Abstract
SPT (serine palmitoyltransferase) catalyses the rate-limiting step for the de novo synthesis of sphingolipids. Mammalian SPT is believed to be a heterodimer composed of two subunits, SPTLC1 and SPTLC2. We reported previously the identification of a new third SPT subunit, SPTLC3. In the present study, we have investigated the structure of the SPT complex in more detail. Pull-down assays with antibodies against SPTLC3 concomitantly co-precipitated SPTLC1 and SPTLC2 in human placenta extracts and SPTLC3 overexpressing human embryonic kidney-293 cells. By size exclusion chromatography, we determined the molecular mass of the functional SPT complex to be approx. 480 kDa. By Blue-native-PAGE experiments we demonstrated that all three SPT subunits (SPTLC1–3) are co-localized within a single SPT complex. On the basis of these results we conclude that the functional SPT is not a dimer, but a higher organized complex, composed of three distinct subunits (SPTLC1, SPTLC2 and SPTLC3) with a molecular mass of 480 kDa. The stoichiometry of SPTLC2 and SPTLC3 in this complex seems not to be fixed and is probably changed dynamically in dependence of the tissue specific SPTLC2 and SPTLC3 expression levels. Based on our own and earlier published data we propose a model of an octameric SPT structure. The observed dynamic composition of the SPT complex could provide a cellular mechanism to adjust SPT activity to tissue specific requirements in sphingolipid synthesis.
Keywords: ceramide, serine palmitoyltransferase (SPT), sphingolipid
Abbreviations: AONS, 8-amino-7-oxononanoate synthase; BN, blue native; ER, endoplasmic reticulum; HEK-293, human embryonic kidney-293; OTG, octyl glucoside; PLP, pyridoxal 5-phosphate; POAS, PLP-dependent α-oxoamine synthase; SPT, serine palmitoyltransferase
INTRODUCTION
Sphingolipids are an ubiquitously distributed class of lipids that can be found in all higher organisms. Sphingoid bases confer important structural properties to membranes and to their partition into microdomains (membrane rafts). They modulate the activities of various enzymes, such as protein kinases, protein phosphatases and phospholipases in cells or cell-free systems [1], and are involved in many cellular events including proliferation, differentiation, senescence, apoptosis and inflammatory responses [2].
De novo sphingolipid biosynthesis is initiated by the condensation of L-serine with palmitoyl-CoA to generate 3-ketodihydrosphingosine. This PLP (pyridoxal 5-phosphate)-dependent reaction is catalysed by the SPT (serine palmitoyltransferase; EC 2.3.1.50). SPT is ubiquitously expressed [3] and essential for embryonic development, since homozygous SPTLC1 and SPTLC2 knockout mice are non-viable [4]. SPT activity is exerted by three distinct subunits, SPTLC1, SPTLC2 [5] and the SPTLC3 identified recently [6]. The amino acid sequences between SPTLC1 and SPTLC2 (or SPTLC3) show a mutual similarity of about 20%, whereas the sequences for SPTLC2 and SPTLC3 show a significantly higher identity of 68% (with 84% similarity) on the amino acid level. SPT belongs to the POAS (PLP-dependent α-oxoamine synthase) family. Other members of this family include 5-amino-levulinic acid synthase, KBL (2-amino-3 ketobutyrate ligase) and AONS (8-amino-7-oxononanoate synthase) [5]. All known members of the POAS family, with the exception of SPT, are soluble homodimers, whereas SPT is composed of distinct subunits and bound to the outer membrane of the ER (endoplasmic reticulum). Based on its similarity to the other POAS members, the active SPT enzyme is considered to be a heterodimer composed of the two subunits, SPTLC1 and SPTLC2 [5]. The crystal structure of AONS indicates that the active site of this enzyme is formed at the interface of the two subunits [7]. However, in contrast to the other members of the POAS family, which have PLP-binding sites on each subunit, SPT contains only one PLP-binding motif, which is located on the SPTLC2 subunit. The SPTLC3 subunit also contains a conserved PLP-binding motif. This raises new questions about the structure and composition of the SPT enzyme. The significant homology between SPTLC2 and SPTLC3 makes it probable that SPTLC3 also forms a heterodimer with SPTLC1. However, a second possibility could be that two SPTLC3 subunits form a homodimer in analogy to the other POAS members or, thirdly, that SPT is in fact not a dimer, but a higher organized complex and composed of several distinct subunits including SPTLC1, SPLTLC2 and SPTLC3. In the present study, we have addressed that issue and we present strong evidence that the native SPT enzyme is a multi-subunit complex, composed of all three subunits, with a molecular mass of 480 kDa.
MATERIALS AND METHODS
General
All chemicals, unless otherwise stated, were purchased from Sigma. The direct TOPO® cloning vector pcDNA3.2 is from Invitrogen. Protein G–agarose was from Roche. Anti V5-Tag monoclonal antibodies were from Serotec. The database search was done with NCBI (National Center for Biotechnology Information) Blast (www.ncbi.nlm.nih.gov/blast/) and subsequent analysis was performed using the BioEdit program (v5.0.9; Department of Microbiologie, North Carolina State University, Raleigh, NC, U.S.A.). The cloning and immunization procedures for the SPT subunits SPTLC1, SPTLC2 and SPTLC3 were described earlier [6].
Transfection
HEK-293 (human embryonic kidney-293) cells were grown in Petri dishes in Dulbecco's modified Eagle's medium with 10% (v/v) foetal calf serum. Transfections were performed using Metafectene™ (Biontex, Munich, Germany) according to the manufacturers’ instructions. The cells where cultured in the presence of G418 to generate a pool of stably transfected cells.
Preparation of human placenta extract
Human placenta tissue, obtained freshly after caesarean section, was cut into small pieces and washed several times in ice-cold PBS. The tissue pieces were homogenized in a blender and filtered through gauze. The filtrate was centrifuged at 2000 g for 5 min at 4°C and then washed twice in PBS, before freezing in liquid N2.
BN (blue native)-PAGE
BN-PAGE was performed according to the method described by Schägger and von Jagow [8], with the following modification. The final volume of the resolving gels was 5.2 ml with a linear gradient from 7–15% (w/v) acrylamide. Cells were lysed on ice in BN lysis buffer [100 mM Bis/Tris, pH 7.0, 0.5 M aminohexanoic acid, 35% (v/v) glycerol, 5 mM MgCl2, 0.5 mM dithiothreitol, benzonase and Complete™ protease inhibitor] containing 0.25–1% SDS (as stated in the text). BN loading buffer [100 mM Bis/Tris, pH 7.0, 0.5 M aminohexanoic acid and 5% (w/v) Coomassie G-250] was added to each sample (20–50 μg of total protein per lane) and loaded on the gel. The cathode buffer was composed of 50 mM Tricine and 15 mM Tris/HCl (pH7.0) at 4°C, with 0.01% Coomassie G-250. Once the running-front was at the middle of the separating gel the cathode buffer was exchanged by the same buffer without Coomassie G-250. For the anode buffer we used 25 mM Bis/Tris (pH 7.0) at 4°C. Proteins were separated on a Bio-Rad MiniProtean System (Gel Size 70×55×1 mm) for 4 h at 150 V.
Immunoprecipitation
Cells were transfected and cultivated as described above and harvested at 70–80% confluence. The medium was removed and the cells washed twice with ice-cold PBS. The cells were lysed in immunoprecipitation buffer (PBS containing 0.2% Triton X-100 and Complete™ protease inhibitor) for 15 min followed by sonication [15 pulses for 0.5 s at 50% power with 0.5 s intervals between pulses using a Sonoplus HD2070 (Bandelin)]. Insoluble cell debris was removed by centrifugation at 13000 g for 5 min at 4°C. The supernatant was incubated overnight with 25 μl of anti-(V5–agarose) (Sigma) with constant rolling at 4°C. In the negative control, the precipitation was blocked by the addition of 5 μg of purified V5-peptide as a competitor. After incubation, the beads were washed three times for 10 min in immunoprecipitation buffer (with constant rolling at 4°C) and the bound SPT complex was eluted by the addition of 10 mg/ml free V5-peptide for 1 h at 4°C. The eluted proteins were separated by SDS/PAGE (12% gels), blotted on to a PVDF membrane and incubated with the appropriated antiserum as stated in the text.
Size-exclusion chromatography
HEK-293 cells and homogenized placenta tissue were lysed in HET buffer (25 mM Hepes, pH 8.0, 5 mM EDTA and 0.2% Triton X-100) for 15 min followed by sonication (15 s pulses at 50% power with 0.5 s intervals between pulses). Insoluble cell debris was removed by centrifugation at 13000 g for 5 min at 4°C. The supernatant (100 μl) was loaded on to a size-exclusion column (Superose 6 10/300GL; Amersham Biosciences) equilibrated in HET buffer. The eluate was fractionated into 20 fractions, which were assayed for SPT activity and the presence of SPTLC1–3 by Western blotting. In some experiments, the SPT complex was chemically cross-linked by the addition of formaldehyde. HEK-293 cells were resuspended in PBS and sonicated (30 s pulses at 50% power with 0.5 s intervals between the pulses). The suspension (1 mg/ml) was treated by the addition of formaldehyde (25 μM) for 1 h at 4°C and then SDS was added to a final concentration of 0.5%. Insoluble cell debris was removed by centrifugation at 13000 g for 5 min at 4°C, and the supernatant (100 μl) was loaded on to the column.
Activity assay
The reaction was carried out in 25 mM Hepes (pH 8.0), 5 mM EDTA, 0.5 mM L-serine, 0.05 mM palmitoyl-CoA, 20 μM pyridoxal-5′-phosphate, 0.2% Triton X-100 and 0.1 μCi L-[U-14C]serine (Amersham Biosciences) for 20 min at 37°C. In the control samples, 40 μM Myrocin (ISP1; Sigma) was added to inhibit SPT activity. The total assay volume was 200 μl. The reaction was stopped by the addition of 400 μl of 0.5 M NH4OH, followed by the addition of 2.25 ml of chloroform/methanol (1:2, v/v). Long-chain bases were extracted by adding 0.75 ml of NH4OH and 0.75 ml of chloroform, followed by vortexing and centrifuging briefly. The upper aqueous layer was aspirated off and the lower layer was washed twice with 2 ml of 0.1 M KCl. 0.5 ml of the lower organic phase was removed and counted using a scintillation counter.
RESULTS
Expression of SPTLC1–3 in HEK-293 cells and placenta extract
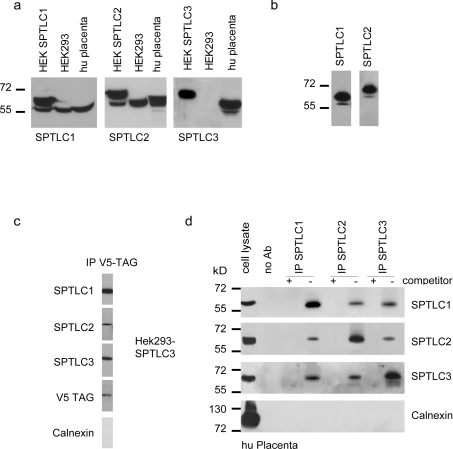
Human SPTLC1, SPTLC2 and SPTLC3 cDNA were amplified from a cDNA library and cloned into a mammalian expression vector (pcDNA3.1) in frame with the C-terminal V5–His-tag. The subunits were expressed individually in HEK-293 cells and selected for Neomycin resistance to generate a pool of stably expressing cells. Upon SDS/PAGE and Western blotting, all subunits were detected by an anti-V5 antibody. The expressed proteins showed no signs of degradation and all subunits were expressed at similar levels [6]. The expression of the individual SPT subunits was counter tested using polyclonal antibodies against SPTLC1, SPTLC2 and SPTLC3 [6]. All antibodies showed specific binding towards their antigen. The transfected and the endogenously expressed subunits can be distinguished by a mass difference of 5 kDa, due to the fused V5–His-tag (Figure 1a). In SPTLC3 expressing HEK-293 cells, only the transfected protein is visible, as SPTLC3 is not endogenously expressed in this cell line [6]. Placenta tissue, on the other hand, expresses high levels of SPTLC3 (Figure 1a), which has been demonstrated previously [6].
Figure 1. Expression and immunoprecipitation of the SPT complex.
(a) Expression of the SPT subunits in HEK-293 cells and human placenta tissue. Total cell extract (50 μg/lane) was separated by SDS/PAGE (10% gels), blotted on to PVDF and probed for the SPT subunits using subunit specific antibodies against SPTLC1, SPTLC2 and SPTLC3. The first lanes show transfected HEK-293 cells overexpressing the respective subunit. All subunits were specifically detected by their respective antibodies. The heterologously expressed subunits migrate to a slightly larger size due to the mass addition of the added V5-tag. The polyclonal antibodies detect both the transfected (upper band) and the endogenous (lower band) subunits. (b) Extracts from SPTLC1 and SPTLC2 overexpressing HEK-293 cells were incubated with the anti-V5 antibody to specifically precipitate the V5-tagged subunits. The precipitate was then separated by SDS/PAGE, blotted on to PVDF and probed with the polyclonal antibody against the respective subunit. In both cases we could see the precipitation of not only the V5-tagged subunit, but also the endogenously encoded untagged subunit. This indicates that the tagged and untagged subunits are located within the same protein complex. This contradicts the concept of a heterodimeric SPT structure. (c) The V5-tagged SPTLC3 subunit from overexpressing HEK-293 cells was captured by an anti-V5 antibody and the precipitate probed for the co-precipitation of SPTLC1 and SPTLC2. Surprisingly, all three SPT subunits could be detected in the precipitate. Calnexin, as an abundant ER protein, was not found in the precipitate, showing the specificity of the assay. (d) In order to confirm these results we repeated this experiment with human placenta tissue. Placenta tissue was shown to natively express high levels of SPTLC3. The positive control (left-hand lanes) shows that all proteins, including calnexin, are expressed in human placenta. The extract was incubated with polyclonal antibodies against SPTLC1, SPTLC2 and SPTLC3, and precipitated by the addition of Protein G–agarose. In the negative control we added an excess (5 μg/ml) of target-specific blocking peptides (immunization peptides) as a competitor. After separation by SDS/PAGE (10% gels) and blotting on to PVDF, the precipitate was cross-probed for the co-precipitation of the other two subunits. For all of the samples we observed the co-precipitation of the other two subunits, which had not been targeted directly. No signal was seen in the presence of the blocking peptides or when the beads were used alone. To further exclude that the observed complex is possibly caused by an unspecific aggregation of ER proteins, we again probed for calnexin, which was not detected. This suggests that all three SPT subunits are located within a single complex. Molecular mass markers given in kDa are shown to the left-hand side of the Figures in (a), (b) and (d). Ab, antibody; hu placenta, human placenta.
SPTLC1, SPTLC2 and SPTLC3 form a single multi-subunit SPT complex
The high similarity in structure and function between SPTLC2 and SPTLC3 [6] raises the question if SPTLC3, like SPTLC2, forms a complex with SPTLC1. To address this issue, we performed pull-down assays with the V5-tagged SPTLC1 and SPTLC2 subunits, using an anti-V5 antibody and probed the precipitate with the respective polyclonal antibodies (Figure 1b). For each subunit we could detect not only the V5-tagged subunit (Figure 1b, upper bands), but also the endogenously encoded untagged subunit (Figure 1b, lower bands) in the precipitate. This indicates that the tagged and untagged SPT subunits are both located within the same complex, which cannot be explained by the concept of a heterodimeric SPT structure.
In a second experiment, we captured the V5-tagged subunit of SPTLC3-overexpressing HEK-293 cells. After washing, the bound SPTLC3 subunit was specifically eluted by the addition of free V5 peptide. The eluates were separated by SDS/PAGE, blotted on to PVDF and probed for the presence of SPTLC1, SPTLC2 and SPTLC3. Surprisingly we observed the concomitant co-precipitation of the SPTLC1 and SPTLC2 subunit when pulling down the V5-tagged SPTLC3 subunit (Figure 1c). This indicates that all three subunits are located within a single complex. To exclude, that these findings are accidentally caused by unspecific interactions of the proteins with the anti-V5 antibody or the beads, we also tested the precipitate for the presence of calnexin. Calnexin is an abundant 90 kDa chaperone with a single transmembrane domain and binds, like SPT, to the ER membrane. We could not detect any calnexin in the precipitate (Figure 1c), which shows a specific precipitation of the target protein. It furthermore largely excludes that the observed co-precipitation of SPTLC1, SPTLC2 and SPTLC3 is due to the formation of detergent-insoluble membrane complexes [9].
However, since native HEK-293 cells do not express SPTLC3, the overexpression of this subunit causes an artificial situation in these cells. Therefore we performed a similar experiment with primary human placenta tissue, which was shown to natively express high levels of SPTLC3 [6]. Each of the SPT subunits was targeted separately by an affinity-purified polyclonal antibody and the precipitates were probed for the co-precipitation of the other subunits. For each antibody a negative control was included to which an excess of free immunisation peptide was added to compete for the antigen binding. As before, we observed, for each subunit, the co-precipitation of the other two subunits independently of the antibody used (Figure 1d). Again, no co-precipitation of calnexin was seen in these samples, which proves the specificity of the method. This result further supports the previous observations that SPT is not a dimeric, but rather a multimeric complex, which is composed of all three SPT subunits.
The active SPT complex has an apparent molecular mass of 480 kDa
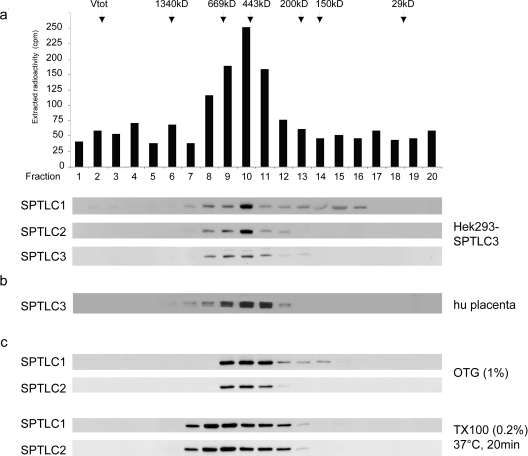
The molecular mass of an SPT dimer, either consisting of SPTLC1 and SPTLC2 or SPTLC1 and SPTLC3 would result in a molecular mass of approx. 120 kDa. To our knowledge, no data on the molecular mass of the functional mammalian SPT enzyme have been published so far. We therefore fractionated SPTLC3-overexpressing HEK-293 cells, as well as human placenta extract, by size-exclusion chromatography. A total of 20 fractions were collected, and each fraction was assayed for SPT activity and probed for SPTLC1, SPTLC2 and SPTLC3 by Western blotting (Figure 2a). SPT activity was detected between fractions 8 and 12, with peak activity observed in fraction 10. This elution volume equals a molecular mass of approx. 480 kDa, as determined by the parallel separation of defined molecular mass markers. In all fractions showing SPT activity, we could observe the parallel elution of all three SPT subunits. The highest concentration was seen in fraction 10, which also showed the highest SPT activity. Whereas SPTLC1 was also found in decreasing amounts between fractions 12 and 16, we did not observe the elution of SPTLC2 or SPTLC3 in earlier or later fractions. This shows the absence of free SPTLC2 or SPTLC3, as well as the absence of larger protein aggregates. In a control experiment, we also separated the soluble SPT (sSPT) isoform from Sphingomonas paucimobilis with the same protocol. This prokaryotic SPT is, in contrast to the mammalian form, a homodimer with two identical subunits [10]. In concordance to its theoretical mass, this prokaryotic SPT eluted at a mass of around 110 kDa. This indicates that the observed mass of 480 kDa for the mammalian complex is not caused by an artefact of our method.
Figure 2. Size exclusion chromatography of the functional SPT complex.
(a) Size-exclusion chromatography of a 0.2% Triton X-100 cell extract obtained from SPTLC3 overexpressing HEK-293 cells. The fractions were assayed for SPT activity and probed for the presence of SPTLC1, SPTLC2 and SPTLC3 on a Western blot. The arrows mark the elution fractions of the marker proteins (masses shown in kDa). The maximal SPT activity eluted in fraction 10, which corresponds to a mass of 460–480 kDa. In all fractions that displayed SPT activity we observed the co-elution of the three SPT subunits. The intensity of the signal correlated with the measured SPT activity, and showed a maximum intensity in fraction 10. This result suggests that the mammalian SPT enzyme has a functional size of 460–480 kDa. (b) Fractionation of a 0.2% Triton X-100 extract from human placenta tissue. The experimental conditions are the same as (a). The elution profile for SPTLC1 and SPTLC2 was identical to (a) (results not shown). (c) To exclude that the observed SPT complex is due to Triton X-100 insoluble complexes, wild-type HEK-293 cells were lysed in either 1% (w/v) OTG or 0.2% Triton X-100 (TX100) with a subsequent incubation at 37°C for 20 min. Both conditions are reported to resolve the formation of detergent insoluble complexes [11]. As before, we observed a parallel elution of the subunits at a molecular mass of approx 480 kDa.
To exclude the possibility that the observed complex is due to the formation of Triton X-100-insoluble protein aggregates, we performed the same experiments with HEK-293 cells, which were lysed in the presence of 1% (w/v) OTG (octylglucoside) or 0.2% Triton X-100, followed by incubation at 37°C for 20 min. Both conditions were reported to prevent the formation of detergent-insoluble membrane complexes [11]. In both experiments we observed the same elution profile as shown in Figure 2(b). The OTG lysed cells showed a sharp elution peak between fractions 9 and 11, whereas the Triton X-100 lysate eluted over a somewhat broader range. However, no smaller complexes or free subunits were detected, which indicates that the observed 480 kDa complex is not caused by an unspecific detergent effect. Interestingly, although these last experiments were done with wild-type HEK-293 cells, which do not natively express SPTLC3, we observed the same elution profile as with SPTLC3-overexpressing cells or placenta extract. This indicates that the presence or absence of SPTLC3 does not alter the size of the SPT complex.
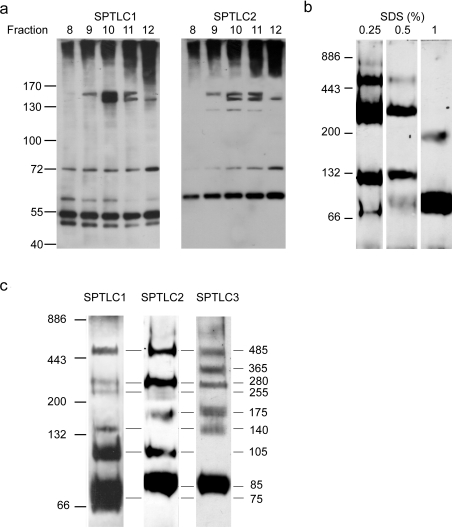
We finally also tried to covalently cross-link the SPT complex. To avoid unspecific cross-links due to the long carbon chains of commonly used bifunctional cross-linkers, we used limiting amounts of formaldehyde instead. After incubating the cells with formaldehyde for 1 h at 4°C, the cells were lysed in SDS and the proteins separated by size-exclusion chromatography. In fractions 9–11, we observed the elution of a stably cross-linked 160 kDa complex, which was positive for SPTLC1 and SPTLC2 (Figure 3a). The signal appears as a double band, which indicates the formation of two slightly different cross-links. A second complex was detected at a mass of 130 kDa, and contained only SPTLC2. This signal probably originates from the cross-linking of two SPTLC2 subunits. Interestingly, a third complex at approx. 72 kDa was also detected, which contained SPTLC1 and SPTLC2. The origin of this smaller complex is not yet known. The free SPT monomers were detected according to their theoretical masses at 55 kDa and 65 kDa for SPTLC1 and SPTLC2 respectively. Owing to the limited spatial resolution and the appearance of a smeary signal in the high-molecular-mass region of the gel, the detection of higher-molecular-mass cross-linked complexes was not possible.
Figure 3. Crosslinking and electrophoretic analysis of the SPT complex.
(a) HEK-293 cells were treated with limiting amounts of formaldehyde for 1 h at 4°C, lysed in 0.5% SDS and fractionated over a size-exclusion column. Fractions 8–12 were separated by SDS/PAGE (7% gels) and immunostained for SPTLC1 and SPTLC2. We observed the formation of a stably cross-linked complex of approx. 160 kDa, which was positive for SPTLC1 and SPTLC2. Due to the limited spatial resolution and the appearance of a smeary signal on top of the gel we could not detect higher molecular mass cross-linked complexes. (b) HEK-293 cells were lysed in BN buffer with increasing amounts of SDS (0.25–1%). The extract was separated by BN-PAGE, blotted on to PVDF and probed for the SPTLC2 subunit. The pattern shows the gradual breakdown of the full SPT complex into several smaller sub-complexes. The intensity of the breakdown correlates with the SDS concentration used in the lysis buffer. (c) BN-PAGE of human placenta extract. The homogenized tissue was lysed in BN-PAGE buffer containing 0.5% SDS and separated by BN-PAGE (7–15% gels) as described above. The gel was blotted and stained separately for SPTLC1, SPTLC2 and SPTLC3. As before, we observed a specific breakdown of the SPT complex into various sub-complexes. Most of those complexes showed a positive signal for two or all three SPT subunits. The masses of these complexes was deduced from the molecular mass of defined marker proteins. The largest complex was detected with a molecular mass of approx 480 kDa, which is in very good agreement with the deduced mass of 480 kDa from the size exclusion chromatography shown in Figure 2. Molecular mass markers are shown to the left-hand side of the Figures in kDa.
Separation of the SPT complex by semi-denaturing BN-PAGE
We therefore tried to further analyse the SPT complex using a modified BN-PAGE system. The cells were lysed in the presence of low amounts of SDS on ice. These semi-denaturing conditions lead to a gradual breakdown of the full size SPT complex into various sub-complexes. These sub-complexes were separated on a BN-PAGE system in the absence of SDS to avoid the further breakdown. Figure 3(b) demonstrates that the extent of this breakdown depends on the amount of SDS that was used in the lysis buffer. Low concentrations of SDS (0.25%) induced primarily larger sub-complexes, whereas higher SDS concentrations (1%) induced a further breakdown of the SPT complex into its monomeric and dimeric components (Figure 3b). The composition of the sub-complexes was analysed by immunostaining the blotted gel with antibodies against SPTLC1, SPTLC2 and SPTLC3. The molecular masses of the sub-complexes were deduced from the migration of defined marker proteins. Figure 3(c) shows the migration pattern of human placenta extract, which was lysed in the presence of 0.5% SDS and Western blotted individually for SPTLC1, SPTLC2 and SPTLC3. We observed a ladder-like migration pattern of the three SPT subunits. The largest signal we could detect had again a molecular mass of 480 kDa and was detected by antibodies against all three subunits. The mass of this complex is identical to the mass of the active SPT complex as it was determined by gel filtration. A second sub-complex with a molecular mass of 280 kDa was also positive for all three subunits. Interestingly a slightly smaller sub-complex at 255 kDa was positive only for SPTLC1 and SPTLC2, but not SPTLC3. The monomers of the individual subunits migrated to a slightly larger apparent size in comparison with the SDS/PAGE results. This discrepancy is probably caused by the absence of denaturants and detergents in the gel system. The observed breakdown of the SPT complex into various sub-complexes indicates, furthermore, that the native SPT complex is composed of more than two subunits.
DISCUSSION
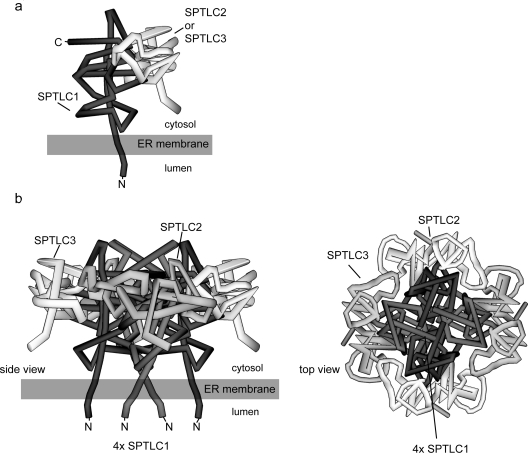
SPT is a member of a subfamily of POAS enzymes. Among the members of this family identified to date, only the eukaryotic (but not the prokaryotic) SPT is a membrane bound enzyme, whereas all other POAS members are soluble homodimers. Also the finding that SPT is composed of distinct subunits makes it different from other members of the POAS family. On the basis of the existence of two SPT subunits, SPTLC1 and SPTLC2, and in concordance to the other members of the POAS family, it has been supposed for a long time that SPT is a heterodimeric molecule. The identification of a third SPT subunit (SPTLC3) introduced a new aspect to this issue. The distinct homology between SPTLC2 and SPTLC3, as well as the presence of a consensus PLP-binding sequence in both proteins makes it probable that, in analogy to SPTLC2, SPTLC3 also interacts with SPTLC1. By testing this hypothesis in two independent pull-down assays with two different tissues (Figures 1c and 1d) we found strong evidence that all three subunits are located within the same complex. Also the fact that the precipitation of the V5-tagged SPTLC1 and SPTLC2 subunits leads to the concomitant precipitation of the untagged endogenous subunits (Figure 1b) indicates that the native SPT complex is, in fact, composed of several subunits. We could exclude, to a large extent, that the observed co-precipitation of the subunits is due to unspecific association or the generation of detergent insoluble complexes, since calnexin, which is an abundant ER membrane protein, was not detected in the precipitate. We also showed that the precipitation was specific for the targeted proteins, since no signal was seen when the antibodies were blocked by the immunization peptides (Figure 1d). The apparent molecular mass of the functional complex was deduced by size-exclusion chromatography to be 480 kDa (Figure 2). This is in very good agreement with the BN-PAGE analysis, where the largest detectable complex also showed a molecular mass of 480 kDa (Figure 3c). The generation of a fragmentation pattern by applying semi-denaturing conditions further supports the idea that the native SPT complex is composed of several subunits. We also demonstrated that the observed 480 kDa complex is not caused by the formation of Triton X-100 insoluble protein complexes. OTG or the incubation of a Triton X-100 extract for 20 min at 37°C prevents the formation of these complexes [11]. Also under these conditions the complex eluted at a mass of 480 kDa. Finally, we could demonstrate that chemical cross-linking of extract from native HEK-293 cells forms a stable 160 kDa complex, which consists of at least SPTLC1 and SPTLC2. Due to the limitation in the resolution of the gel we could not detect cross-linked complexes of a higher molecular mass. However, the fact that this 160 kDa complex was found in fractions 9–11, which equals a molecular mass of approx. 480 kDa, indicates that this cross-link is a fragment of an originally larger complex. The hypothesis that SPT is in fact a multi-subunit complex was put forward very recently by Hojjati and colleagues [4]. Based on the molecular mass of the complex, the native SPT holoenzyme could be composed of 6–8 subunits. This leads to questions about the composition and structure of this multi-subunit complex. The AONS homodimer contains two symmetrical active sites, which are located at the monomer–monomer interface [7,12]. In SPT, the PLP-binding domain is only present on SPTLC2 and SPTLC3, but not on the SPTLC1 subunit. An SPT dimer would hence contain one active site only. Based on the three-dimensional AONS structure, Gable and colleagues modelled a putative structure of a SPTL1–SPTLC2 dimer [13]. Although only one PLP-site is present in this model, they showed that residues of the SPTLC1 subunit are also involved in the formation of the active site. In analogy with the other members of the POAS family, it is also likely that the active site of the soluble, homodimeric prokaryotic SPT is formed at the interface of the two subunits. However, it must be emphasized that the mammalian form, in contrast to the prokaryotic SPT, is a membrane bound heterodimer. Earlier epitope-tagging experiments on the hamster SPTLC1 subunit showed that SPTLC1 is an integral membrane protein with a single transmembrane domain and that the orientation of its N-terminus is towards the ER lumen [14]. From this topological model the catalytic site of SPT can be deduced to be oriented towards the cytosolic space. In this context SPTLC1 could act as an anchor which binds SPTLC2 and/or SPTLC3 to the cytosolic side of the ER membrane. The cytosolic orientation of the catalytic site seems to be relevant, since the cytosol is the major pool of serine and palmitoyl-CoA. On the basis of these earlier findings, our own observations and the presumption that the active site is formed at the interface of the monomers, we developed a theoretical model of the SPT structure (Figure 4). In this model we claim that the full SPT is built by the association of four dimers. The dimers are formed by the interaction of SPTLC1 and SPTLC2 or SPTLC1 and SPTLC3 (Figure 4a). Those dimeric substructures are then clustered together into one octameric macromolecule (Figure 4b). The SPTLC1 subunits are hereby N-terminally bound to the ER membrane, whereas the C-terminal part forms a cytosol-oriented active site by interacting with the opposing SPTLC2 or SPTLC3 subunit. The fact that we observe the identical elution profiles for SPTLC3-overexpressing and SPTLC3-deficient wild-type HEK-293 cells indicates that within this complex the presence of SPTLC2 and SPTLC3 are interchangeable. The similar structure and the existence of a PLP-binding domain in both proteins make it possible that SPTLC2 could be replaced by SPTLC3 and vice versa. Such a dynamic exchange would explain why we observe the same elution profile in wild-type and SPTLC3 overexpressing HEK-293 cells (Figure 2). Tissues with high levels of SPTLC3 expression, such as the placenta, are therefore mainly forming SPTLC1–SPTLC3 dimers, whereas intermediate expression levels of SPTLC2 or SPTLC3 would result in the mixed presence of both subunits in the complex. Since SPTLC2 and SPTLC3 have almost the same molecular mass, the different complexes will not be distinguishable by mass alone. Currently, the biochemical differences between SPTLC2 and SPTLC3 are not clear. However, the fact that cells have a set of two distinct, but functionally redundant, subunits seems to be beneficial for the cell. The pronounced expression difference for SPTLC2 and SPTLC3 between various tissues [6] speaks for a tissue- and cell-type-specific role for these isoforms, probably as an adaptation to the tissue specific requirements of sphingolipid synthesis.
Figure 4. Theoretical model of the SPT complex structure.
The model is based on the following assumptions. First, that the N-terminal end of SPTLC1 is bound to the ER membrane; secondly, that SPTLC2 and SPTLC3 are not interacting directly with the ER membrane; thirdly, that the active site of SPT is formed at the interface between two monomers; and, finally, that SPTLC2 can be dynamically replaced by SPTLC3 within the complex. (a) The SPTLC2 or SPTLC3 monomers form a dimeric base structure with SPTLC1, which is then assembled to its final octameric state. (b) Putative model of the octameric SPT complex. In the conformation shown, two SPTLC1–SPTLC2 and two SPTLC1–SPTLC3 dimers are assembled together to form an octameric circular structure. The stoichiometry of SPTLC2 and SPTLC3 within the complex depends on their individual cellular expression levels. The final complex contains four active sites, which are located at the monomer–monomer interface. The N-termini of the SPTLC1 subunits are bound to the ER membrane in a manner that the catalytic head group protrudes towards the cytosolic compartment of the cell.
In conclusion, our results provide strong evidence that SPT is not a dimer, but is composed of several subunits. The molecular mass of the complex was determined to be approx. 480 kDa by size-exclusion chromatography and native protein electrophoresis. Based on these findings we propose a model in which the native SPT enzyme is an octamer composed of four SPTLC1–SPTLC2 or SPTLC1–SPTLC3 dimers. We also propose that the presence of SPTLC2 and SPTLC3 within this complex is dynamically interchangeable and depends on the fluctuating expression levels for SPTLC2 and SPTLC3 in the individual tissues. However, further experiments are necessary to validate this model.
Acknowledgments
We thank the Hartmann Müller Stiftung, the Herzog Egli Foundation, the EMDO Foundation and the Foundation for Scientific Research (University of Zurich) for financially supporting this work.
References
- 1.Hannun Y. A., Luberto C., Argraves K. M. Enzymes of sphingolipid metabolism: from modular to integrative signaling. Biochemistry. 2001;40:4893–4903. doi: 10.1021/bi002836k. [DOI] [PubMed] [Google Scholar]
- 2.Hannun Y. A., Obeid L. M. The ceramide-centric universe of lipid-mediated cell regulation: stress encounters of the lipid kind. J. Biol. Chem. 2002;277:25847–25850. doi: 10.1074/jbc.R200008200. [DOI] [PubMed] [Google Scholar]
- 3.Merrill A. H., Jr, Nixon D. W., Williams R. D. Activities of serine palmitoyltransferase (3-ketosphinganine synthase) in microsomes from different rat tissues. J. Lipid Res. 1985;26:617–622. [PubMed] [Google Scholar]
- 4.Hojjati M. R., Li Z., Jiang X. C. Serine palmitoyl-CoA transferase (SPT) deficiency and sphingolipid levels in mice. Biochim. Biophys. Acta. 2005;1737:44–51. doi: 10.1016/j.bbalip.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 5.Hanada K. Serine palmitoyltransferase, a key enzyme of sphingolipid metabolism. Biochim. Biophys. Acta. 2003;1632:16–30. doi: 10.1016/s1388-1981(03)00059-3. [DOI] [PubMed] [Google Scholar]
- 6.Hornemann T., Richard S., Rutti M. F., Wei Y., von Eckardstein A. Mammalian serine-palmitoyltransferase: cloning and initial characterisation of a new subunit. J. Biol. Chem. 2006;281:37275–37281. doi: 10.1074/jbc.M608066200. [DOI] [PubMed] [Google Scholar]
- 7.Alexeev D., Alexeeva M., Baxter R. L., Campopiano D. J., Webster S. P., Sawyer L. The crystal structure of 8-amino-7-oxononanoate synthase: a bacterial PLP-dependent, acyl-CoA-condensing enzyme. J. Mol. Biol. 1998;284:401–419. doi: 10.1006/jmbi.1998.2086. [DOI] [PubMed] [Google Scholar]
- 8.Schagger H., von Jagow G. Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal. Biochem. 1991;199:223–231. doi: 10.1016/0003-2697(91)90094-a. [DOI] [PubMed] [Google Scholar]
- 9.Sprong H., Degroote S., Nilsson T., Kawakita M., Ishida N., van der Sluijs P., van Meer G. Association of the Golgi UDP-galactose transporter with UDP-galactose:ceramide galactosyltransferase allows UDP-galactose import in the endoplasmic reticulum. Mol. Biol. Cell. 2003;14:3482–3493. doi: 10.1091/mbc.E03-03-0130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ikushiro H., Hayashi H., Kagamiyama H. Bacterial serine palmitoyltransferase: a water-soluble homodimeric prototype of the eukaryotic enzyme. Biochim. Biophys. Acta. 2003;1647:116–120. doi: 10.1016/s1570-9639(03)00074-8. [DOI] [PubMed] [Google Scholar]
- 11.Brown D. A., Rose J. K. Sorting of GPI-anchored proteins to glycolipid-enriched membrane subdomains during transport to the apical cell surface. Cell. 1992;68:533–544. doi: 10.1016/0092-8674(92)90189-j. [DOI] [PubMed] [Google Scholar]
- 12.Webster S. P., Alexeev D., Campopiano D. J., Watt R. M., Alexeeva M., Sawyer L., Baxter R. L. Mechanism of 8-amino-7-oxononanoate synthase: spectroscopic, kinetic, and crystallographic studies. Biochemistry. 2000;39:516–528. doi: 10.1021/bi991620j. [DOI] [PubMed] [Google Scholar]
- 13.Gable K., Han G., Monaghan E., Bacikova D., Natarajan M., Williams R., Dunn T. M. Mutations in the yeast LCB1 and LCB2 genes, including those corresponding to the hereditary sensory neuropathy type I mutations, dominantly inactivate serine palmitoyltransferase. J. Biol. Chem. 2002;277:10194–10200. doi: 10.1074/jbc.M107873200. [DOI] [PubMed] [Google Scholar]
- 14.Yasuda S., Nishijima M., Hanada K. Localization, topology, and function of the LCB1 subunit of serine palmitoyltransferase in mammalian cells. J. Biol. Chem. 2003;278:4176–4183. doi: 10.1074/jbc.M209602200. [DOI] [PubMed] [Google Scholar]