Abstract
Previously, we found that bombesin receptor subtype 3 (BRS-3) significantly increased in an ozone-stressed airway hyperresponsiveness animal model and resulted in induced wound repair and protection from acute lung injury. In the present study, we determined molecular mechanisms of BRS-3 regulation in human BECs (bronchial epithelial cells) in response to ozone stress. Ten oligonucleotide probes corresponding to various regions of the BRS-3 promoter were used in EMSA (electrophoretic mobilityshift assays). Four were found to have an enhanced mobility shift with extracts from ozone-stressed cells. On the basis of the assay of mutated probes binding with extracts and antibody supershift, they were verified as MTF-1 (metal-regulatory-element-binding transcription factor-1), PPARα (peroxisome-proliferator-activated receptor α), AP-2α (activator protein 2α) and HSF-1 (heat-shock factor 1). Next, ChIP (chromatin immunoprecipitation) assay, site-directed mutagenesis technology and antisense oligonucleotide technology were used to observe these transcription factors associated with the BRS-3 promoter. Only AP-2α and PPARα increased ozone-inducible DNA binding on the BRS-3 promoter and BRS-3 expression. The time courses of AP-2α and PPARα activation, followed by BRS-3 expression, were also examined. It was shown that ozone-inducible BRS-3 expression and AP-2α- and PPARα-binding activity correlated over a 48 h period. The translocation of PPARα was observed by immunofluorescence assay, which showed that PPARα nuclear translocation increased after ozone exposure. Our data suggest that AP-2α and PPARα may be especially involved in this ozone-inducible up-regulation mechanism of BRS-3 expression.
Keywords: activator protein 2α (AP-2α), airway hyperresponsiveness, bombesin receptor subtype-3 (BRS-3), human bronchial epithelial cell, ozone, peroxisome-proliferator-activated receptor α (PPARα)
Abbreviations: AHR, airway hyperresponsiveness; AP-2, activator protein 2; ASO, antisense oligonucleotide; BEC, bronchial epithelial cell; BLP, bombesin-like peptide; BRS-3, bombesin receptor subtype 3; ChIP, chromatin immunoprecipitation; DMEM, Dulbecco's modified Eagle's medium; EMSA, electrophoretic mobility-shift assay; FAM, 5-carboxyfluorescein; FBS, fetal bovine serum; FR, flanking region; β-gal, β-galactosidase; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; GRP, gastrin-releasing peptide; HLF, human lung fibroblast; HSF-1, heat-shock factor 1; MTF-1, metal-regulatory-element-binding transcription factor-1; NMB, neuromedin B; PPAR, peroxisome-proliferator-activated receptor; RXR, retinoid X receptor; TAMRA, 6-carboxytetramethylrhodamine
INTRODUCTION
Bombesin is a 14 amino acid peptide first isolated from frog skin in 1971 [1]. With the use of antibodies against amphibian bombesin, BLP (bombesin-like peptide) immunoreactivity was identified in mammalian brain, gut and lung [2]. The highest BLP levels were observed in human fetal lung, localized to the pulmonary neuroendocrine cells [3]. Two major pulmonary BLPs were subsequently determined to be GRP (gastrin-releasing peptide) and NMB (neuromedin B), which contribute to diverse biological functions in the central nervous system and peripheral tissues, including thermoregulation, satiety, control of circadian rhythm, stimulation of gastrointestinal hormone release and macrophage activation, which are important developmental effects and potent growth effects [4–7]. To date, three mammalian bombesin-like receptors have been cloned: GRP/bombesin-preferring receptor, NMB receptor and the orphan bombesin receptor subtype-3 (BRS-3). These receptors are seven-transmembrane-spanning, G-protein-coupled receptors primarily leading to the activation of multiple cell signalling pathways [8–10].
Unlike GRP and NMB receptors, which have a widespread distribution in the central nervous system and peripheral tissues, the distribution of BRS-3 is more limited, having been identified in secondary spermatocytes, pregnant uterus, certain brain regions, human lung tissues, adipose tissue and epidermal cancer cell lines [11–15]. Its expression level in the majority of normal tissues is extremely low, whereas in the developing lung [4,16] and certain lung carcinomas, it is high [17]. Although in a recent study, BRS-3-deficient mice were reported to develop obesity and impaired glucose metabolism [18], suggesting a role for BRS-3 in the regulation of energy metabolism, there is very little known about the biological function of BRS-3 activation. This is in great part because the natural ligand of BRS-3 has not yet been identified.
In contrast, there is accumulating evidence that BLPs and their receptors play an important role in pathological conditions of the lung, including chronic inflammatory lung disease, lung cancer, bronchopulmonary dysplasia and acute lung injury [19–21]. Furthermore, results from our previous experiments demonstrated that the expression of BRS-3 mRNA was significantly up-regulated in an ozone-stressed AHR (airway hyperresponsiveness) animal model and resulted in wound repair and protection from lung injury [22]. Thus these novel insights linked BRS-3 expression with mediating recovery from acute lung injury in the AHR model, an important mechanism that is believed to be significant in the pathophysiology of bronchial asthma.
We therefore felt prompted to gain further insight into the regulation of BRS-3 gene expression in human BECs (bronchial epithelial cells). Ten oligonucleotide probes covering all potential nuclear-factor-binding sites were designed according to the BRS-3 promoter and the nuclear factors involved in induced expression of BRS-3 in response to ozone were screened using EMSA (electrophoretic mobility-shift assay), which were verified by ChIP (chromatin immunoprecipitation). Further experiments determined that PPARα (peroxisome-proliferator-activated receptor α) and AP-2α (activator protein 2α) ASOs (antisense oligonucleotides) or site-directed mutagenesis of PPARα- and AP-2α-binding sites down-regulated ozone-induced BRS-3 overexpression. Our data suggest that the effects of BRS-3 on the recovery from acute lung injury in the AHR model are mediated, at least in part, through transcription factor PPARα and AP-2α recruitment.
MATERIALS AND METHODS
Chemicals
All chemicals were purchased from Sigma unless stated otherwise. Modifying enzymes were from New England Biolabs.
Cell culture
Human BECs (derived from normal humans) and HLFs (human lung fibroblasts) were incubated in DMEM (Dulbecco's modified Eagle's medium)/Ham's F12 (1:1) containing 100 IU/ml penicillin, 100 μg/ml streptomycin and 10% (v/v) heat-inactivated FBS (fetal bovine serum) at 37 °C in an atmosphere containing 5% CO2.
EMSA and supershift assay
After exposing cells to 1.5 p.p.m. ozone for 30 min under culture conditions, nuclear extracts were prepared on ice with ice-cold reagents. The oligonucleotide probes used in EMSAs were designed according to a computer-based search with the software Transcription Element Search System (http://www.cbil.upenn.edu/cgibin/tess/tess) encompassing putative binding sites in the BRS-3 promoter. Nuclear-extract-binding reactions were performed at 4 °C for 15 min using 10 μg of the extract and 1 μl of biotin-labelled oligonucleotide (TaKaRa) in 20 μl of binding buffer. Protein–oligonucleotide probe complexes were resolved using native 5% polyacrylamide gels and run in 0.5× TBE (Tris/borate/EDTA) buffer for 1 h, then transferred on to nitrocellulose membranes. Protein–oligonucleotide probe complexes were detected by Chemiluminescent Nucleic Acid Detection Module (Pierce). For competition experiments, unlabelled competitor oligonucleotides were pre-incubated at 100-fold excess with the labelled probe. The nuclear factors in the retarded bands were verified using the assay of mutated probes binding with extracts and antibody supershift analysis.
Chromatin immunoprecipitation
Cells were formaldehyde-cross-linked for 10 min as described previously [23]. For every 2 mg of protein extract processed for ChIP, 10 μg of mouse IgG bound to Protein A–Sepharose (GE Healthcare) was used to pre-clear for 1 h with rocking at 4 °C. The extracts were immunoprecipitated with 1 μg of specific antibodies or mouse IgG (Santa Cruz Biotechnology) by rocking overnight at 4 °C. Immunocomplexes were washed eight times as described in [23], and the protein was degraded in digestion buffer at 56 °C overnight and then incubated at 65 °C for 30 min. The DNA was phenol/chloroform-extracted, and ethanol-precipitated. The nuclear-factor-binding sites of BRS-3 were amplified by PCR with the primers 5′-ACTTTGGGAGGCTGAGG-3′ (forward; nucleotides − 970 to − 953) and 5′-TTCGGGTTCACTGGAC-3′ (reverse; nucleotides − 625 to − 609) (362 bp product) for MTF-1 (metal-regulatory-element-binding transcription factor-1) and PPARα sites of BRS-3; 5′-GGTCCCAAGATTCCC-3′ (forward; nucleotides − 320 to − 305) and 5′-ATCCGTAACCTGCACT-3′ (reverse; nucleotides − 74 to − 58) (263 bp product) for AP-2α and HSF-1 (heat-shock factor 1) sites of BRS-3.
ASO design
Four ASOs were designed according to mRNA sequences of MTF-1 (5′-GTCTGGACTGTGTTCCCCCAT-3′ against nucleotides 1–21), PPARα (5′-TAACGGGCTCTCTAGATCGCC-3′ against nucleotides 19–39), AP-2α (5′-CAGCTGGGGCAACCGTGCCGT-3′ against nucleotides 55–72) and HSF-1 (5′-GGGGCCCACGGGCAGATCCAT-3′ against nucleotides 1–21). The sequence of the nonsense oligonucleotide was 5′-TCTGGACTGTCCCCCATTTCT-3′ and all ASOs were synthesized (TaKaRa) as 21-base phosphorothioate oligonucleotides. BECs were transfected with an ASO and Lipofectin (Invitrogen) mixture for 4 h in serum-free DMEM, and then cultured in normal growth media for 40 h. The effects of the ASOs were measured by EMSA and Western blotting after ozone exposure for 30 min and further culture for 4 h.
Western blotting
Samples of 50 μg of whole-cell lysates were used per lane for Western blot analysis. Anti-AP-2α and anti-PPARα monoclonal antibodies (Santa Cruz Biotechnology) were diluted at 1:500. Horseradish-peroxidase-conjugated affinity-purified secondary antibody (Boster) was used at a 1:2000 dilution.
Real-time PCR
RNA was extracted from BECs using TRIzol® reagent and reverse transcription was performed with AMV (avian myelobastosis virus) reverse transcriptase (Qiagen). PCR was then carried out by using primers and Taqman probes, which were labelled at their 5′-end with the reporter dye FAM (5-carboxyfluorescein) and at their 3′-end with the quencher TAMRA (6-carboxytetramethylrhodamine) (TaKaRa): 5′-GGCTCAAAGGCAGCCTCACT-3′ (forward; nucleotides 152–171), 5′-AGTCTTCAGGATGGCATTGG-3′ (reverse; nucleotides 622–641) and 5′-FAM-TGCCAGTGGATGCAACTCAC-TAMRA-3′ for BRS-3, and 5′-CCACTCCTCCACCTTTGAC-3′, 5′-ACCCTGTTGCTGTAGCCA-3′ and 5′-FAM-TTGCCCTCAACGACCACTTGTC-TAMRA-3′ for GAPDH (glyceraldehyde-3-phosphate dehydrogenase). Briefly, 2 μl (out of 20 μl) of the reverse-transcribed reaction mixture was added to 50 μl of PCR mixture for 50 cycles. Each cycle consisted of 94 °C for 10 s, 53 °C for 30 s and 72 °C for 40 s using Taq polymerase. Negative controls consisted of an equal volume of water substituted for the volume of RNA in the reverse transcription reaction. Normalization of mRNA expression data was achieved by comparing the copy numbers of target mRNAs with that of human GAPDH mRNA for each run.
Site-directed mutagenesis
Genomic DNA was isolated from BECs and 1261 bp of the 5′-FR (flanking region) of BRS-3 was amplified by PCR with the primers 5′-GGGGTACCCCAATGATGATTTTCATTCTTAAAAGTTCTTT-3 (forward; nucleotides –1261 to − 1232; KpnI site is underlined) and 5′-CTAGCTCGTTCTTCTGAAGACTGTGTCTTTAATATTTC-3′ (reverse; nucleotides –1 to − 31; SacI site is underlined). The PCR product was digested with KpnI and SacI and ligated into the KpnI/SacI sites of plasmid pGL3-Basic (Promega) resulting in the human BRS-3 promoter–luciferase reporter pGL3/FR/luc. The authentic plasmid was verified by restriction enzyme mapping and direct DNA sequencing. Next, mutants of four transcription-factor-binding sites were generated in the luciferase reporter plasmid pGL3/FR/luc using mutated primers and muta-direct™ enzyme following the recommendations of the manufacturer (TaKaRa). Mutants were selected by mutazyme™ enzyme (TaKaRa) and the correct DNA sequence was verified by direct DNA sequencing. Cells were transfected using a mixture of Lipofectin, plasmid DNA and control plasmid pSV-β-Gal (Promega), following the manufacturer's instructions, for 4 h in serum-free DMEM, and then cultured in normal growth medium for 40 h. The cells were stressed by ozone for 30 min and cultured for an additional 4 h before harvesting for measurement of luciferase and β-gal (β-galactosidase) enzyme activities. Data are expressed as luciferase activity normalized to β-gal expression to standardize variations in transfection efficiency.
Immunofluorescence
The translocation of PPARα was assayed by immunofluorescence. Cells were fixed for 1 h with 4% (w/v) formaldehyde in PBS and permeabilized for 1 h with 4% (w/v) formaldehyde/0.2% (v/v) Tween 20 in PBS at room temperature (25 °C). Cells were briefly rehydrated with 0.2% (v/v) Tween 20 in PBS before blocking overnight in a solution containing 50% (v/v) FBS, 6% (w/v) dried skimmed milk powder, 3% (w/v) BSA, 0.2% (v/v) Tween 20 and 0.02% (w/v) sodium azide. Cells were incubated with primary antibodies (Santa Cruz Biotechnology) overnight at 4 °C, and washed extensively with 0.2% (v/v) Tween 20 in PBS before and after incubation with secondary antibodies for 30 min to 1 h at room temperature. Cells were mounted in 30% (v/v) glycerol in PBS and analysed by fluorescence microscopy.
Statistical analyses
Numerical data were analysed using an unpaired Student's t test. Results are expressed as means±S.D. P<0.05 was considered to be statistically significant.
RESULTS
Identification of transcription factors binding to the human BRS-3 promoter region
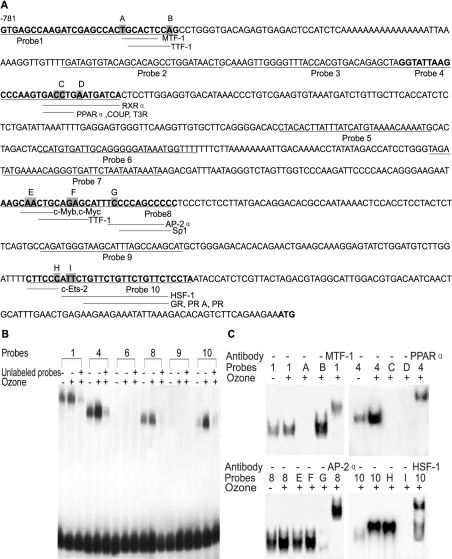
Ten oligonucleotides were designed corresponding to putative cis-regulatory binding sites in the BRS-3 promoter (Figure 1A) and were used in EMSAs to determine transcription factor binding in BECs. EMSAs with primers 1, 4, 8 and 10 that included competition with 100-fold excess of unlabelled primers revealed DNA–protein binding complexes as shown in Figure 1(B).
Figure 1. Screen of transcription factors binding to the human BRS-3 promoter region.
(A) Depicted is the 781 bp DNA sequence of the human BRS-3 5′-FR. The translation start codon ATG is in bold and underlined. Probes 1–10 used for EMSA are indicated by underlined sequences. Transverse lines represent possible nuclear-factor-binding sites in corresponding probes. The shaded residues represent the mutated sites in different mutated probes, which are listed alphabetically. (B) Specific DNA–protein complex binding to the human BRS-3 promoter was demonstrated using probes 1, 4, 8 and 10 as competition with unlabelled primer significantly reduced binding intensity. (C) Transcription factors MTF-1, PPARα, AP-2α and HSF-1 were identified as binding proteins to these four specific DNA sequences with EMSAs and supershift assays.
Therefore we analysed further the nature of protein binding to these genomic DNA sequences represented in primers 1, 4, 8 and 10 by inducing specific mutations (Figure 1A) followed by EMSA to investigate protein–DNA binding. Mutation A within the MTF-1 consensus site in primer 1, but not mutation B, resulted in the loss of the specific band (Figure 1C). Mutations C and D within the putative RXRα (retinoid X receptor α) and RXRα/PPARα/COUP (chicken ovalbumin upstream promoter)/T3R (tri-iodothyronine receptor)-binding sites also resulted in the loss of the specific band (Figure 1C), suggesting that the binding protein complex may be RXRα or a RXRα–PPARα dimer. Of mutations E, F and G in primer 8, only the mutation in the putative AP-2α-binding site resulted in the loss of the specific retarded band (Figure 1C). Of mutations H and I in primer 10, affecting the consensus binding sites for c-Ets-2 and HSF-1 respectively, loss of the specific band was detected only using mutation I (Figure 1C). In additional supershift experiments with specific antibodies and control antibody against those transcription factors identified in EMSAs and mutational analysis, the four DNA-binding proteins were characterized independently as zinc-finger MTF-1, RXRα–PPARα (using an anti-PPARα antibody), AP-2α and HSF-1, as demonstrated by supershifted specific DNA–protein complexes (Figure 1C). Next, we investigated the four nuclear factors that bound to BRS-3 promoters in vivo, with or without ozone stress, using ChIP, which suggests an interaction between PPARα or AP-2α and BRS-3 (Figure 2).
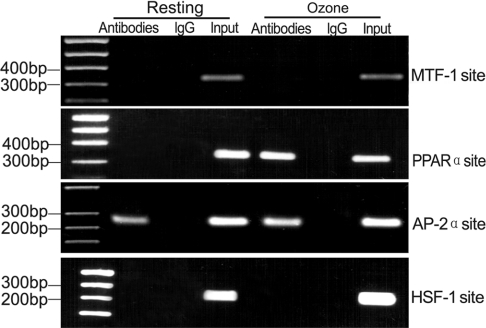
Figure 2. ChIP of four nuclear factors bound to the human BRS-3 gene.
Cross-linked chromatin prepared from resting BECs or ozone-stressed BECs was immunoprecipitated with four kinds of specific antibodies or non-immune IgG as indicated.
Inhibition of transcription factors and human BRS-3 expression subsequent to ASO treatment
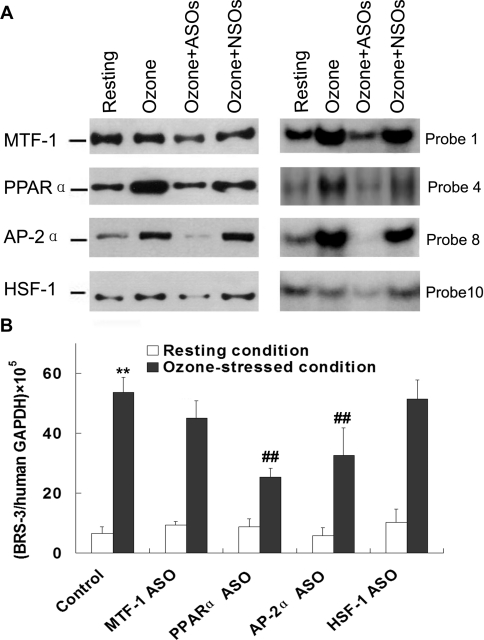
ASOs targeting the four transcription factors of interest were designed, and treatment efficiency was assessed by EMSAs and Western blotting. The results showed that all four ASOs caused substantial reduction in protein expression levels and their DNA-binding ability in ozone-stressed BECs, while the nonsense oligonucleotide had no effect (Figure 3A).
Figure 3. Inhibition of transcription factors and human BRS-3 expression subsequent to ASO treatment.
(A) The protein expression levels of MTF-1, PPARα, AP-2α and HSF-1 in ozone-stressed BECs were assessed by Western blotting (left-hand panel) and the DNA-binding activity by EMSA (right-hand panel) with and without ASO treatment. NSO, nonsense oligonucleotide. (B) Human BRS-3 mRNA expression was significantly induced by ozone exposure. The inducible expression was substantially reduced after ASO treatment specifically targeting PPARα and AP-2α. Results are means±S.D. for four experiments. **P<0.01 compared with resting BEC group; ##P<0.01 compared with ozone-stressed BEC group without ASO treatment.
Moreover, we determined human BRS-3 mRNA expression by real-time PCR in BECs after ASO treatment. Results showed that BRS-3 mRNA expression was significantly up-regulated (approx. 50-fold) in ozone-stressed BEC (Figure 3B), whereas pre-treatment with ASOs targeting PPARα and AP-2α yielded significant reduction of ozone-stress-induced BRS-3 expression (Figure 3B). Pre-treatment with ASOs targeting MTF-1 and HSF-1 had no effect on BRS-3 expression in ozone-stressed BECs and human BRS-3 expression under resting conditions was not affected by pre-treatment with ASOs (Figure 3B).
Transcriptional regulation of human BRS-3 in BECs and HLFs
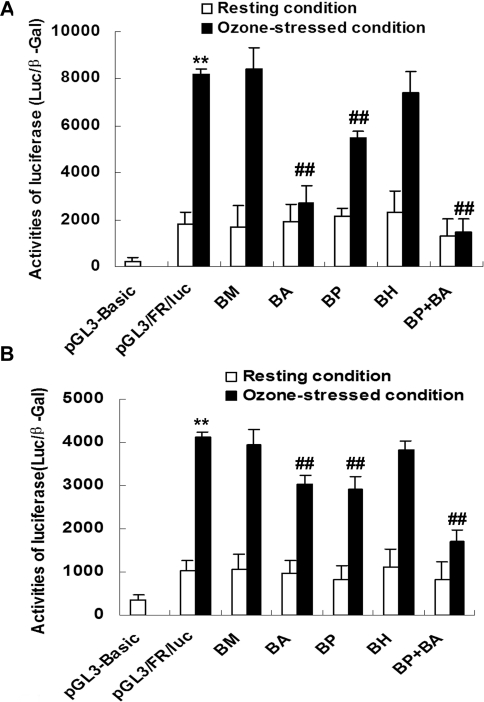
To test the biological effect of transcription factors MTF-1, PPARα, AP-2α and HSF-1 binding to the human BRS-3 promoter region, we cloned a 1261 bp fragment of the human BRS-3 5′-FR (− 1261/− 1) into plasmid pGL3-Basic, yielding the human BRS-3 promoter–luciferase reporter pGL3/FR/luc. By site-directed mutagenesis of putative transcription-factor-binding sites within pGL3/FR/luc, we generated mutants BM (mutation from G to A at − 758 of MTF-1 sites), BP (mutation from G to A at − 615 of PPARα sites), BA (mutation from C to T at − 305 and − 269 of AP-2α sites) and BH (mutation from T to G at − 108 of HSF-1 sites). Mutant BP+BA refers to plasmid pGL3/FR/luc in which both PPARα and AP-2α sites were mutated. Under resting conditions in transient transfection experiments, none of the transcription-factor-binding site mutants in pGL3/FR/luc resulted in any significant changes when compared with the plasmid harbouring the wild-type genomic sequence (Figure 4). Results from similar transfection experiments under ozone-stressed conditions demonstrated robust 4-fold activation (P<0.01) of the human BRS-3 promoter–luciferase reporter in both BECs and HLFs (Figure 4). Furthermore, using reporter plasmid mutants of the AP-2α sites or PPARα sites (BA and BP) in BECs under ozone stress a reduction in human BRS-3 promoter activity of almost 90% and 33% were detected when compared with promoter activity under ozone stress respectively (P<0.01; Figure 4A). Plasmid BP+BA containing mutants of both AP-2α and PPARα sites completely blocked the ozone-induced human BRS-3 promoter activation, while reporter plasmid mutants of the MTF-1 or HSF-1 sites (BM and BH) did not alter on the ozone-stress-induced response (Figure 4). Similar experiments in HLFs showed very similar results compared with BECs, albeit the magnitude of reduction in ozone-stress-induced activation was not as pronounced (Figure 4B).
Figure 4. Transcriptional activation of the human BRS-3 promoter–luciferase reporter in BECs (A) and HLFs (B).
Results are means±S.D. relative luciferase activities (normalized to β-gal) from at least five independent transient transfection experiments each performed in triplicate. Mutations BA, BP and BP+BA, but not BM and BH, of the pGL3/FR/luc reporter resulted in reduction of ozone-induced BRS-3 promoter activation (**P<0.01 compared with pGL3/FR/luc under resting conditions; ##P<0.01 compared with wild-type pGL3/FR/luc under ozone-stressed conditions).
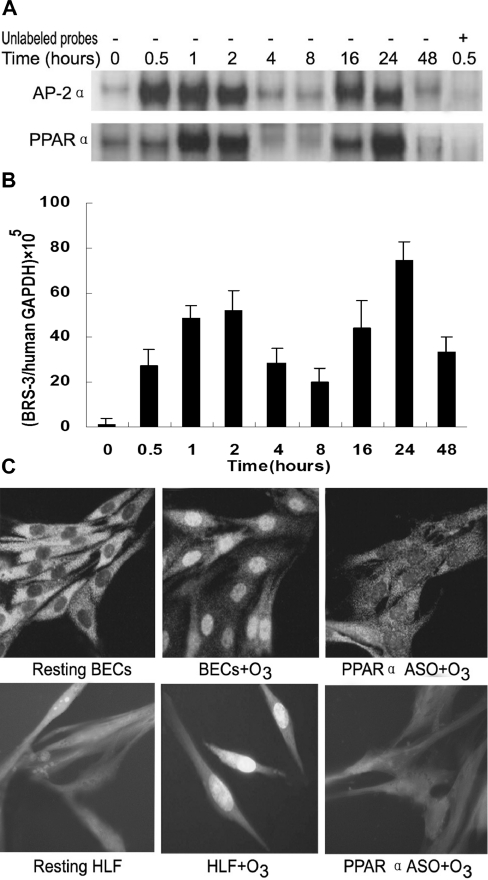
Time course of AP-2α, PPARα and human BRS-3 expression and PPARα nuclear translocation
To delineate further the potential role of PPARα and AP-2α in BRS-3 regulation in BECs, a 48 h time course of PPARα and AP-2α DNA-binding activity and BRS-3 expression was examined after 30 min of ozone stress using EMSA (with probes 1 and 4 respectively) and real-time PCR respectively. Nuclear translocation of PPARα was detected by immunofluorescence microscopy. The results showed that the ozone-induced activation of PPARα and AP-2α exhibited a biphasic pattern over a 48 h period (Figure 5A). The first activation of AP-2α occurred within 30 min of ozone exposure and had decreased at 4 h. The second activation was observed 16 h after ozone exposure followed by decreased activation at 48 h after exposure. A virtually identical PPARα DNA-binding activation pattern, albeit slightly lagging behind the one of AP-2α, was observed after ozone exposure (Figure 5A). Meanwhile, BRS-3 mRNA expression as measured by real-time PCR also followed precisely this biphasic pattern of PPARα and AP-2α activation. A gradual increase of human BRS-3 expression occurred until it reached a first peak of an approx. 50-fold increase over resting conditions at 2 h, and the second peak of approx. 70-fold increase was detectable 24 h after ozone exposure (Figure 5B). Using a PPARα-specific antibody, we demonstrated further that PPARα nuclear translocation occurred as early as 30 min after ozone exposure in BECs and HLFs (Figure 5C). The translocation effects induced by ozone can be blocked by PPARα ASO, indicating independently its activation via ozone exposure.
Figure 5. Time course of human BRS-3 mRNA expression and DNA–protein binding activities of AP-2α and PPARα.
(A) The time course of AP-2α and PPARα DNA binding to BRS-3 promoter sequence under ozone stress was determined by EMSA. (B) Expression of human BRS-3 mRNA was assayed by real-time PCR. Results are means±S.D. for four independent experiments. (C) Nuclear translocation of PPARα after ozone exposure in BECs was demonstrated by immunofluorescence. Magnification × 400.
DISCUSSION
There is a growing body of experimental and clinical evidence that BLPs play an important role in normal lung development and in several pathological conditions in the lung, and the results of our previous experiments showed that the expression of BRS-3 mRNA resulted in wound repair and protection from lung injury [22], so there is significant academic value and clinical application prospects to determine the regulation and control mechanisms of BRS-3 gene expression.
The human BRS-3 gene is located at Xq26-q28 and contains a 1.261 kb 5′-FR and three exons. Transcription factors tightly regulate gene expression in response to intra- and extra-cellular stimuli, and they often play a central role in determining the cell fate. In the present study, ten oligonucleotide probes corresponding to various regions of the BRS-3 promoter were designed, and the spectrum of potential nuclear factors involved in induced expression of BRS-3 in response to ozone were screened using EMSA. The ChIP assay, ASO assay and promoter-reporter assay demonstrated that AP-2α and PPARα are involved in the ozone-inducible BRS-3 transcriptional regulation in BECs. Similar experiments in HLFs showed very similar results compared with those in BECs, indicating these findings are not limited to BECs.
AP-2 consists of a family of five isoforms designated AP-2α, AP-2β, AP-2γ, AP-2δ and AP-2ζ, with a unique modular structure consisting of an N-terminal proline- and glutamine-rich transactivational domain and a complex helix–span–helix motif which is necessary and sufficient for dimerization and site-specific DNA binding [24–26]. AP-2 mediates transcriptional activation in response to two signal transduction pathways: the phorbol ester/diacylglycerol-inducible protein kinase C pathway and the cAMP-dependent protein kinase A pathway [25]. The expression of AP-2α is associated with the embryonic differentiation and cancer [27,28]. Also, AP-2α has been shown to regulate neuropeptide receptors [29,30] and neuropeptide genes [31,32]. However, the activation and translocation of AP-2α remained unclear. PPAR is a ligand-activated transcription factor and a member of the nuclear hormone receptor superfamily [33]. PPAR modulates target gene expression in response to ligand activation after heterodimerization with retinoid X receptor and binding to peroxisome-proliferator-responsive elements [33]. Three different PPAR isoforms, each encoded by distinct genes, have been identified and designated PPARα, PPARβ and PPARγ [33]. The functional activity of PPAR and its translocation from plasma into the nucleus were modulated by phosphorylation [34]. Accumulating evidence supports a link between PPARα and diabetes, obesity, dyslipidaemia and inflammation [35,36]. In the present study, we observed that PPARα translocation increased after 30 min of ozone exposure, indicating that PPARα may modulate BRS-3 expression under ozone stress. Through further studies, we found that ozone-inducible AP-2α and PPARα activation exhibited a biphasic pattern during a 48 h observation period and the expression of BRS-3 mRNA was consistent with PPARα and AP-2α activation, indicating that AP-2α and PPARα jointly regulate the expression of BRS-3 in response to ozone. However, how the early signals are transmitted to AP-2α and PPARα and how the early signal molecules assemble with AP-2α and PPARα in response to ozone require further studies.
In conclusion, the present study demonstrates that robust transcriptional BRS-3 up-regulation occurs during acute ozone-induced AHR and is mediated at least in part by ozone-induced recruitment of PPARα and AP-2α to the human BRS-3 promoter, thereby ameliorating the effects of acute lung injury.
Acknowledgments
This work was supported by grant #30470755 from National Natural Science Foundation of China and grant #04JJ3095 from Hunan Natural Science Foundation.
References
- 1.Anastasi A., Erspamer V., Bucci M. Isolation and structure of bombesin and alytesin, 2 analogous active peptides from the skin of the European amphibians Bombina and Alytes. Experientia. 1971;27:166–167. doi: 10.1007/BF02145873. [DOI] [PubMed] [Google Scholar]
- 2.Walsh J. H., Reeve J. R., Jr Mammalian bombesin-like peptides: neuromodulators of gastric function and autocrine regulators of lung cancer growth. Peptides. 1985;6(suppl. 3):63–68. doi: 10.1016/0196-9781(85)90352-3. [DOI] [PubMed] [Google Scholar]
- 3.Shan L., Emanuel R. L., Dewald D., Torday J. S., Asokanathan N., Wada K., Wada E., Sunday M. E. Bombesin-like peptide receptor gene expression, regulation, and function in fetal murine lung. Am. J. Physiol. Lung Cell. Mol. Physiol. 2004;286:L165–L173. doi: 10.1152/ajplung.00436.2002. [DOI] [PubMed] [Google Scholar]
- 4.Lemaire I., Jones S., Khan M. F. Bombesin-like peptides in alveolar macrophage: increased release in pulmonary inflammation and fibrosis. Neuropeptides. 1991;20:63–72. doi: 10.1016/0143-4179(91)90041-g. [DOI] [PubMed] [Google Scholar]
- 5.Moody T. W., Merali Z. Bombesin-like peptides and associated receptors within the brain: distribution and behavioral implications. Peptides. 2004;25:511–520. doi: 10.1016/j.peptides.2004.02.012. [DOI] [PubMed] [Google Scholar]
- 6.Ohki-Hamazaki H., Iwabuchi M., Maekawa F. Development and function of bombesin-like peptides and their receptors. Int. J. Dev. Biol. 2005;49:293–300. doi: 10.1387/ijdb.041954ho. [DOI] [PubMed] [Google Scholar]
- 7.Aguayo S. M., Schuyler W. E., Murtagh J. J., Jr, Roman J. Regulation of lung branching morphogenesis by bombesin-like peptides and neutral endopeptidase. Am. J. Respir. Cell Mol. Biol. 1994;10:635–642. doi: 10.1165/ajrcmb.10.6.8003340. [DOI] [PubMed] [Google Scholar]
- 8.Garcia L. J., Pradhan T. K., Weber H. C., Moody T. W., Jensen R. T. The gastrin-releasing peptide receptor is differentially coupled to adenylate cyclase and phospholipase C in different tissues. Biochim. Biophys. Acta. 1997;1356:343–354. doi: 10.1016/s0167-4889(97)00007-4. [DOI] [PubMed] [Google Scholar]
- 9.Hou W., Tsuda T., Jensen R. T. Neuromedin B activates phospholipase D through both PKC-dependent and PKC-independent mechanisms. Biochim. Biophys. Acta. 1998;1391:337–350. doi: 10.1016/s0005-2760(98)00014-9. [DOI] [PubMed] [Google Scholar]
- 10.Ryan R. R., Weber H. C., Mantey S. A., Hou W., Hilburger M. E., Pradhan T. K., Coy D. H., Jensen R. T. Pharmacology and intracellular signaling mechanisms of the native human orphan receptor BRS-3 in lung cancer cells. J. Pharmacol. Exp. Ther. 1998;287:366–380. [PubMed] [Google Scholar]
- 11.Aoki K., Sun Y. J., Aoki S., Wada K., Wada E. Cloning, expression, and mapping of a gene that is upregulated in adipose tissue of mice deficient in bombesin receptor subtype-3. Biochem. Biophys. Res. Commun. 2002;290:1282–1288. doi: 10.1006/bbrc.2002.6337. [DOI] [PubMed] [Google Scholar]
- 12.Cullen A., Emanuel R. L., Torday J. S., Asokananthan N., Sikorski K. A., Sunday M. E. Bombesin-like peptide and receptors in lung injury models: diverse gene expression, similar function. Peptides. 2000;21:1627–1638. doi: 10.1016/s0196-9781(00)00294-1. [DOI] [PubMed] [Google Scholar]
- 13.Fathi Z., Corjay M. H., Shapira H., Wada E., Benya R., Jensen R., Viallet J., Sausville E. A., Battey J. F. BRS-3: a novel bombesin receptor subtype selectively expressed in testis and lung carcinoma cells. J. Biol. Chem. 1993;268:5979–5984. [PubMed] [Google Scholar]
- 14.Gorbulev V., Akhundova A., Grzeschik K. H., Fahrenholz F. Organization and chromosomal localization of the gene for the human bombesin receptor subtype expressed in pregnant uterus. FEBS. Lett. 1994;340:260–264. doi: 10.1016/0014-5793(94)80150-9. [DOI] [PubMed] [Google Scholar]
- 15.Jennings C. A., Harrison D. C., Maycox P. R., Crook B., Smart D., Hervieu G. J. The distribution of the orphan bombesin receptor subtype 3 in the rat CNS. Neuroscience. 2003;120:309–324. doi: 10.1016/s0306-4522(03)00260-4. [DOI] [PubMed] [Google Scholar]
- 16.Emanuel R. L., Torday J. S., Mu Q., Asokananthan N., Sikorski K. A., Sunday M. E. Bombesin-like peptides and receptors in normal fetal baboon lung: roles in lung growth and maturation. Am. J. Physiol. Lung. Cell. Mol. Physiol. 1999;277:L1003–L1017. doi: 10.1152/ajplung.1999.277.5.L1003. [DOI] [PubMed] [Google Scholar]
- 17.Reubi J. C., Wenger S., Schmuckli-Maurer J., Schaer J.-C., Gugger M. Bombesin receptor subtypes in human cancers: detection with the universal radioligand 125I-[D-TYR6, β-ALA11, PHE13, NLE14] bombesin(6–14) Clin. Cancer Res. 2002;8:1139–1146. [PubMed] [Google Scholar]
- 18.Ohki-Hamazaki H., Watase K., Yamamoto K., Ogura H., Yamano M., Yamada K., Maeno H., Imaki J., Kikuyama S., Wada E., Wada K. Mice lacking bombesin receptor subtype-3 develop metabolic defects and obesity. Nature. 1997;390:165–169. doi: 10.1038/36568. [DOI] [PubMed] [Google Scholar]
- 19.Cullen A., Van Marter L. J., Allred E. N., Moore M., Parad R. B., Sunday M. E. Urine bombesin-like peptide elevation precedes clinical evidence of bronchopulmonary dysplasia. Am. J. Respir. Crit. Care Med. 2002;165:1093–1097. doi: 10.1164/ajrccm.165.8.2108044. [DOI] [PubMed] [Google Scholar]
- 20.Subramaniam M., Sugiyama K., Coy D. H., Kong Y., Miller Y. E., Weller P. F., Wada K., Wada E., Sunday M. E. Bombesin-like peptides and mast cell responses: relevance to bronchopulmonary dysplasia? Am. J. Respir. Crit. Care Med. 2003;168:601–611. doi: 10.1164/rccm.200212-1434OC. [DOI] [PubMed] [Google Scholar]
- 21.Sunday M. E., Yoder B. A., Cuttitta F., Haley K. J., Emanuel R. L. Bombesin-like peptide mediates lung injury in a baboon model of bronchopulmonary dysplasia. J. Clin. Invest. 1998;102:584–594. doi: 10.1172/JCI2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tan Y. R., Qi M. M., Qin X. Q., Xiang Y., Li X., Wang Y., Qu F., Liu H. J., Zhang J. S. Wound repair and proliferation of bronchial epithelial cells enhanced by bombesin receptor subtype 3 activation. Peptides. 2006;27:1852–1858. doi: 10.1016/j.peptides.2005.12.012. [DOI] [PubMed] [Google Scholar]
- 23.Szak S. T., Mays D., Pietenpol J. A. Kinetics of p53 binding to promoter sites in vivo. Mol. Cell. Biol. 2001;21:3375–3386. doi: 10.1128/MCB.21.10.3375-3386.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Williams T., Tjian R. Characterization of a dimerization motif in AP-2 and its function in heterologous DNA-binding proteins. Science. 1991;251:1067–1071. doi: 10.1126/science.1998122. [DOI] [PubMed] [Google Scholar]
- 25.Imagawa M., Chiu R., Karin M. Transcription factor AP-2 mediates induction by two different signal-transduction pathways: protein kinase C and cAMP. Cell. 1987;51:251–260. doi: 10.1016/0092-8674(87)90152-8. [DOI] [PubMed] [Google Scholar]
- 26.Wang H. V., Vaupel K., Buettner R., Bosserhoff A. K., Moser M. Identification and embryonic expression of a new AP-2 transcription factor, AP-2ϵ. Dev. Dyn. 2004;231:128–135. doi: 10.1002/dvdy.20119. [DOI] [PubMed] [Google Scholar]
- 27.Batsche E., Muchardt C., Behrens J., Hurst H. C., Cremisi C. RB and c-Myc activate expression of the E-cadherin gene in epithelial cells through interaction with transcription factor AP-2. Mol. Cell. Biol. 1998;18:3647–3658. doi: 10.1128/mcb.18.7.3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rietveld L. E., Koonen-Reemst A. M., Sussenbach J. S., Holthuizen P. E. Dual role for transcription factor AP-2 in the regulation of the major fetal promoter P3 of the gene for human insulin-like growth factor II. Biochem. J. 1999;338:799–806. [PMC free article] [PubMed] [Google Scholar]
- 29.Chen S., Shi H., Liu X., Segaloff D. L. Multiple elements and protein factors coordinate the basal and cyclic adenosine 3′,5′-monophosphate-induced transcription of the lutropin receptor gene in rat granulosa cells. Endocrinology. 1999;140:2100–2109. doi: 10.1210/endo.140.5.6722. [DOI] [PubMed] [Google Scholar]
- 30.Woltje M., Kraus J., Hollt V. Regulation of mouse δ-opioid receptor gene transcription: involvement of the transcription factors AP-1 and AP-2. J. Neurochem. 2000;74:1355–1362. doi: 10.1046/j.1471-4159.2000.0741355.x. [DOI] [PubMed] [Google Scholar]
- 31.Cheng Y. H., Handwerger S. AP-2α modulates human corticotropin-releasing hormone gene expression in the placenta by direct protein-protein interaction. Mol. Cell. Endocrinol. 2002;191:127–136. doi: 10.1016/s0303-7207(02)00081-3. [DOI] [PubMed] [Google Scholar]
- 32.Katsel P. L., Greenstein R. J. Identification of overlapping AP-2/NF-κB-responsive elements on the rat cholecystokinin gene promoter. J. Biol. Chem. 2001;276:752–758. doi: 10.1074/jbc.M007553200. [DOI] [PubMed] [Google Scholar]
- 33.Schachtrup C., Emmler T., Bleck B., Sandqvist A., Spener F. Functional analysis of peroxisome proliferator responsive element motifs in genes of fatty acid-binding proteins. Biochem. J. 2004;382:239–245. doi: 10.1042/BJ20031340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gardner O. S., Dewar B. J., Graves L. M. Activation of mitogen-activated protein kinases by peroxisome proliferator-activated receptor ligands: an example of nongenomic signaling. Mol. Pharmacol. 2005;68:933–941. doi: 10.1124/mol.105.012260. [DOI] [PubMed] [Google Scholar]
- 35.Trifilieff A., Bench A., Hanley M., Bayley D., Campbell E., Whittaker P. PPAR-α and -γ but not -δ agonists inhibit airway inflammation in a murine model of asthma: in vitro evidence for an NF-κB-independent effect. Br. J. Pharmacol. 2003;139:163–171. doi: 10.1038/sj.bjp.0705232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ikeda I., Konno R., Shimizu T., Ide T., Takahashi N., Kawada T., Nagao K., Inoue N., Yanagita T., Hamada T., et al. Campest-5-en-3-one, an oxidized derivative of campesterol, activates PPARα, promotes energy consumption and reduces visceral fat deposition in rats. Biochim. Biophys. Acta. 2006;1760:800–807. doi: 10.1016/j.bbagen.2006.02.017. [DOI] [PubMed] [Google Scholar]