particular, several lines of evidence suggest that activation of procaspase 2 may occur upstream of cytochrome c release and procaspase 9 activation [5–7]. Other studies have shown that fully processed or cleaved purified recombinant caspase 2 protein has an ability to induce MOMP (mitochondrial outer membrane permeabilization) [8–10], an effect that may require BID (BH3 interacting domain death agonist) in certain settings [9,11,12]. Additionally, previous findings have demonstrated that the activation of procaspase 2 occurs in a large caspase-activating complex [13], termed the PIDDosome, which includes RAIDD (RIP-associated ICH-1/CED-3-homologous protein with a death domain) and PIDD (p53-induced protein with a death domain) [14]. However, unlike initiator caspases 8, 9 and 10, caspase 2 cannot cleave effector caspases directly [9,15]. Further, procaspase 2 was shown to be a substrate for caspase 3 in vitro [16,17], although it is currently unknown whether caspase 3-mediated cleavage yields catalytically active caspase 2. Thus, caspase 2, which shares the highest degree of sequence similarity of any mammalian caspase with the Caenorhabditis elegans cell death protease CED-3 [18], would seem to retain features of both initiator and effector caspases.
The aim of the present study was to determine whether procaspase 2 is cleaved and/or exhibits activity as an upstream initiator or downstream effector caspase during DNA-damage-induced apoptosis. We used an approach to silence APAF1 and thereby abolish apoptosome-mediated procaspase 9 activation in Jurkat T-lymphocytes. The results indicated that Jurkat cells and MEFs (mouse embryonic fibroblasts) lacking APAF1 expression were resistant to ETOP- or MITOX-induced apoptosis, failing to undergo MOMP or activate/cleave any initiator or effector procaspase. In contrast, Apaf-1-deficient Jurkat cells were able to cleave procaspases, including caspase 2, and underwent apoptosis when incubated with anti-Fas antibody (CH11). Combined, our findings indicate that Apaf-1 is essential for all procaspase activation and the promotion of mitochondrial apoptotic events in response to DNA damage.
EXPERIMENTAL
Cell culture
Wild-type Jurkat T-lymphocytes (clone E6.1) were cultured in RPMI 1640 complete medium supplemented with 10% (v/v) heat-inactivated fetal bovine serum, 2% (w/v) glutamine, 100 units/ml penicillin and 100 μg/ml streptomycin at 37 °C in a humidified 5% CO2 incubator. For transfected Jurkat cells, 1 mg/ml Geneticin (Invitrogen) was substituted for penicillin and streptomycin. Wild-type and APAF1−/− MEFs [19] were cultured in Dulbecco's modified Eagle's medium supplemented with 10% (v/v) heat-inactivated fetal bovine serum, 2% (w/v) glutamine, 100 units/ml penicillin and 100 μg/ml streptomycin at 37 °C in a humidified 5% CO2 incubator. Cells were maintained in an exponential growth phase for all experiments. Apoptosis was induced with ETOP (10–50 μM) (Sigma–Aldrich), MITOX (100 nM) (Sigma–Aldrich), or anti-Fas antibody (50 ng/ml) (clone CH11, Upstate Biotechnology, Charlottesville, VA, U.S.A.).
Subcellular fractionation
Cells (106) were washed in PBS, resuspended in 50 μl of buffer (140 mM mannitol, 46 mM sucrose, 50 mM KCl, 1 mM KH2PO4, 5 mM MgCl2, 1 mM EGTA and 5 mM Tris/HCl, pH 7.4) supplemented with a mixture of protease inhibitors (Complete™ Mini-EDTA Free; Roche) and permeabilized with 3–10 μg of digitonin on ice for 10 min. Plasma membrane permeabilization was monitored by Trypan Blue staining, and cell suspensions were centrifuged at 12000 g for 10 min at 4 °C. Supernatant and pellet fractions were subjected to Western blot analysis.
Western blotting
Pelleted cells (5×106) were resuspended and lysed in 200 μl of ice-cold lysis buffer (10 mM Tris/HCl, pH 7.4, 10 mM NaCl, 3 mM MgCl2, 1 mM EDTA, 0.1% Nonidet P40) supplemented with a mixture of protease inhibitors (Complete™ Mini EDTA-Free). Protein concentrations were determined using the BCA (bicinchoninic acid) assay (Pierce) and equal amounts were mixed with Laemmli's loading buffer [62.5 mM Tris/HCl, pH 6.8, 2% (w/v) SDS, 25% (v/v) glycerol, 0.01% Bromophenol Blue and 5% (v/v) 2-mercaptoethanol]. Western blot analysis was carried out as described previously [7]. The antibodies used were rabbit anti-AIF (apoptosis-inducing factor; Cell Signaling Technology), mouse anti-Apaf-1 (clone 94408, R&D Systems, Minneapolis, MN, U.S.A.), rabbit anti-BID (Cell Signaling), mouse anti-caspase 2 (clone 35; BD Pharmingen), rat anti-caspase 2 (clone 11B46; Axxora, San Diego, CA, U.S.A.), rabbit anti-caspase 3 (clone 8G10; Cell Signaling), rabbit anti-caspase 6 (Cell Signaling), rabbit anti-caspase 7 (Cell Signaling), mouse anti-caspase 8 (clone 1C12; Cell Signaling), rabbit anti-caspase 9 (Cell Signaling), mouse-specific rabbit anti-caspase 9 (Cell Signaling), mouse-specific rabbit anti-cleaved caspase 9 (Asp353) (Cell Signaling), mouse anti-cytochrome c (clone 7H8.2C12; BD Pharmingen), rabbit anti-G3PDH (Trevigen, Gaithersburg, MD, U.S.A.) and mouse anti-Smac (second mitochondria-derived activator of caspase)/DIABLO [direct IAP (inhibitor of apoptosis protein)-binding protein with low pI; Cell Signaling].
Flow cytometry for cell death and ΔΨ (mitochondrial membrane potential) measurements
PS (phosphatidylserine) exposure on the outer leaflet of the plasma membrane was detected using the Annexin V–FITC Apoptosis Detection Kit II (BD PharMingen) according to the manufacturer's instructions. In brief, approx. 5×105 cells were pelleted following drug or anti-Fas treatment and washed in PBS. Next, the cells were resuspended in 100 μl of binding buffer containing annexin V–FITC and PI (propidium iodide). Prior to flow cytometric analysis, 400 μl of binding buffer (10 mM Hepes, pH 7.4, 140 mM NaCl and 2.5 mM CaCl2) was added to the cells. For ΔΨ determination, the MitoProbe DiIC1(5) Kit (Invitrogen, Molecular Probes, Carlsbad, CA, U.S.A.) was used. Briefly, 106 cells were pelleted following drug treatment and resuspended in 1 ml of warm PBS. Next, 5 μl of 10 μM DiIC1(5) was added to the cells and incubated in a humidified 5% CO2 incubator at 37 °C for 15 min. Cells were pelleted, resuspended in 500 μl of PBS and analysed by flow cytometry.
RNAi (RNA interference)
The vector-based pSUP (pSUPER.neo) RNAi system (OligoEngine, Seattle, WA, U.S.A.) was used to suppress the APAF1 gene (RefSeq Accession AF013263) expression. The gene-specific targeting insert specifies a 19-nucleotide sequence corresponding to nucleotides 875–893 (5′-AGGCAGATTTGCCAGAACA-3′) downstream of the transcription start site, which is separated by a 9-nucleotide non-complementary spacer (5′-TTCAAGAGA-3′) from the reverse complement of the same 19-nucleotide sequence. The sequence was ligated into the BglII and XhoI sites of the pSUP vector, which was subsequently transformed into TOP10 competent Escherichia coli cells (Invitrogen) according to the manufacturer's instructions. Several clones were obtained, and the correct insert was verified by sequence analysis. Wild-type Jurkat T-lymphocytes (107) were transfected with 20 μg of plasmid DNA (pSUP or pSUP-APAF1) by electroporation using a Bio-Rad Gene Pulser Xcell system (0.4-cm cuvette, 300 V and 950 μF). Cells were allowed to recover in RPMI 1640 complete growth medium for 48 h at 37 °C in a humidified 5% CO2 incubator. Selection of transfected cells was performed in the presence of 1 mg/ml Geneticin for several weeks, at which time serial dilutions were performed to obtain single-cell clones of APAF1-silenced cells. Clones 4 and 6 were used for the present study.
Measurement of caspase activity
Cells (105) were pelleted and washed once with ice-cold PBS. For DEVD-AMC (Asp-Glu-Val-Asp-7-amino-4-methylcoumarin) cleavage, cells were resuspended in 25 μl of PBS, added to a microtitre plate, and combined with substrate dissolved in a standard reaction buffer [100 mM Hepes, pH 7.25, 10% (w/v) sucrose, 10 mM DTT (dithiothreitol) and 0.1% CHAPS]. For VDVAD-AMC (Val-Asp-Val-Ala-Asp-AMC) or LEHD-AMC (Lys-Glu-His-Asp-AMC) cleavage, cells were resuspended in 25 μl of PBS, added to a microtitre plate, and combined with substrate dissolved in a standard reaction buffer [100 mM Mes, pH 6.5, 10% (w/v) polyethylene glycol, 10 mM DTT and 0.1% CHAPS]. Cleavage of the fluorogenic peptide substrates was monitored by AMC production in a FLx800 Multi-detection Microplate Reader (BioTek Instruments, Winooski, VT, U.S.A.) using 355 nm excitation and 460 nm emission wavelengths.
Microscopy
Cells were viewed using a Nikon TE2000S inverted microscope and a 40× objective. Images were acquired using a CoolSNAP ES camera (Roper Scientific, Ottobrunn, Germany) and MetaMorph software (Molecular Devices, Union City, CA, U.S.A.), and saved as TIFF files.
RESULTS AND DISCUSSION
Drug-induced caspase activation and apoptosis in wild-type Jurkat cells
As mentioned, a role for caspase 2 as an initiator caspase has been suggested for mitochondria-mediated apoptosis induced by genotoxic stress [5,7,14,20,21]. In addition, in vitro studies using recombinant proteins have demonstrated that cleaved caspase 2 can cause MOMP directly [8–10] or by cleaving the BH3-only protein BID [9,11,12], which moves to mitochondria and facilitates the release of cytochrome c and Smac/DIABLO. However, whether interchain cleavage and activation of procaspase 2 occur early or late within the caspase cascade following DNA damage in intact cells remains unclear and was a primary focus of the current study.
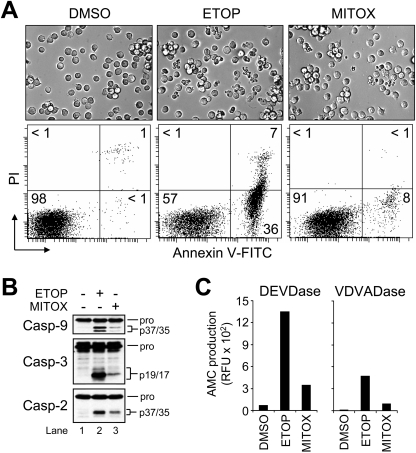
Jurkat T-lymphocytes (clone E6.1) were used, as these cells are a well-characterized and frequently used model for investigating mechanisms of mitochondria- and receptor-mediated apoptosis. As shown in Figure 1(A), cell shrinkage and plasma membrane blebbing were induced when wild-type Jurkat T-cells were treated with a clinically relevant concentration of ETOP (10 μM) or MITOX (100 nM) for 6 h. Likewise, drug-treated cells externalized PS on the plasma membrane (ETOP, approx. 43%; MITOX, approx. 10%) as measured by annexin V–FITC fluorescence (Figure 1A). Western blot analysis of cell lysates obtained at 6 h post-drug treatment revealed that ETOP induced the processing of procaspases 9, 3 and 2 more than MITOX (Figure 1B, compare lane 2 with lane 3). Similarly, caspase 3-like (DEVDase) and caspase 2-like (VDVADase) activities were much more robust in cells that had been treated with ETOP than MITOX (Figure 1C). Although procaspase 9 was cleaved (Figure 1B, lanes 2 and 3), only a slight increase in caspase 9-like (LEHDase) activity was observed in cells treated with either drug (results not shown). This was somewhat expected, however, as caspase 9 was shown previously to exhibit little activity on synthetic substrates [22].
Figure 1. Topo2 inhibitors induce procaspase activation and apoptosis in wild-type Jurkat cells.
(A) Wild-type Jurkat cells (106/ml) were cultured with DMSO, 10 μM ETOP or 100 nM MITOX for 6 h and processed for light microscopy (40× objective) or the quantification of cell death by flow cytometric analysis of annexin V–FITC and PI staining as described in the Experimental section. Quadrants are defined as: live (lower left), early apoptotic (lower right), late apoptotic (upper right) and necrotic (upper left). Numbers refer to the percentage of cells in each quadrant. (B) and (C) Duplicate aliquots of cells were harvested and lysed for Western blotting or caspase activity measurements as described in the Experimental section. Enzyme activity was monitored by the production of AMC.
Stable knockdown of APAF1 prevents drug-induced apoptosis in Jurkat cells
Activation of multiple caspases is a general phenomenon of cells undergoing apoptosis. Recent work has shown that fully cleaved caspase 2 can induce cytochrome c release directly or by cleaving the BH3-only protein BID. This has fuelled speculation that caspase 2 resides upstream of caspase 9 within the mitochondrial apoptotic pathway. An unresolved issue, however, is whether interchain cleavage or activation of procaspase 2 precedes mitochondrial apoptotic events in intact cells. To investigate this matter, we used a vector-based system that directs the synthesis of siRNAs (small interfering RNAs) to produce stable silencing of APAF1 in Jurkat cells. We reasoned that such cells would allow us to determine unambiguously whether interchain cleavage of procaspase 2 takes place upstream or downstream of apoptosome-mediated procaspase 9 activation. In other words, if it were the case that procaspase 2 is activated and/or cleaved as a legitimate initiator caspase during DNA-damage-induced apoptosis [5,14], then its activation and cleavage should proceed unimpaired in cells deficient of Apaf-1.
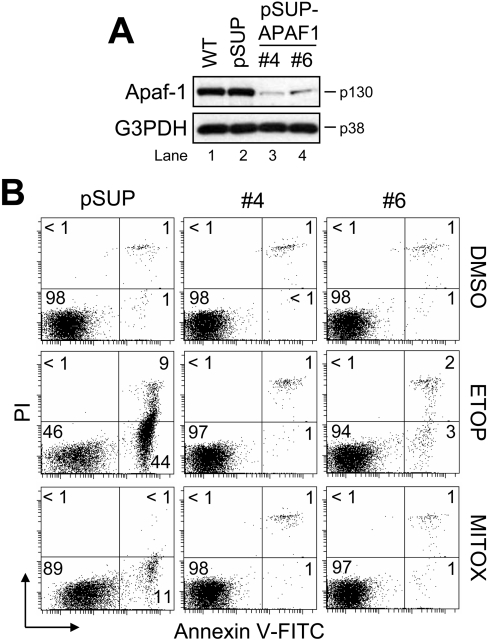
For stable siRNA-mediated silencing of APAF1, we used pSUP [23]. The specific APAF1-targeting insert specifies a 19-nucleotide sequence corresponding to nucleotides 875–893 (5′-AGGCAGATTTGCCAGAACA-3′) downstream of the transcription start site. Wild-type Jurkat cells were transfected with either the control vector (pSUP) or pSUP-APAF1 and selected with Geneticin. Subsequently, pSUP-APAF1 cells were serially diluted to obtain individual APAF1 knockdown clones. Two such clones (4 and 6) were used for the present study, and the suppression of Apaf-1 was confirmed by Western blot analysis (Figure 2A, compare lanes 1 and 2 with lanes 3 and 4).
Figure 2. Apaf-1-deficient Jurkat cells are resistant to drug-induced apoptosis.
(A) Wild-type (WT), control vector-transfected (pSUP), or single-cell APAF1 knockdown (pSUP-APAF1) Jurkat clones were harvested and lysed for Western blotting. G3PDH was used as a loading control. (B) Cells (106/ml) were cultured with DMSO, 10 μM ETOP or 100 nM MITOX for 6 h and processed for cell death determination by flow cytometric analysis of annexin V–FITC and PI staining. Quadrants are defined as: live (lower left), early apoptotic (lower right), late apoptotic (upper right) and necrotic (upper left). Numbers refer to the percentage of cells in each quadrant.
To test whether Apaf-1 was required for drug-induced apoptosis in Jurkat cells, we treated the different clones with 10 μM ETOP or 100 nM MITOX for 6 h and evaluated them for differences in PS exposure. As expected, pSUP cells readily underwent apoptosis (cf. Figure 1A) in response to ETOP (approx. 53%) and MITOX (approx. 12%) (Figure 2B). Strikingly, clone 4 was totally resistant to apoptosis induced by either drug, whereas clone 6, which retains slightly more Apaf-1 than clone 4 (Figure 2A, compare lane 4 with lane 3), displayed relatively little apoptosis (approx. 5%) in response to ETOP. Taken together, these data indicate that Apaf-1 is a prerequisite for drug-induced apoptosis, as suggested previously [24].
Apaf-1 is essential for drug-induced procaspase 2 activation in Jurkat cells
Previous evidence collected largely using gel filtration approaches and/or immortalized cell lines expressing endogenous PIDD, including Jurkat cells [14], suggested that procaspase 2 activation occurs in an Apaf-1-independent, apoptosome-like protein complex [13] called the PIDDosome [14], which, in addition to procaspase 2, includes RAIDD and PIDD. Other findings have demonstrated that cleaved caspase 2 can induce MOMP in vitro, an effect that seems to require processing of the zymogen to its long and short subunits [8–12] but may be independent of any associated proteolytic activity [8–10]. However, information on the ordering of procaspase 2 cleavage relative to established mitochondrial apoptotic events and the activation of other procaspases under more natural conditions is currently lacking.
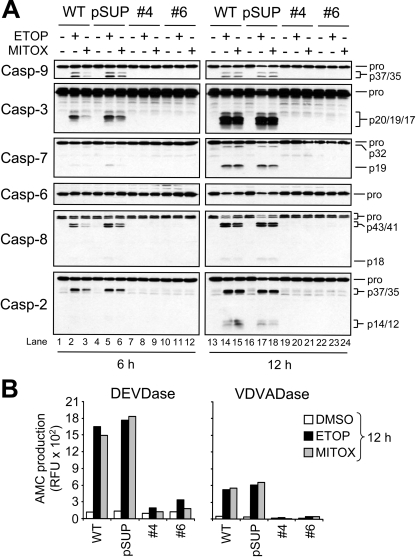
Treatment of WT or pSUP Jurkat cells with 10 μM ETOP or 100 nM MITOX for 6 h led to the cleavage of procaspases 9, 3, 7, 8 and 2 (Figure 3A, lanes 1–6). In contrast, neither ETOP nor MITOX was able to induce proteolysis of any procaspase, including caspase 2, in the APAF1 knockdown clones (Figure 3A, lanes 7–12). However, because accumulating evidence indicates dimerization, and not proteolytic cleavage, of initiator procaspases is required for their activation, we were interested to determine whether procaspase 2 cleavage would occur at a later time-point [13,14,25–28]. We reasoned that even if procaspase 2 had been activated by dimerization in a manner resembling procaspases 8 and 9 [25–28], with time cleavage of procaspase 2 would occur as the dimers self-processed. At the same time, we were interested to see whether procaspase 8 would be cleaved at either time-point, as it was reported that ETOP can activate this protease by causing an up-regulation of Fas-ligand, which, in turn, can interact with the Fas receptor [29–31]. At 12 h post-drug treatment, more extensive cleavage of caspases 3, 7, 8 and 2 was observed in wild-type and pSUP cells, as well as the partial disappearance of the pro-form of caspase 6 (Figure 3A, lanes 13–18). Remarkably, neither procaspase 2 nor any other procaspase had undergone detectable proteolytic processing by 12 h in the Apaf-1-deficient Jurkat clones (Figure 3A, lanes 19–24), which corresponds to a time-point where approx. 85% of the wild-type and pSUP cells were undergoing apoptosis (results not shown). Furthermore, caspase activity measurements revealed that drug treatment for 6 h (results not shown) or 12 h led to little or no increase in caspase 3-like (DEVDase) and caspase 2-like (VDVADase) activity in the APAF1 knockdown cells (Figure 3B). Combined, these findings suggest that procaspase 2 proteolysis and elevations in caspase 2-like activity following genotoxic stress occur largely, if not exclusively, downstream of Apaf-1-mediated procaspase 9 activation in Jurkat cells.
Figure 3. Drug-induced activation of procaspases does not occur in APAF1-silenced Jurkat cells.
(A) Wild-type (WT), control-vector-transfected (pSUP) and Apaf-1-deficient (clones 4 and 6) cells (106/ml) were cultured with or without 10 μM ETOP or 100 nM MITOX for 6 and 12 h, harvested and lysed for Western blotting. (B) Duplicate aliquots of cells at the 12 h time-point were harvested for caspase activity determination. Enzyme activity was monitored by the production of AMC. RFU, relative fluorescence units.
ETOP-induced apoptosis and procaspase activation are inhibited in APAF1−/− MEFs
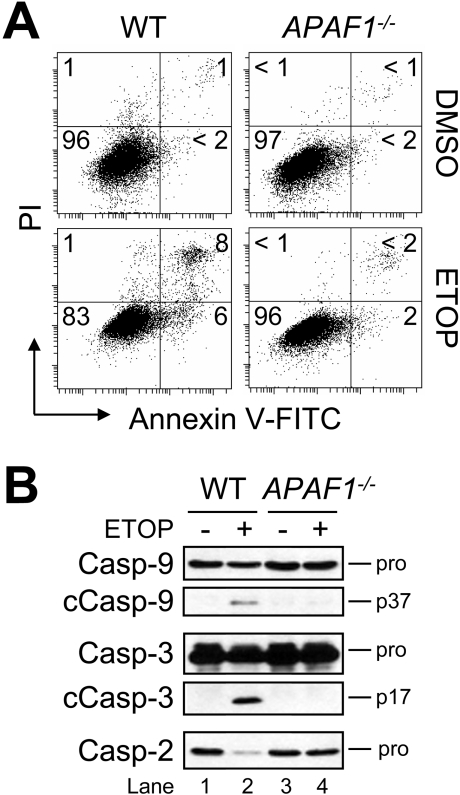
Next, to determine whether these findings were unique to Jurkat cells or had broader significance, we treated wild-type and APAF1−/− MEFs with 50 μM ETOP for 24 h and assessed differences in PS exposure. As shown in Figure 4(A), the results indicated that fibroblasts lacking APAF1 were resistant to drug treatment, whereas wild-type MEFs exhibited approx. 14% apoptosis. Further, despite being far less sensitive than Jurkat cells to ETOP-induced apoptosis (cf. Figures 1 and 2), wild-type MEFs, nevertheless, displayed cleavage of procaspases 9, 3 and 2 (Figure 4B, lanes 1 and 2), whereas drug-treated APAF1-deficient cells did not (Figure 4B, lanes 3 and 4). Taken together, these data provide additional support for a model of drug-induced apoptosis that places the proteolysis of all apoptotic procaspases, including caspase 2, downstream of Apaf-1.
Figure 4. Suppression of drug-induced procaspase activation and apoptosis in APAF1−/− MEFs.
(A) Wild-type (WT) and APAF1−/− MEFs were cultured with DMSO or 50 μM ETOP for 24 h and processed for cell death determination by flow cytometric analysis of annexin V–FITC and PI staining. Quadrants are defined as: live (lower left), early apoptotic (lower right), late apoptotic (upper right) and necrotic (upper left). Numbers refer to the percentage of cells in each quadrant. (B) Duplicate aliquots of cells were harvested and lysed for Western blotting. cCasp, cleaved caspase.
Drug-induced mitochondrial events of apoptosis are impaired in Apaf-1-deficient cells
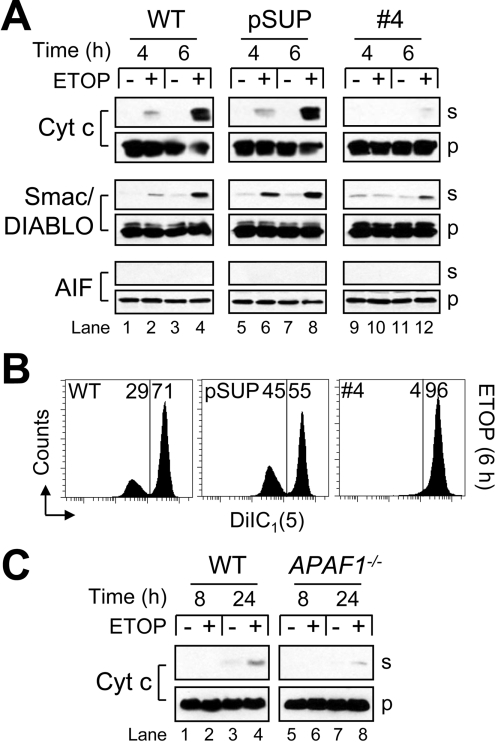
Some evidence in the literature suggests that mitochondria may act to amplify, rather than initiate, caspase activity in response to γ-radiation [32]. Because procaspase 2 was not processed in cells lacking Apaf-1 (Figures 3 and 4), and because previous results have shown that fully cleaved caspase 2 can induce MOMP directly [8–10] or by cleaving BID to tBID (truncated BID) [9,11,12], we next investigated whether cytochrome c release and other features of MOMP were altered in Apaf-1-deficient cells. When wild-type or pSUP cells were treated with 10 μM ETOP, the release of cytochrome c and Smac/DIABLO was observed after 4 h and had increased dramatically by 6 h (Figure 5A, lanes 1–8). Strikingly, APAF1 knockdown Jurkat cells released far less of both proteins after 4 and 6 h (Figure 5A, lanes 9–12). At no point was AIF found to be present in the cytosol of any ETOP-treated cells (Figure 5A). Furthermore, a partial loss of ΔΨ was observed in drug-treated wild-type and pSUP, but not Apaf-1-deficient, Jurkat cells at 6 h (Figure 5B). This finding would appear to be in agreement with earlier reports indicating that the loss of ΔΨ during apoptosis depends upon the activation of caspases and the cleavage of the respiratory complex I subunit NDUFS1 [NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75 kDa (NADH-coenzyme Q reductase)] [33,34]. However, another possible explanation for the loss of ΔΨ only occurring in wild-type and pSUP cells is that, because these cells released far more cytochrome c into the cytosol than APAF1 knockdown cells (Figure 5A), the electron transport chain was inhibited in a subpopulation of wild-type and pSUP cells, which, in turn, led to a suppression of respiration-dependent ΔΨ (Figure 5B).
Figure 5. Impairment of mitochondrial apoptotic events in Apaf-1deficient cells.
(A) Wild-type (WT), control-vector-transfected (pSUP) and Apaf-1-deficient cells (106/ml) were cultured in the presence or absence of 10 μM ETOP for 4 or 6 h and harvested for subcellular fractionation as described in the Experimental section. Supernatant (s) and pellet (p) cell fractions were analysed by Western blotting. (B) Duplicate aliquots of cells at 6 h were processed for ΔΨ determination as described in the Experimental section. Reduced DiIC1(5) fluorescence is indicative of a loss of ΔΨ, and numbers refer to the percentage of cells in each region. (C) WT and APAF1−/− MEFs were cultured in the presence or absence of 50 μM ETOP for 8 or 24 h and harvested for subcellular fractionation. Supernatant (s) and pellet (p) cell fractions were analysed by Western blotting.
We also evaluated ETOP-treated wild-type and APAF1−/− MEFs for differences in cytochrome c release. As shown in Figure 5(C), no release had occurred in either cell type at 8 h, whereas the wild-type cells had released noticeably more cytochrome c than APAF1−/− fibroblasts at 24 h. The reason this difference was not more pronounced probably reflects the overall reduced sensitivity of MEFs to ETOP-induced apoptosis as compared with Jurkat cells. Overall, these data indicate that events residing downstream of Apaf-1 are required to promote cytochrome c release in drug-treated cells, suggesting that initial cytochrome c release may not irreversibly commit a cell to die, as has been reported previously [35,36].
Apaf-1 is not required for receptor-mediated procaspase processing and apoptosis in Jurkat cells
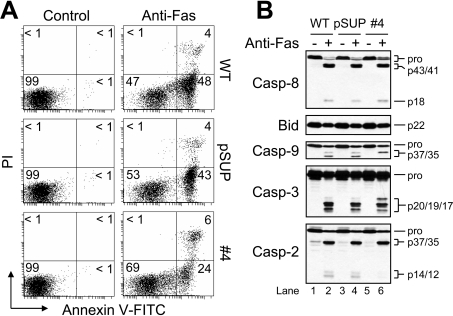
Because nearly all drug-induced proteolysis of procaspases and apoptosis were blocked in the APAF1 knockdown Jurkat clones (Figures 2 and 3), and because Jurkat cells are considered to be of type II origin [37,38], we were interested to test these cells’ susceptibility to receptor-mediated apoptosis. As shown in Figure 6(A), wild-type, pSUP and clone 4 cells underwent approx. 52%, approx. 47% and approx. 30% apoptosis respectively, when incubated in the presence of anti-Fas (50 ng/ml) for 6 h. Additionally, anti-Fas induced cleavage of the BH3-only protein BID and procaspases 8, 3 and 2 in wild-type, pSUP and Apaf-1-deficient cells (Figure 6B). Procaspase 9 was also cleaved, although yielding two bands (p37/35) in wild-type and pSUP cells, and only one band (p37) in APAF1 knockdown cells (Figure 6B, lanes 2, 4 and 6). The probable explanation for this is that apoptosome-mediated activation and cleavage of procaspase 9, which requires Apaf-1, occurs after Asp315 and generates a 35 kDa fragment, whereas caspase 3-mediated proteolysis of procaspase 9 occurs after Asp330 and produces a 37 kDa fragment [39]. The pattern of procaspase 3 processing also differed among wild-type, pSUP and Apaf-1-deficient cells in that the 20-kDa active intermediate fragment [40] was observed exclusively in cells lacking Apaf-1 (Figure 6B). A possible explanation for this finding is that, because apoptosome-mediated caspase 9 activation does not occur in these cells, cleavage of procaspase 3 occurs less efficiently as only caspase 8 contributes to the effect. By comparison, cleavage of procaspase 3 would be expected to involve both caspase 8 and caspase 9 in wild-type and pSUP cells upon treatment with anti-Fas. Taken together, Apaf-1-deficient cells exhibited less apoptosis and less cleavage of caspases as compared with wild-type and pSUP cells in response to anti-Fas. However, the extent of protection was small, indicating that Apaf-1 is dispensable for procaspase processing and apoptosis induced by anti-Fas antibody relative to ETOP.
Figure 6. Apaf-1 is not necessary for receptor-mediated apoptosis of Jurkat cells.
(A) Wild-type (WT), control-vector-transfected (pSUP) and Apaf-1-deficient cells (106/ml) were cultured with agonistic anti-Fas antibody (50 ng/ml) for 6 h and processed for cell death determination by flow cytometry. Quadrants are defined as: live (lower left), early apoptotic (lower right), late apoptotic (upper right) and necrotic (upper left). Numbers refer to the percentage of cells in each quadrant. (B) Duplicate aliquots of cells were harvested and lysed for Western blotting.
Concluding remarks
Part of what makes caspase 2 enigmatic is that, although it is a CARD (caspase recruitment domain)-containing caspase (similar to caspase 9), its substrate specificity is closer to that of non-CARD-containing effector caspases, such as caspases 3 and 7 [18,41,42]. Further, caspase 2 appears to be a putative initiator caspase that cannot cleave and activate an effector procaspase directly, and proteolysis of procaspase 2 is thought to occur, at least in part, by caspase 3-mediated cleavage [16,17,43]. However, it remains unknown whether procaspase 2 is in fact activated upon cleavage by caspase 3. More recently, it was reported that activation of procaspase 2 occurs in a large protein complex termed the PIDDosome, which also contains the putative adaptor protein RAIDD [14]. In that study, the authors demonstrated that PIDDosome formation could occur in Jurkat cells [14], despite the fact that Jurkat cells are heterozygous for a mutation in p53 that introduces a premature stop codon at Arg196 [44]. Additional evidence supporting a PIDDosome-based mechanism for procaspase 2 activation indicated that apoptosis induced by PIDD overexpression is associated with an early activation of procaspase 2 [45]. However, it should be noted that a different study found that procaspase 2 is recruited to a large activating complex that is sufficient for its activation, but does not include either RAIDD or Apaf-1 [13]. This finding would seem to rule out the possibility that the requirement of Apaf-1 for interchain cleavage of procaspase 2 in our system is due to a direct interaction between these proteins [46]. A more recent study presented complementary findings to those described here, where the authors used a cell- permeable, biotinylated pan-caspase inhibitor (bVAD-fmk) to show that procaspase 9 and 8, but not procaspase 2, are the apical caspases activated during ETOP-induced apoptosis [46]. In light of these published observations and our current findings, it could be that the ability of fully processed caspase 2 to induce MOMP [8–12] is more important as part of an amplification mechanism than as a true initiating event of mitochondria-mediated apoptosis.
Interestingly, during the completion of our work, Lakhani and colleagues published data complementary to our findings [47]. In particular, they reported that caspases 3 and 7 are key mediators of mitochondrial apoptotic events as evidenced by the fact that Bax translocation and cytochrome c release were delayed in CASP3−/−/CASP7−/− MEFs treated with UV radiation [47]. Interestingly, data contained in the paper's supporting online material illustrate that neither procaspase 2 nor 8 underwent cleavage in doubly deficient cells. On the whole, these observations raise the distinct possibility that certain caspase events, considered traditionally to be downstream of MOMP, are in fact needed to participate in a feed-forward amplification loop to drive the efficient release of cytochrome c. In other words, it seems plausible that, although the release of cytochrome c and Smac/DIABLO is an important initiating event of apoptosis, this may not always be a ‘lethal blow’ to a cell in situations where the initial death signal is not amplified by downstream caspases. Additional studies are ongoing in our laboratory to test this possibility, as well as to investigate other poorly defined aspects of genotoxic drug-induced apoptotic signalling.
Acknowledgments
We thank Pengli Bu and Shary N. Shelton (University of Kansas Medical Center) for valuable help in making the pSUP and pSUP-APAF1 cells, Dr Joyce Slusser (University of Kansas Medical Center) for skillful assistance with flow cytometry, Dr Cristina Pop and Dr Fiona Scott (Burnham Institute for Medical Research, La Jolla, CA, U.S.A.) for valuable discussions, and Dr Chunying Du (Stowers Institute for Medical Research, Kansas City, MO, U.S.A.) for kindly providing wild-type and APAF1−/− MEFs. This work was supported by NIH (National Institutes of Health) grants K22 ES011647 (to J. D. R.) and P20 RR016475 from the INBRE (IDeA Networks of Biomedical Research Excellence) programme of the NCRR (National Center for Research Resources). The Flow Cytometry Core is supported in part by NIH grant P20 RR016443 from the NCRR.
References
- 1.Wang J. C. Cellular roles of DNA topoisomerases: a molecular perspective. Nat. Rev. Mol. Cell Biol. 2002;3:430–440. doi: 10.1038/nrm831. [DOI] [PubMed] [Google Scholar]
- 2.Wilstermann A. M., Osheroff N. Stabilization of eukaryotic topoisomerase II–DNA cleavage complexes. Curr. Top. Med. Chem. 2003;3:321–338. doi: 10.2174/1568026033452519. [DOI] [PubMed] [Google Scholar]
- 3.Froelich-Ammon S. J., Osheroff N. Topoisomerase poisons: harnessing the dark side of enzyme mechanism. J. Biol. Chem. 1995;270:21429–21432. doi: 10.1074/jbc.270.37.21429. [DOI] [PubMed] [Google Scholar]
- 4.Li P., Nijhawan D., Budihardjo I., Srinivasula S. M., Ahmad M., Alnemri E. S., Wang X. Cytochrome c and dATP-dependent formation of Apaf-1/caspase-9 complex initiates an apoptotic protease cascade. Cell. 1997;91:479–489. doi: 10.1016/s0092-8674(00)80434-1. [DOI] [PubMed] [Google Scholar]
- 5.Lassus P., Opitz-Araya X., Lazebnik Y. Requirement for caspase-2 in stressinduced apoptosis before mitochondrial permeabilization. Science. 2002;297:1352–1354. doi: 10.1126/science.1074721. [DOI] [PubMed] [Google Scholar]
- 6.Paroni G., Henderson C., Schneider C., Brancolini C. Caspase-2 can trigger cytochrome c release and apoptosis from the nucleus. J. Biol. Chem. 2002;277:15147–15161. doi: 10.1074/jbc.M112338200. [DOI] [PubMed] [Google Scholar]
- 7.Robertson J. D., Enoksson M., Suomela M., Zhivotovsky B., Orrenius S. Caspase-2 acts upstream of mitochondria to promote cytochrome c release during etoposide-induced apoptosis. J. Biol. Chem. 2002;277:29803–29809. doi: 10.1074/jbc.M204185200. [DOI] [PubMed] [Google Scholar]
- 8.Enoksson M., Robertson J. D., Gogvadze V., Bu P., Kropotov A., Zhivotovsky B., Orrenius S. Caspase-2 permeabilizes the outer mitochondrial membrane and disrupts the binding of cytochrome c to anionic phospholipids. J. Biol. Chem. 2004;279:49575–49578. doi: 10.1074/jbc.C400374200. [DOI] [PubMed] [Google Scholar]
- 9.Guo Y., Srinivasula S. M., Druilhe A., Fernandes-Alnemri T., Alnemri E. S. Caspase-2 induces apoptosis by releasing proapoptotic proteins from mitochondria. J. Biol. Chem. 2002;277:13430–13437. doi: 10.1074/jbc.M108029200. [DOI] [PubMed] [Google Scholar]
- 10.Robertson J. D., Gogvadze V., Kropotov A., Vakifahmetoglu H., Zhivotovsky B., Orrenius S. Processed caspase-2 can induce mitochondria-mediated apoptosis independently of its enzymatic activity. EMBO Rep. 2004;5:643–648. doi: 10.1038/sj.embor.7400153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bonzon C., Bouchier-Hayes L., Pagliari L. J., Green D. R., Newmeyer D. D. Caspase-2-induced apoptosis requires bid cleavage: a physiological role for bid in heat shock-induced death. Mol. Biol. Cell. 2006;17:2150–2157. doi: 10.1091/mbc.E05-12-1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao Z., Shao Y., Jiang X. Essential roles of the Bcl-2 family of proteins in caspase-2-induced apoptosis. J. Biol. Chem. 2005;280:38271–38275. doi: 10.1074/jbc.M506488200. [DOI] [PubMed] [Google Scholar]
- 13.Read S. H., Baliga B. C., Ekert P. G., Vaux D. L., Kumar S. A novel Apaf-1-independent putative caspase-2 activation complex. J. Cell Biol. 2002;159:739–745. doi: 10.1083/jcb.200209004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tinel A., Tschopp J. The PIDDosome, a protein complex implicated in activation of caspase-2 in response to genotoxic stress. Science. 2004;304:843–846. doi: 10.1126/science.1095432. [DOI] [PubMed] [Google Scholar]
- 15.Baliga B. C., Read S. H., Kumar S. The biochemical mechanism of caspase-2 activation. Cell Death Differ. 2004;11:1234–1241. doi: 10.1038/sj.cdd.4401492. [DOI] [PubMed] [Google Scholar]
- 16.Li H., Bergeron L., Cryns V., Pasternack M. S., Zhu H., Shi L., Greenberg A., Yuan J. Activation of caspase-2 in apoptosis. J. Biol. Chem. 1997;272:21010–21017. doi: 10.1074/jbc.272.34.21010. [DOI] [PubMed] [Google Scholar]
- 17.Slee E. A., Harte M. T., Kluck R. M., Wolf B. B., Casiano C. A., Newmeyer D. D., Wang H. G., Reed J. C., Nicholson D. W., Alnemri E. S., et al. Ordering the cytochrome c-initiated caspase cascade: hierarchical activation of caspases-2, -3, -6, -7, -8, and -10 in a caspase-9-dependent manner. J. Cell Biol. 1999;144:281–292. doi: 10.1083/jcb.144.2.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lamkanfi M., Declercq W., Kalai M., Saelens X., Vandenabeele P. Alice in caspase land. A phylogenetic analysis of caspases from worm to man. Cell Death Differ. 2002;9:358–361. doi: 10.1038/sj.cdd.4400989. [DOI] [PubMed] [Google Scholar]
- 19.Honarpour N., Du C., Richardson J. A., Hammer R. E., Wang X., Herz J. Adult Apaf-1-deficient mice exhibit male infertility. Dev. Biol. 2000;218:248–258. doi: 10.1006/dbio.1999.9585. [DOI] [PubMed] [Google Scholar]
- 20.Seth R., Yang C., Kaushal V., Shah S. V., Kaushal G. P. p53-dependent caspase-2 activation in mitochondrial release of apoptosis-inducing factor and its role in renal tubular epithelial cell injury. J. Biol. Chem. 2005;280:31230–31239. doi: 10.1074/jbc.M503305200. [DOI] [PubMed] [Google Scholar]
- 21.Lin C. F., Chen C. L., Chang W. T., Jan M. S., Hsu L. J., Wu R. H., Tang M. J., Chang W. C., Lin Y. S. Sequential caspase-2 and caspase-8 activation upstream of mitochondria during ceramide- and etoposide-induced apoptosis. J. Biol. Chem. 2004;279:40755–40761. doi: 10.1074/jbc.M404726200. [DOI] [PubMed] [Google Scholar]
- 22.Ryan C. A., Stennicke H. R., Nava V. E., Burch J. B., Hardwick J. M., Salvesen G. S. Inhibitor specificity of recombinant and endogenous caspase-9. Biochem. J. 2002;366:595–601. doi: 10.1042/BJ20020863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brummelkamp T. R., Bernards R., Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- 24.Yoshida H., Kong Y. Y., Yoshida R., Elia A. J., Hakem A., Hakem R., Penninger J. M., Mak T. W. Apaf1 is required for mitochondrial pathways of apoptosis and brain development. Cell. 1998;94:739–750. doi: 10.1016/s0092-8674(00)81733-x. [DOI] [PubMed] [Google Scholar]
- 25.Pop C., Timmer J., Sperandio S., Salvesen G. S. The apoptosome activates caspase-9 by dimerization. Mol. Cell. 2006;22:269–275. doi: 10.1016/j.molcel.2006.03.009. [DOI] [PubMed] [Google Scholar]
- 26.Boatright K. M., Renatus M., Scott F. L., Sperandio S., Shin H., Pedersen I. M., Ricci J. E., Edris W. A., Sutherlin D. P., Green D. R., Salvesen G. S. A unified model for apical caspase activation. Mol. Cell. 2003;11:529–541. doi: 10.1016/s1097-2765(03)00051-0. [DOI] [PubMed] [Google Scholar]
- 27.Muzio M., Stockwell B. R., Stennicke H. R., Salvesen G. S., Dixit V. M. An induced proximity model for caspase-8 activation. J. Biol. Chem. 1998;273:2926–2930. doi: 10.1074/jbc.273.5.2926. [DOI] [PubMed] [Google Scholar]
- 28.Stennicke H. R., Deveraux Q. L., Humke E. W., Reed J. C., Dixit V. M., Salvesen G. S. Caspase-9 can be activated without proteolytic processing. J. Biol. Chem. 1999;274:8359–8362. doi: 10.1074/jbc.274.13.8359. [DOI] [PubMed] [Google Scholar]
- 29.Friesen C., Herr I., Krammer P. H., Debatin K. M. Involvement of the CD95 (APO-1/FAS) receptor/ligand system in drug-induced apoptosis in leukemia cells. Nat. Med. 1996;2:574–577. doi: 10.1038/nm0596-574. [DOI] [PubMed] [Google Scholar]
- 30.Kasibhatla S., Brunner T., Genestier L., Echeverri F., Mahboubi A., Green D. R. DNA damaging agents induce expression of Fas ligand and subsequent apoptosis in T lymphocytes via the activation of NF-κB and AP-1. Mol. Cell. 1998;1:543–551. doi: 10.1016/s1097-2765(00)80054-4. [DOI] [PubMed] [Google Scholar]
- 31.Petak I., Tillman D. M., Harwood F. G., Mihalik R., Houghton J. A. Fas-dependent and -independent mechanisms of cell death following DNA damage in human colon carcinoma cells. Cancer Res. 2000;60:2643–2650. [PubMed] [Google Scholar]
- 32.Marsden V. S., Ekert P. G., Van Delft M., Vaux D. L., Adams J. M., Strasser A. Bcl-2-regulated apoptosis and cytochrome c release can occur independently of both caspase-2 and caspase-9. J. Cell Biol. 2004;165:775–780. doi: 10.1083/jcb.200312030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ricci J. E., Gottlieb R. A., Green D. R. Caspase-mediated loss of mitochondrial function and generation of reactive oxygen species during apoptosis. J. Cell Biol. 2003;160:65–75. doi: 10.1083/jcb.200208089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ricci J. E., Munoz-Pinedo C., Fitzgerald P., Bailly-Maitre B., Perkins G. A., Yadava N., Scheffler I. E., Ellisman M. H., Green D. R. Disruption of mitochondrial function during apoptosis is mediated by caspase cleavage of the p75 subunit of complex I of the electron transport chain. Cell. 2004;117:773–786. doi: 10.1016/j.cell.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 35.Goldstein J. C., Waterhouse N. J., Juin P., Evan G. I., Green D. R. The coordinate release of cytochrome c during apoptosis is rapid, complete and kinetically invariant. Nat. Cell Biol. 2000;2:156–162. doi: 10.1038/35004029. [DOI] [PubMed] [Google Scholar]
- 36.Waterhouse N. J., Goldstein J. C., von Ahsen O., Schuler M., Newmeyer D. D., Green D. R. Cytochrome c maintains mitochondrial transmembrane potential and ATP generation after outer mitochondrial membrane permeabilization during the apoptotic process. J. Cell Biol. 2001;153:319–328. doi: 10.1083/jcb.153.2.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scaffidi C., Fulda S., Srinivasan A., Friesen C., Li F., Tomaselli K. J., Debatin K. M., Krammer P. H., Peter M. E. Two CD95 (APO-1/Fas) signaling pathways. EMBO J. 1998;17:1675–1687. doi: 10.1093/emboj/17.6.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scaffidi C., Schmitz I., Zha J., Korsmeyer S. J., Krammer P. H., Peter M. E. Differential modulation of apoptosis sensitivity in CD95 type I and type II cells. J. Biol. Chem. 1999;274:22532–22538. doi: 10.1074/jbc.274.32.22532. [DOI] [PubMed] [Google Scholar]
- 39.Srinivasula S. M., Ahmad M., Fernandes-Alnemri T., Alnemri E. S. Autoactivation of procaspase-9 by Apaf-1-mediated oligomerization. Mol. Cell. 1998;1:949–957. doi: 10.1016/s1097-2765(00)80095-7. [DOI] [PubMed] [Google Scholar]
- 40.Liu H., Chang D. W., Yang X. Interdimer processing and linearity of procaspase-3 activation: a unifying mechanism for the activation of initiator and effector caspases. J. Biol. Chem. 2005;280:11578–11582. doi: 10.1074/jbc.M414385200. [DOI] [PubMed] [Google Scholar]
- 41.Garcia-Calvo M., Peterson E. P., Rasper D. M., Vaillancourt J. P., Zamboni R., Nicholson D. W., Thornberry N. A. Purification and catalytic properties of human caspase family members. Cell Death Differ. 1999;6:362–369. doi: 10.1038/sj.cdd.4400497. [DOI] [PubMed] [Google Scholar]
- 42.Hofmann K., Bucher P., Tschopp J. The CARD domain: a new apoptotic signalling motif. Trends Biochem. Sci. 1997;22:155–156. doi: 10.1016/s0968-0004(97)01043-8. [DOI] [PubMed] [Google Scholar]
- 43.O'Reilly L. A., Ekert P., Harvey N., Marsden V., Cullen L., Vaux D. L., Hacker G., Magnusson C., Pakusch M., Cecconi F., et al. Caspase-2 is not required for thymocyte or neuronal apoptosis even though cleavage of caspase-2 is dependent on both Apaf-1 and caspase-9. Cell Death Differ. 2002;9:832–841. doi: 10.1038/sj.cdd.4401033. [DOI] [PubMed] [Google Scholar]
- 44.Cheng J., Haas M. Frequent mutations in the p53 tumor suppressor gene in human leukemia T-cell lines. Mol. Cell Biol. 1990;10:5502–5509. doi: 10.1128/mcb.10.10.5502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Berube C., Boucher L. M., Ma W., Wakeham A., Salmena L., Hakem R., Yeh W. C., Mak T. W., Benchimol S. Apoptosis caused by p53-induced protein with death domain (PIDD) depends on the death adapter protein RAIDD. Proc. Natl. Acad. Sci. U.S.A. 2005;102:14314–14320. doi: 10.1073/pnas.0506475102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tu S., McStay G. P., Boucher L. M., Mak T., Beere H. M., Green D. R. In situ trapping of activated initiator caspases reveals a role for caspase-2 in heat shock-induced apoptosis. Nat. Cell Biol. 2006;8:72–77. doi: 10.1038/ncb1340. [DOI] [PubMed] [Google Scholar]
- 47.Lakhani S. A., Masud A., Kuida K., Porter G. A., Jr., Booth C. J., Mehal W. Z., Inayat I., Flavell R. A. Caspases 3 and 7: key mediators of mitochondrial events of apoptosis. Science. 2006;311:847–851. doi: 10.1126/science.1115035. [DOI] [PMC free article] [PubMed] [Google Scholar]