Abstract
Pi (inorganic phosphate) limitation severely impairs plant growth and reduces crop yield. Hence plants have evolved several biochemical and morphological responses to Pi starvation that both enhance uptake and conserve use. The mechanisms involved in Pi sensing and signal transduction are not completely understood. In the present study we report that a previously uncharacterized transcription factor, BHLH32, acts as a negative regulator of a range of Pi starvation-induced processes in Arabidopsis. In bhlh32 mutant plants in Pi-sufficient conditions, expression of several Pi starvation-induced genes, formation of anthocyanins, total Pi content and root hair formation were all significantly increased compared with the wild-type. Among the genes negatively regulated by BHLH32 are those encoding PPCK (phosphoenolpyruvate carboxylase kinase), which is involved in modifying metabolism so that Pi is spared. The present study has shown that PPCK genes are rapidly induced by Pi starvation leading to increased phosphorylation of phosphoenolpyruvate carboxylase. Furthermore, several Arabidopsis proteins that regulate epidermal cell differentiation [TTG1 (TRANSPARENT TESTA GLABRA1), GL3 (GLABRA3) and EGL3 (ENHANCER OF GL3)] positively regulate PPCK gene expression in response to Pi starvation. BHLH32 can physically interact with TTG1 and GL3. We propose that BHLH32 interferes with the function of TTG1-containing complexes and thereby affects several biochemical and morphological processes that respond to Pi availability.
Keywords: Arabidopsis, basic helix–loop–helix (bHLH) protein, gene expression, inorganic phosphate (Pi) starvation, phosphoenolpyruvate carboxylase kinase (PPCK), root hair formation
Abbreviations: AtPT1, phosphate transporter 1; bHLH, basic helix–loop–helix; CPC, CAPRICE; Ct, threshold cycle value; EGL3, ENHANCER OF GLABRA3; GFP, green fluorescent protein; GL3, GLABRA3; GST, glutathione S-transferase; LPI, low Pi insensitive; MS, Murashige–Skoog; PEPC, phosphoenolpyruvate carboxylase, PEPCK, PEPC kinase; PHR1, PHOSPHATE STARVATION RESPONSE 1; Pi, inorganic phosphate; RT, reverse transcriptase; SQD1, UDP-sulfoquinovose synthase 1; TTG1, TRANSPARENT TESTA GLABRA1; WER, WEREWOLF
INTRODUCTION
Phosphorus is an essential macronutrient for the growth, development and reproduction of plants. It is absorbed by plants almost entirely in the form of Pi (inorganic phosphate), whose availability frequently limits crop yield [1]. Although Pi is relatively abundant in many soils, much of it is unavailable for uptake because it forms insoluble complexes with cations. Plants respond to Pi limitation in several ways [1–4]. They enhance Pi uptake by modifying root architecture and growth, including an increase in the number and length of root hairs, expression of Pi transporters, induction of endogenous and secreted phosphatases and RNases, and exudation of organic acids. They also limit the metabolic requirements for Pi by engaging alternative respiratory pathways and bypassing Pi-requiring steps, and by increasing the accumulation of anthocyanins. To achieve these responses, the expression of many genes are altered in Pi starvation. Microarray experiments have identified genes showing different response kinetics to Pi starvation [5–7]. For example, ‘early’ responses (observed after 4 h of Pi starvation) can be distinguished from ‘late’ responses (observed after 100 h) [5]. Many of the ‘early’ responsive genes are also sensitive to other types of stress [5].
Over the last five years, some of the molecular mechanisms through which cellular Pi status is sensed in plants have been revealed, but the picture is far from complete. The Arabidopsis gene PHR1 (PHOSPHATE STARVATION RESPONSE 1) encodes a single MYB transcription factor required for the induction of a subset of the genes whose expression is activated in Pi starvation [8]. Genes that are controlled by PHR1 include the Pi transporter AtPT1 (ARAth, Pht1;1), the acid phosphatase AtACP5 and the ribonuclease AtRNS1. PHR1 is thought to be a positive regulator of some, but not all, Pi starvation-induced gene expression. Mutations in Arabidopsis PSR1 (PHOSPHATE STARVATION RESPONSE 1) reduce the activities of Pi starvation-inducible isoforms of ribonuclease and acid phosphatase, whereas PDR2 (PHOSPHATE DEFICIENCY RESPONSE 2) is thought to link Pi-sensing to root development [9,10], but neither gene has been characterized. Recently several lpi (low Pi insensitive) mutants have been reported [11]. In a similar manner to PHR1, the LPI genes affect only a subset of Pi starvation responses; for LPI genes, these include root development and expression of some Pi starvation-induced genes, but neither anthocyanin accumulation nor root hair formation. In Pi starvation some of the effects of PHR1 are mediated by expression of the microRNA miR399. This suppresses expression of UBC24 (an E2 ubiquitin-conjugating enzyme), the product of the PHO2 gene, and leads to over-accumulation of Pi [12–15]. The targets of the ubiquitin-conjugating enzyme, and the functional effects of their ubiquitination, are not known, but the miR399/PHO2 pathway is thought to affect the allocation of Pi within the plant [14].
The phr1 mutation does not affect the increased number and length of root hairs found in Pi starvation [8]. Recently it was shown that the Arabidopsis SUMO E3 ligase AtSIZ1 negatively regulates some Pi starvation responses such as increased root hair number and length that do not involve PHR1 [16]. Furthermore, mutations in the inositol polyphosphate kinase gene AtIPK1 also impinge on Pi status signalling to root hair development: mutants have an elevated Pi content but fail to repress root hair growth fully in Pi sufficiency [17]. As well as its role as a negative regulator, AtSIZ1 also positively regulates some Pi starvation responses in which PHR1 is involved, and can SUMOylate PHR1 in vitro [16]. There is also evidence for negative regulation in the induction of the Pi transporter gene AtPT2 (ARAth, Pht1;4) in Pi starvation [18]. Thus the available data suggest that plants respond to Pi starvation through multiple different sensing and signalling mechanisms and that several components remain to be identified.
The increased formation of root hairs in Pi starvation is particularly striking because it can increase the total root surface area by an order of magnitude, and is more sensitive to Pi limitation than to deficiencies in most other nutrients [19]. Root epidermal cells differentiate in a position-dependent manner, with hair cells overlying two cortical cell files and non-hair cells overlying a single cortical file [20]. Much information is now available about genes involved in these differentiation events. GL3 (GLABRA3) and EGL3 (ENHANCER OF GL3) encode bHLH (basic helix–loop–helix) proteins that together specify non-hair cell fate. GL3 and EGL3 act redundantly in a complex with the WD40 protein TTG1 (TRANSPARENT TESTA GLABRA1) and the R2R3 MYB protein WER (WEREWOLF). This complex functions by inducing expression of the homeodomain protein GL2 (GLABRA2) in non-hair cells. Hair cell fate is specified by the single MYB proteins CPC (CAPRICE) and TRY (TRIPTYCHON), negative regulators of GL2 [21,22]. Surprisingly, expression of GL3 and EGL3 occurs primarily in developing hair cells. The current model [23] involves movement of GL3 and EGL3 from hair to non-hair cells, where the TTG1–GL3–EGL3–WER complex induces expression of GL2 and CPC, and reciprocal movement of CPC to hair cells where it reduces expression of GL2 but increases expression of GL3 and EGL3. Hence the specification of non-hair versus hair cell fate depends on the cellular distribution of TTG1, GL3, EGL3 and CPC. Any factor that perturbs this balance could affect root hair formation either positively or negatively. Pi starvation leads to ectopic development of hairs in the non-hair position [24], though, in addition, a change in anatomy leads to an increase in hair cell files [25]. However, the mechanisms by which Pi starvation is first sensed and then subverts the normal differentiation programme are not known.
One of the metabolic responses to Pi starvation is thought to be the bypass of the pyruvate kinase step in glycolysis via induction of PEPC (phosphoenolpyruvate carboxylase) [1,3,4,26]. PEPC is itself regulated by phosphorylation, which activates the enzyme by reducing its sensitivity to feedback inhibition by malate [27,28]. Control of PEPC is mainly exerted at the level of expression of PPCK (PEPC kinase) genes [29,30]. In the present study we show that expression of the PPCK1 and PPCK2 genes in Arabidopsis is rapidly induced in Pi starvation, consistent with a role for activation of flux through PEPC as an early part of the Pi starvation response. This has led us to identify several genes that affect Pi starvation responses, some of which have previously been implicated in controlling epidermal cell differentiation and gene expression. One newly identified component, BHLH32, functions as a negative regulator of a range of biochemical and morphological processes that are induced in Pi starvation.
EXPERIMENTAL
Plant growth conditions
Arabidopsis suspension culture cells were derived from the Landsberg erecta ecotype [31]. Cells were grown photomixotrophically in 500 ml flasks containing 200 ml of culture medium [1×MS (Murashige–Skoog) salts with minimal organics, 0.5 mg/l α-naphthaleneacetic acid, 0.05 mg/l kinetin (all obtained from Sigma) and 3% (w/v) sucrose (pH 5.8)]. Suspension cultures were grown at 20 °C in a continuous low fluence rate of white light (20 μmol·m−2·s−1) with constant shaking (110 rev./min) and were subcultured every week by a 1:10 dilution [32]. For studies of the effects of Pi, 7-day-old cells were collected and then cultured in MS micronutrient medium (6.2 mg/l H2BO3, 0.025 mg/l CoCl2, 0.025 mg/l CuSO4, 27.8 mg/l FeSO4, 16.9 mg/l MgSO4, 0.25 mg/l NaMoO3, 0.83 mg/l KI and 8.6 mg/l ZnSO4; all from Sigma) containing 50 mM sucrose, 5 mM KNO3 and KH2PO4 as indicated. Cells were then collected, frozen in liquid nitrogen and stored at −80°C.
For seedling culture, the phr1 and bhlh32 (SALK_013517) lines were in the Columbia background, while gl3, egl3 and ttg1 were in the Landsberg erecta background. The Salk_013517 T-DNA insert line (obtained from the Nottingham Arabidopsis Stock Centre) was self-fertilized to obtain a homozygous line as judged by PCR. Seeds were sterilized and then germinated in a 250 ml flask of 100 ml 1/5×MS macronutrients with 1.25 mM KH2PO4, 1×MS micronutrient medium containing 50 mM sucrose and 2.5 mM Mes buffered to pH 6.0 by addition of Bis/Tris propane (Sigma). Seedlings were grown at 22°C in a continuous low fluence rate of white light (20 μmol·m−2·s−1) with constant shaking (110 rev./min). Seven days after germination seedlings were transferred to 1/5×MS without KH2PO4, 1×MS micronutrient medium containing 50 mM sucrose and 2.5 mM Mes (pH 6.0) for 12 h or as indicated. The whole seedlings were harvested for gene expression analysis.
For analysis of root hair development, seeds were germinated on 1×MS complete medium, 3% sucrose, 2.5 mM Mes (pH 5.7), 0.8% agar for 5 days, then transferred to 1/20×MS macronutrients, 1×MS micronutrients, 3% sucrose, 2.5 mM Mes (pH 5.7), 1.2% agar with a total Pi concentration of either 1.25 mM or 0.06 mM for 3 days. Roots were photographed on agar.
RNA extraction, RT (reverse transcriptase)-PCR and real-time quantitative PCR
Total RNA was extracted using RNeasy plant mini kits (Qiagen) and then treated with DNase (DNA-free; Ambion) to remove contaminating genomic DNA. Total RNA (2.5 μg) was used to synthesize first strand cDNA and subsequently PCR was performed using specific primers for PPCK1, PPCK2 and ACTIN2 as described previously [32]: primers for AtPT1 (At5g43350), forward 5′-acagatatcgagcttgagga-3′, reverse 5′-ttggcgaagaagaaagtaag-3′; primers for AtPT2 (At2g38940), forward 5′-cgatcattccacttccttct-3′, reverse 5′-acagcttttggctcatgtcc-3′; primers for SQD1 (UDP-sulfoquinovose synthase; At4g33030), forward 5′-ctactgctctccacttgtcc-3′, reverse 5′-tttggagttccatactcacc-3′; primers for BHLH32 (At3g25710), forward 5′-gtctctcctcctccttccatgg-3′, reverse 5′-cttgacttaattaactaattataacatcg-3′. All PCR was performed using the same programme: 94°C for 2 min, 25 cycles of 94°C for 30 s, 60°C for 30 s, 72°C for 1 min; and a final extension of 72°C for 5 min.
Real-time quantitative PCR was performed in 25 μl reaction mixtures containing 5 μl of 10-fold diluted first-strand cDNA, 0.3 μM of each primer and 1×QuantiTect SYBR Green PCR Master Mix (Qiagen). The following primers were used: PPCK1 forward (5′-gaagattctcaaccgaacaagcatt-3′), PPCK1 reverse (5′-ttggtaggctggcggaatcta-3′); PPCK2 forward (5′-tgtatcatcggaagcgaaggatt-3′), PPCK2 reverse (5′-ttgctgaaacacacaactacgtttg-3′). The relative expression level was calculated using the comparative Ct (threshold cycle value) method. The expression level of UBQ10 (At4g05320) was measured using the primers described in [14] and was taken as the housekeeping control. Fold changes (2−ΔΔCt) were expressed relative to wild-type seedlings without Pi starvation (zero time).
Antibody production and Western blot analysis
The soybean GmPPCK2 gene [33] was cloned into a modified version of pPICZα (Invitrogen) which places FLAG and His10 tags at the N-terminus [34] and was expressed in Pichia pastoris SMD116H cells (Invitrogen). The protein was purified from the extracellular fluid using chromatography on Phenyl Sepharose [35] and Chelating Sepharose Fast Flow (Amersham Biosciences), and antisera were raised in rabbits. PPCK was detected after SDS/PAGE as described in [36].
Analytical methods
For measurements of anthocyanin and Pi content, seedlings were grown on agar plates with 1/2×MS complete medium for 5 days and then transferred to plates plus (0.625 mM) or minus (0.04 mM) Pi for 7 days. Whole seedlings were extracted. The contents of anthocyanins and Pi were measured as described in [37] and [17] respectively. Crude extracts of seedlings or suspension cultured cells were prepared and used for determination of the L-malate sensitivity of PEPC as described previously [30]. Nuclei were isolated from 7-day-old seedlings using the CelLytic™ PN (plant nuclei) isolation kit (Sigma) according to the manufacturer's instructions. The GFP (green fluorescent protein)–BHLH32 fusion was detected using anti-GFP as the primary antibody as described in [36].
Complementation of bhlh32
A full-length cDNA encoding BHLH32 was amplified by PCR (forward primer 5′-caccatgtacgcaatgaaagaagaag-3′, reverse primer 5′-tcccattttggatccctaattaactaaccc-3′) and cloned into the Gateway entry vector pENTR™/SD/D-TOPO (Invitrogen). The fragment was transferred into the binary vector pGWB6 (Invitrogen) to yield the caulifower mosaic virus 35S promoter:GFP–BHLH32 fusion using the Gateway LR recombination reaction. The construct was stably transformed into bhlh32 mutant plants and homozygous lines were selected.
Clone construction, transformation and yeast-two-hybrid analysis
Full-length cDNAs encoding PHR1, BHLH32, GL3, EGL3 and TTG1 were amplified by PCR. Primers for PHR1 were: forward 5′-ctagggaattcatggaggctcgtcc-3′, reverse 5′-tccgagggatcccagcaaagacca-3′; primers for BHLH32, forward 5′-acgaattcatgtacgcaatgaaagaagaag-3′, reverse 5′-tcccattttggatccctaattaactaaccc-3′; primers for GL3, forward 5′-gggagctcgaattcgccatggctaccggacaaaacaga-3′, reverse 5′-ggtctagaggatcctcaacagatccatgcaaccc-3′; primers for EGL3, forward 5′-atggggatgaagaattcatggcaaccggag-3′, reverse 5′-ctaaaccttggatccttaacatatccatgc-3′; and primers for TTG1, forward 5′-cacacgaattcatggataattcagctcc-3′, reverse 5′-aggcctagcctgcaggaattatgtagcg-3′. Products were cloned into pTOPO-TA (Invitrogen). The full-length cDNAs for PHR1, BHLH32, GL3 and EGL3 were then released from their respective pTOPO clones as EcoRI/BamHI fragments, except for TTG1 which was released as an EcoRI/PstI fragment, and the fragments were ligated into the corresponding sites of the vectors pGBKT7 or pGADT7 (BD Biosciences). Pairs of different combinations of these pGBKT7 and pGADT7 vectors were co-transformed into yeast strain AH109 competent cells (BD Biosciences) and plated onto synthetic dropout medium lacking leucine, tryptophan, and histidine and supplemented with X-α-gal to investigate the interaction of these hybrid proteins.
α-Galactopyranosidase activity was assayed with p-nitrophenyl α-galactopyranoside (Sigma) according to a protocol provided with the Matchmaker system (BD Biosciences). One unit of α-galactopyranosidase was defined as the amount of enzyme that hydrolyses 1 μmol PNPG (p-nitrophenyl β-D-glucopyranoside) to p-nitrophenol and D-galactose in 1 min at 30°C in acetate buffer (pH 4.5).
In vitro pulldown assay
BHLH32, GL3 and TTG1 were recombined in-frame to pGBKT7 as described above. They were used as templates in a coupled transcription–translation system (TNT®T7; Promega) to synthesize protein to be used as prey. BHLH32 was also recombined in-frame to pET-41a(+) (Novagen) to produce GST (glutathione S-transferase)-tagged protein as a bait. Subsequently the pulldown assay was performed according to the manufacturer's protocol for the MagneGST™ Pull-Down System (Promega).
RESULTS AND DISCUSSION
PEPCK gene expression responds rapidly to Pi starvation
In experiments on PPCK expression in an Arabidopsis cell culture [31] we noted that expression increased towards the end of each 7-day growth period and decreased following transfer to fresh medium. Systematic investigation of this phenomenon showed that depletion of Pi in the medium led to induction of PPCK expression. The effect of exposing Arabidopsis cells to reduced Pi concentrations for 3 h is illustrated by semi-quantitative RT-PCR in Figure 1(A). Expression of PPCK2 changes dramatically over the range 0–2.4 mM Pi, whereas there is a lesser change of expression of PPCK1. To test whether the Pi starvation-induced expression of PPCK genes affected the phosphorylation state of PEPC, we measured the sensitivity of PEPC to inhibition by malate (a well-established indicator of the phosphorylation state) [29]. The increase in PPCK expression in response to Pi starvation was accompanied by a decrease in the malate sensitivity of PEPC (Figure 1B). These data indicate that Pi starvation increases the phosphorylation state of PEPC, thus demonstrating the functional effect of increased PPCK expression.
Figure 1. Pi starvation induces PPCK gene expression and increases the phosphorylation state of PEPC.
(A) and (B) 7-day-old cells were transferred to medium containing the indicated concentration of KH2PO4 for 3 h. (A) Cells were collected, RNA was isolated and gene expression was analysed by RT-PCR. (B) Cells were treated as in (A). The malate sensitivity of PEPC in cell extracts is expressed as the percentage inhibition given by 1 mM L-malate. Error bars are S.D. for n=5 measurements in one representative experiment. Relative to the value at 2.4 mM Pi, malate sensitivity is significantly reduced at 0.6 mM Pi and at zero Pi (P<0.01 and P<0.001 respectively, by Student's t test. (C) 7-day-old seedlings were transferred to MS micronutrient medium containing 50 mM sucrose and 5 mM KNO3 without Pi for the times indicated. Seedlings were collected, RNA was isolated and gene expression was analysed by RT-PCR. (D) Left-hand panel, seedlings were harvested after growth for 7 days with 1.25 mM Pi or after a further 3 h without Pi and PPCK protein was detected by Western blot analysis following SDS/PAGE of extracts (15 μg protein); right-hand panel, purified tagged soybean PPCK resolved on SDS/PAGE and stained with Coomassie blue. The Mr of this protein is slightly bigger than those of the two Arabidopsis PPCKs.
The rapid induction of PPCK gene expression in response to Pi starvation can also be detected using seedlings in liquid culture. Increased expression of PPCK genes was detectable within 1 h of the transfer of seedlings to Pi-deficient medium (Figure 1C), and an increase in the abundance of PPCK protein was evident by 3 h after transfer as judged by Western blot analysis (Figure 1D; the purified tagged soybean PPCK, against which the antiserum used in the present study was raised, is shown in the right-hand panel). The phosphate transporter gene AtPT2 was also rapidly induced, whereas AtPT1 was not affected in the time scale used. SQD1, which is involved in sulfolipid synthesis [38], also showed no increase in expression within 3 h of Pi starvation. This is in agreement with data from real-time quantitative RT-PCR experiments using hydroponically grown Arabidopsis, in which expression of SQD1 was not induced after 4 h of Pi starvation [5]. In that study [5] it was reported that many of the genes that respond rapidly to Pi starvation are also induced by other stresses, and it was argued that rapid responses to Pi starvation are actually stress responses. However PPCK expression in Arabidopsis is not increased by such stresses as drought, high light or cold ([32]; Z. H. Chen and H. G. Nimmo, unpublished work). Moreover, as is the case for several genes induced by Pi starvation [39,40], repression of PPCK expression by Pi was replicated by the non-metabolizable analogue phosphite (results not shown), indicating that Pi status affects PPCK expression rapidly through a signalling system that detects Pi and phosphite rather than through direct effects on metabolism or a stress response. Thus the induction of PPCK expression and the resultant phosphorylation of PEPC seem to be two of the fastest known responses specifically to Pi starvation.
These data thus define a new, rapid response to Pi starvation in Arabidopsis, namely the induction of PPCK gene expression. The consequence is increased phosphorylation of PEPC, one of the key enzymes involved in central metabolism in plants. These findings significantly extend the microarray experiments that showed induction of the PPCK genes in Pi starvation [7] by emphasizing the speed and the functional consequence of the response. There have been several reports that genes encoding plant PEPCs are induced in Pi starvation [7,26,41–43], including the three Arabidopsis ppc genes that encode the phosphorylatable, ‘higher plant-type’ PEPCs [44]. The proposed function of the induction of PEPC in Pi starvation is to provide a bypass of the pyruvate kinase step in glycolysis, reduce the requirement for adenine nucleotides and thus ‘spare’ Pi for use in essential processes. Several other metabolic adaptations to Pi starvation are thought to provide the same general function of sparing Pi [1,3,4,26]. Hence it seems probable that the induction of PPCK expression and the resulting phosphorylation of PEPC serve to provide a rapid metabolic response to Pi starvation.
Identification of transcription factors required for the Pi starvation-induced expression of PPCK genes
The use of seedlings in liquid culture allowed us to test the involvement of several gene products in the response to Pi starvation using a number of well-defined mutants. Figure 2(A) shows results from quantitative RT-PCR experiments. In the phr1 mutant the induction of PPCK1 and PPCK2 expression in Pi starvation is markedly reduced but not eliminated. Another well-known effect of Pi starvation is to increase anthocyanin synthesis, and many regulatory genes for this pathway have been identified. Several of these regulators are additionally involved in controlling epidermal cell fate [45–47]. Figure 2(A) shows that the effect of Pi starvation on PPCK expression is compromised in ttg1, gl3 and egl3 mutant seedlings. However it is unaffected in hy5 (elongated hypocotyl5) mutant seedlings (results not shown). Figure 2(B) shows that the total Pi content of wild-type, ttg1, gl3 and egl3 seedlings is barely affected by 12 h in Pi -deficient medium. PPCK expression is clearly sensitive to a small pool of Pi rather than to total Pi content. The effects of mutations in TTG1, GL3 and EGL3 on induction of PPCK expression by Pi starvation are not simply due to a change in total Pi content.
Figure 2. Gene expression in mutants under Pi starvation.
Seedlings (7 days) were transferred to 1/5×MS without KH2PO4, 1×MS micronutrient medium containing 50 mM sucrose and 2.5 mM Mes (pH 6.0) for 12 h prior to harvesting. (A) Quantitative real-time PCR analysis of PPCK transcript abundance relative to that of UBQ10, values are means±S.D. (n=4). (B) Total Pi contents, means±S.D. (n=4). WT, wild-type.
TTG1 is a pleiotropic gene encoding a WD40 repeat protein involved in root hair initiation, trichome development, production of seed coat mucilage and anthocyanin synthesis [45]. GL3 and EGL3 are bHLH transcription factors that can interact with TTG1 in heterotrimeric TTG1–bHLH–MYB complexes to control various processes in the epidermis [47,48]. Since bHLH proteins are well-known for their ability to form heterodimers, we sought to identify other bHLHs with a role in Pi starvation. Microarray experiments had indicated that expression of BHLH32 is induced after 48 h of Pi starvation [6]. We therefore obtained a T-DNA insertion bhlh32 mutant from the Salk collection and isolated a homozygous line completely devoid of BHLH32 transcripts (Figure 3A). Figure 3(B) shows the response of bhlh32 to Pi starvation compared with the wild-type as time-courses of PPCK expression measured using quantitative RT-PCR following removal of Pi from the medium. Notably, the expression of PPCK2 in Pi-sufficient medium (zero time in Figure 3B) is some 4-fold higher in bhlh32 than in the wild-type, whereas that of PPCK1 is some 3-fold higher. In line with this, the malate sensitivity of PEPC is significantly lower in Pi-sufficient bhlh32 seedlings than in the wild-type (results not shown). However, the inductive effect of Pi starvation on PPCK expression is seen in both the wild-type and bhlh32. Thus BHLH32 acts as a negative regulator of PPCK expression in Pi sufficiency, but does not directly affect induction in Pi starvation.
Figure 3. Increased expression of PPCK and DFR genes in bhlh32.
(A) Seedlings were harvested 3 h after transfer to medium with (+) or without (−) Pi. Expression of BHLH32 and DFR was assessed by RT-PCR. The T-DNA insertion in bhlh32 (Salk_013517) is immediately after the end of the first exon of At3g25710 (results not shown). (B) Seedlings were transferred to medium without Pi for the indicated times. PPCK expression was assessed by quantitative real-time PCR as described for Figure 2. WT, wild-type.
TTG1 acts in the root, seed and shoot epidermis in conjunction with specific transcription factors to control either cell fate or biosynthetic processes [45,46,48]. Pi starvation affects both root biochemistry and root development. For example, it causes increases in secretion of organic acids, root/shoot ratio, root hair proliferation and length and lateral root number. It is therefore not surprising that TTG1 is involved in at least some of these responses, and our results show that a subset of bHLH proteins act with TTG1 in specific responses to Pi starvation. The morphological and biochemical responses of roots to Pi starvation may be linked by the involvement of these proteins.
BHLH32 affects root hair development, anthocyanin formation and Pi content
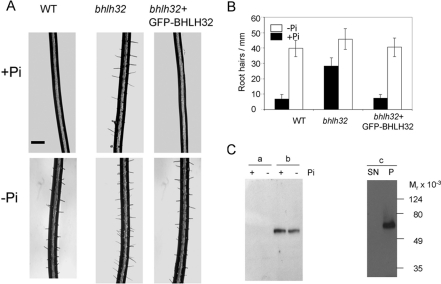
Because of the well-established connection between Pi nutrition and root hair development [2], we studied root hair production in bhlh32. Figures 4(A) and 4(B) clearly show that, in contrast to the wild-type, root hair formation in bhlh32 is not suppressed by growth in the presence of high levels of Pi. This phenotype is reversed by expression of a GFP–BHLH32 fusion protein from the cauliflower mosaic virus 35S promoter. The location of the fusion protein was examined first by centrifugation of seedling extracts and subsequently using isolated nuclei. Figure 4(C) shows that the fusion protein was predominantly nuclear, and was not extracted by high salt, suggesting that it was tightly bound to DNA. The bhlh32 mutant had no obvious growth phenotype apart from this effect on root hairs.
Figure 4. Expression of GFP–BHLH32 corrects the root hair phenotype of bhlh32.
Seedlings were grown as described in the Experimental section. (A) Representative images of Col0, bhlh32 and bhlh32 transformed with the GFP–BHLH32 fusion. The scale bar represents 100 μm. (B) Root hairs were counted from 0.75 mm to 1.75 mm from the root tip, values are means±S.D. (n=5). The bhlh32 line contains significantly more root hairs in the presence of Pi than the wild-type or bhlh32 expressing GFP–BHLH32 (P<0.001, Student's t test). (C) Western blot analysis of the location of GFP–BHLH32. Left-hand panel, seedlings of bhlh32 transformed with the GFP–BHLH32 fusion were grown as described in the Experimental section and harvested before (+Pi) or after (−Pi) 3 h of Pi starvation. Extracts were centrifuged at 12000 g and equal amounts (15 μg protein) were analysed by SDS/PAGE and Western blot analysis with an anti-GFP antibody. Lanes a, supernatants; lanes b, pellets. Right-hand panel, nuclei from seedlings in Pi-sufficient medium were prepared and subjected to high salt extraction (see the Experimental section); SN and P represent the soluble and particulate material from this extraction respectively.
When grown in Pi-sufficient conditions, bhlh32 contained significantly more total Pi and more anthocyanin than the wild-type (Figure 5), whereas phr1, in agreement with previous work [8], showed the opposite effect. We also monitored the expression of DFR, which encodes dihydroflavonol reductase, the key step in the formation of anthocyanins. Figure 3(A) shows that DFR expression in Pi-sufficient conditions was substantially increased in the bhlh32 mutant. Thus BHLH32 acts as a negative regulator of three processes, PPCK expression, root hair formation and anthocyanin production, all of which have in common the property that they respond to Pi starvation. Furthermore, complexes containing TTG1 were required for correct functioning of all of these processes.
Figure 5. bhlh32 contains increased anthocyanin and Pi.
Seedlings were grown as described in the Experimental section. (A) Anthocyanin content and (B) Pi content, values are means±S.D. (n=4). Relative to the wild-type, bhlh32 contains significantly more anthocyanin at both 0.625 mM Pi and 0.04 mM Pi, P<0.001 and P<0.01 respectively, and significantly more Pi at both concentrations, P<0.001 and P<0.05 respectively.
Physical interactions between transcription factors involved in Pi starvation
Short-term Pi starvation of seedlings (1–2 h) did not affect the transcript abundance of PHR1, TTG1, GL3, EGL3 and BHLH32 (results not shown). We therefore reasoned that early responses to Pi starvation may affect the location or activity of complexes involving some or all of these factors rather than the absolute abundance of the proteins, although induction of BHLH32 [6] may be involved in longer-term effects. As a first step towards identifying such complexes we have used the yeast-two-hybrid system. Figures 6(A) and 6(B) show that BHLH32 interacted physically with itself, with TTG1 and with GL3. However it did not appear to interact with either PHR1 or EGL3. Our data confirm the already reported interactions of TTG1 with GL3 and with EGL3 and the formation of GL3–EGL3 heterodimers [47,48]. PHR1 and GL3 both result in auto-activation when fused to the DNA-binding domain (results not shown) so we were unable to test whether they interacted. However, PHR1 did not seem to interact with either TTG1 or EGL3 (Figures 6A and 6B).
Figure 6. Physical interactions between components involved in the Pi starvation response.
(A) and (B) show interactions analysed in the yeast-two-hybrid system. (A) The indicated combinations of BD (DNA-binding domain) and AD (activation domain) vectors were co-transformed into yeast strain AH109 and plated as described in the Experimental section. Positive interactions are shown by the growth of blue colonies. (B) The strength of positive interactions was assessed by α-galactopyranosidase activity assays. Values are means±S.D. from four individual yeast colonies. (C) In vitro pulldown assay. BHLH32 was expressed as a GST-fusion protein in E. coli and immobilized onto glutathione-agarose beads and then incubated with 35S-labelled proteins expressed using the in vitro transcription–translation system. As a negative control, beads were loaded with GST. Bound proteins were resolved by SDS/PAGE and visualized using a Fuji phosphorimager.
To confirm these interactions using another method, we expressed a GST–BHLH32 fusion protein in Escherichia coli and immobilized it onto glutathione-agarose beads. We then used the beads to probe 35S-labelled proteins expressed by in vitro transcription/translation. Figure 6(C) shows that labelled BHLH32, GL3 and TTG1 were all captured by immobilized GST–BHLH32 but not by immobilized GST, thus confirming the results of the yeast-two-hybrid experiments.
The involvement of transcription factors in Pi starvation
The only transcription factor previously implicated in Pi starvation in Arabidopsis is the single MYB protein PHR1 [8]. This is involved in the induction of PPCK genes in Pi starvation but, in addition, it is clear that TTG1, GL3 and EGL3 are also required for full induction (Figure 2). In other systems, the role of TTG1 seems to be in a complex with bHLH and R2R3 MYB proteins [47]. In these complexes the bHLH interacts with both TTG1 and the MYB, which do not interact with each other. PHR1 does not contain the bHLH-interaction domain and we have no evidence to show that it interacts with any of the other factors that we have implicated in Pi starvation responses. We therefore suggest that the involvement of PHR1 in Pi starvation is quite separate to that of a TTG1–bHLH–MYB complex. This is reminiscent of seed mucilage production which is regulated by both TTG1-dependent and TTG1-independent pathways [49].
The three bHLH proteins previously implicated as positive factors in TTG1-dependent pathways, GL3, EGL3 and TT8 (TRANSPARENT TESTA8), show significant amino acid sequence similarity. As judged by yeast-two-hybrid analysis of truncated fragments, they all possess an N-terminal MYB-interacting domain, a central TTG1-interacting domain and a C-terminal bHLH domain [47,48]. BHLH32 is a much smaller protein, apparently lacking a MYB-interacting domain, though it can clearly bind to TTG1 and to some bHLHs, including itself (Figure 6). The function of BHLH32 in Arabidopsis has not previously been reported. Our data show that it negatively regulates root hair formation, anthocyanin accumulation and PPCK expression, all of which are induced by Pi starvation (Figures 3–5). In addition Pi uptake and storage are de-regulated in the bhlh32 mutant. However BHLH32 is not apparently required itself for the induction of root hair production, anthocyanin formation and PPCK expression (Figures 3–5).
How could BHLH32 affect such disparate processes, and could any of its effects be secondary? It is noteworthy that the bhlh32 mutant contains elevated levels of Pi. This could simply reflect increased root hair formation, although disruption of Pi homoeostasis may also be involved. However, while the increased Pi in bhlh32 would be expected to reduce the expression of PPCK and DFR, and the accumulation of anthocyanins, the opposite is observed. Hence BHLH32 must affect these processes quite independently of Pi content.
TTG1–bHLH–MYB complexes are involved in the induction of both DFR and PPCK genes in Pi starvation, though in the latter case PHR1 is also involved. However the underlying mechanisms are not understood. Our data demonstrate that BHLH32 can interact physically with TTG1 and with GL3. We therefore suggest that BHLH32 can interfere with TTG1-containing complexes such that its loss in the bhlh32 mutant partly mimics the effects of Pi starvation and thereby increases the expression of DFR and PPCK even in Pi sufficiency. This is compatible with the nuclear localization of the GFP–BHLH32 fusion protein (Figure 4). The events underlying the effects of Pi starvation on root hair formation are not understood, and this system is more complex because it involves intercellular movement of proteins [23]. Any factor that affects the cellular distribution of TTG1, GL3, EGL3 and CPC could affect root hair formation either positively or negatively. Hence it is possible that in root epidermal cells BHLH32 can interact with TTG1 and bHLHs and affect root hair formation in such a way that it is increased in the bhlh32 mutant.
BHLH32 thus seems to act as a negative regulator of several genes in Pi-sufficient conditions, in line with the concept proposed in [18]. This role of BHLH32 is quite different from that of PHR1, which is viewed as a positive regulator of Pi starvation responses [8]. The combination of negative regulation in the absence of the stimulus (i.e. Pi sufficiency) and positive regulation in response to the stimulus (Pi starvation) resembles the mechanism of plant responses to other environmental stimuli, including light, where positively acting photoreceptor signalling pathways overcome negative regulation of photomorphogenesis in darkness by components such as DET1 (DE-ETIOLATED 1) and COP1 (CONSTITUTIVE PHOTOMORPHOGENIC locus 1) [50].
Acknowledgments
This work was supported by the BBSRC (Biotechnology and Biological Sciences Research Council). We thank Dr Javier Paz-Ares (Departmento de Genetica Molecular de Plantas, Centro Nacional de Biotecnologia, Madrid, Spain) and Dr Alan Lloyd (The Institute for Cellular and Molecular Biology, University of Texas at Austin, Austin, Texas, U.S.A.) for the gift of phr1 and egl3 seed respectively. We thank Janet Laird for excellent technical assistance.
References
- 1.Raghothama K. G. Phosphate acquisition. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1999;50:665–693. doi: 10.1146/annurev.arplant.50.1.665. [DOI] [PubMed] [Google Scholar]
- 2.Bates T. R., Lynch J. P. Stimulation of root hair elongation in Arabidopsis thaliana by low phosphorus availability. Plant Cell Environ. 1996;19:529–538. [Google Scholar]
- 3.Vance C. P., Uhde-Stone C., Allan D. L. Phosphorus acquisition and use: critical adaptations by plants for securing a non-renewable resource. New Phytol. 2003;157:423–447. doi: 10.1046/j.1469-8137.2003.00695.x. [DOI] [PubMed] [Google Scholar]
- 4.Ticconi C. A., Abel S. Short on phosphate: plant surveillance and countermeasures. Trends Plant Sci. 2004;9:548–555. doi: 10.1016/j.tplants.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 5.Hammond J. P., Bennett M. J., Bowen H. C., Broadley M. R., Eastwood D. C., May S. T., Rahn C., Swarup R., Woolaway K. E., White P. J. Changes in gene expression in Arabidopsis shoots during phosphate starvation and the potential for developing smart plants. Plant Physiol. 2003;132:578–596. doi: 10.1104/pp.103.020941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu P., Ma L., Hu X., Wang M., Wu Y., Liu F., Deng X. W. Phosphate starvation triggers distinct alterations of genome expression in Arabidopsis roots and leaves. Plant Physiol. 2003;132:1260–1271. doi: 10.1104/pp.103.021022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Misson J., Raghothama K. G., Jain A., Jouhet J., Block M. S., Bligny R., Ortet P., Creff A., Somerville S., Rolland N., et al. A genome-wide transcriptional analysis using Arabidopsis thaliana Affymetrix gene chips determined plant responses to phosphate starvation. Proc. Natl. Acad. Sci. U.S.A. 2005;102:11934–11939. doi: 10.1073/pnas.0505266102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rubio V., Linhares F., Solano R., Martín A. C., Iglesias J., Leyva A., Paz-Ares J. A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev. 2001;15:2122–2133. doi: 10.1101/gad.204401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen D. L., Delatorre C. A., Bakker A., Abel S. Conditional identification of phosphate-starvation-response mutants in Arabidopsis thaliana. Planta. 2000;211:13–22. doi: 10.1007/s004250000271. [DOI] [PubMed] [Google Scholar]
- 10.Ticconi C. A., Delatorre C. A., Lahner B., Salt D. E., Abel S. Arabidopsis pdr2 reveals a phosphate-sensitive checkpoint in root development. Plant J. 2004;37:801–814. doi: 10.1111/j.1365-313x.2004.02005.x. [DOI] [PubMed] [Google Scholar]
- 11.Sánchez-Calderón L., López-Bucio J., Chacón-López A., Gutiérrez-Ortega A., Hernández-Abreu E., Herrera-Estrella L. Characterization of low phosphorus insensitive mutants reveals a crosstalk between low phosphorus-induced determinate root development and the activation of genes involved in the adaptation of Arabidopsis to phosphorus deficiency. Plant Physiol. 2006;140:879–889. doi: 10.1104/pp.105.073825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fujii H., Chiou T. Z., Lin S.-I., Aung K., Zhu J-K. A miRNA involved in phosphate-starvation response in Arabidopsis. Curr. Biol. 2005;15:2038–2043. doi: 10.1016/j.cub.2005.10.016. [DOI] [PubMed] [Google Scholar]
- 13.Aung K., Lin S.-I., Wu C.-C., Huang Y.-T., Su C.-L., Chiou T.-Z. pho2, a phosphate overaccumulator, is caused by a nonsense mutation in a microRNA399 target gene. Plant Physiol. 2006;141:1000–1011. doi: 10.1104/pp.106.078063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bari R., Pant B. D., Stitt M., Scheible W.-R. PHO2, microRNA399, and PHR1 define a phosphate-signaling pathway in plants. Plant Physiol. 2006;141:988–999. doi: 10.1104/pp.106.079707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chiou T.-Z., Aung K., Lin S.-I., Wu C.-C., Chiang S.-F., Su C.-L. Regulation of phosphate homeostasis by microRNA in Arabidopsis. Plant Cell. 2006;18:412–421. doi: 10.1105/tpc.105.038943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miura K., Rus A., Sharkhuu A., Yokoi S., Karthikeyan A. S., Raghothama K. G., Baek D., Koo Y. D., Jin J. B., Bressan R. A., Yun D-J., Hasegawa P. M. The Arabidopsis SUMO E3 ligase SIZ1 controls phosphate deficiency responses. Proc. Natl. Acad. Sci. U.S.A. 2005;102:7760–7765. doi: 10.1073/pnas.0500778102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stevenson-Paulik J., Bastidas R. J., Chiou S-T., Frye R. A., York J. D. Generation of phytate-free seeds in Arabidopsis through disruption of inositol polyphsosphate kinases. Proc. Natl. Acad. Sci. U.S.A. 2005;102:12612–12617. doi: 10.1073/pnas.0504172102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mukatira U. T., Liu C., Varadarajan D. K., Raghothama K. G. Negative regulation of phosphate starvation-induced genes. Plant Physiol. 2001;127:1854–1862. [PMC free article] [PubMed] [Google Scholar]
- 19.Gilroy S., Jones D. L. Through form to function: root hair development and nutrient uptake. Trends Plant Sci. 2000;5:56–60. doi: 10.1016/s1360-1385(99)01551-4. [DOI] [PubMed] [Google Scholar]
- 20.Schiefelbein J. W. Constructing a plant cell. The genetic control of root hair development. Plant Physiol. 2000;124:1525–1531. doi: 10.1104/pp.124.4.1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schiefelbein J. Cell-fate specification in the epidermis: a common patterning mechanism in the root and shoot. Curr Opin. Plant Biol. 2003;6:74–78. doi: 10.1016/s136952660200002x. [DOI] [PubMed] [Google Scholar]
- 22.Serna L. A network of interacting factors triggering different cell fates. Plant Cell. 2004;16:2258–2263. doi: 10.1105/tpc.104.160931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bernhardt C., Zhao M., Gonzalez A., Lloyd A., Schiefelbein J. The bHLH genes GL3 and EGL3 participate in an intercellular regulatory circuit that controls cell patterning in the Arabidopsis root epidermis. Development. 2005;132:291–298. doi: 10.1242/dev.01565. [DOI] [PubMed] [Google Scholar]
- 24.Müller M., Schmidt W. Environmentally induced plasticity of root hair development in Arabidopsis. Plant Physiol. 2004;134:409–419. doi: 10.1104/pp.103.029066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ma Z., Bielenberg D. G., Brown K. M, Lynch J. P. Regulation of root hair density by phosphorus availability in Arabidopsis thaliana. Plant Cell Environ. 2001;24:459–467. [Google Scholar]
- 26.Plaxton W. C., Carswell M. C. Metabolic aspects of the phosphate starvation response in plants. In: Lerner H. R., editor. Plant Responses to Environmental Stresses: From Phytohormones to Genome Reorganization. New York: Marcel Dekker; 1999. pp. 349–372. [Google Scholar]
- 27.Vidal J., Chollet R. Regulatory phosphorylation of C4 PEP carboxylase. Trends Plant Sci. 1997;2:230–237. [Google Scholar]
- 28.Nimmo H. G. The regulation of phosphoenolpyruvate carboxylase in CAM plants. Trends Plant Sci. 2000;5:75–80. doi: 10.1016/s1360-1385(99)01543-5. [DOI] [PubMed] [Google Scholar]
- 29.Nimmo H. G. Control of the phosphorylation of phosphoenolpyruvate carboxylase in higher plants. Arch. Biochem. Biophys. 2003;414:189–196. doi: 10.1016/s0003-9861(03)00115-2. [DOI] [PubMed] [Google Scholar]
- 30.Hartwell J., Gill A., Nimmo G. A., Wilkins M. B., Jenkins G. I., Nimmo H. G. Phosphoenolpyruvate carboxylase kinase is a novel protein kinase regulated at the level of expression. Plant J. 1999;20:333–342. doi: 10.1046/j.1365-313x.1999.t01-1-00609.x. [DOI] [PubMed] [Google Scholar]
- 31.May M. J., Leaver C. J. Oxidative stimulation of glutathione synthesis in Arabidopsis thaliana suspension cultures. Plant Physiol. 1993;103:621–627. doi: 10.1104/pp.103.2.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fontaine V., Hartwell J., Jenkins G. I., Nimmo H. G. Arabidopsis thaliana contains two phosphoenolpyruvate carboxylase kinase genes with different expression patterns. Plant Cell Environ. 2002;25:115–122. [Google Scholar]
- 33.Sullivan S., Jenkins G. I., Nimmo H. G. Roots, cycles and leaves. Expression of the phosphoenolpyruvate carboxylase kinase gene family in soybean. Plant Physiol. 2004;135:2078–2087. doi: 10.1104/pp.104.042762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fraser N. J. Expression and functional purification of a glycosylation deficient version of the human adenosine 2a receptor for structural studies. Protein Expr. Purif. 2006;49:129–137. doi: 10.1016/j.pep.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 35.Nimmo G. A., Wilkins M. B., Nimmo H. G. Partial purification and characterization of a protein inhibitor of phosphoenolpyruvate carboxylase kinase. Planta. 2001;213:250–257. doi: 10.1007/s004250000501. [DOI] [PubMed] [Google Scholar]
- 36.Chen Z. H., Walker R. P., Tecsi L. I., Lea P. J., Leegood R. C. Phosphoenolpyruvate carboxykinase in cucumber plants is increased both by ammonium and by acidification, and is present in the phloem. Planta. 2004;219:48–58. doi: 10.1007/s00425-004-1220-y. [DOI] [PubMed] [Google Scholar]
- 37.Swain T., Hillis H. E. Phenolic constituents of Prunus domestica. I. Quantitative analysis of phenolic constituents. J. Sci. Food Agr. 1959;10:63–68. [Google Scholar]
- 38.Essigman B., Güler S., Narang R. A., Linke D., Benning C. Phosphate availability affects the thylakoid lipid composition and the expression of SQD1, a gene required for sulfolipid biosynthesis in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 1998;95:1950–1955. doi: 10.1073/pnas.95.4.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carswell M. C., Grant B. R., Plaxton W. C. Disruption of the phosphate-starvation response of oilseed rape suspension cells by the fungicide phosphonate. Planta. 1997;203:67–74. doi: 10.1007/s00050166. [DOI] [PubMed] [Google Scholar]
- 40.Ticconi C. A., Delatorre C. A., Abel S. Attenuation of phosphate starvation responses by phosphite in Arabidopsis. Plant Physiol. 2001;127:963–972. [PMC free article] [PubMed] [Google Scholar]
- 41.Johnson J. F., Allan D. L., Vance C. P. Phosphorus stress-induced proteoid roots show altered metabolism in Lupinus albus. Plant Physiol. 1994;104:657–665. doi: 10.1104/pp.104.2.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moraes T. F., Plaxton W. C. Purification of phosphoenolpyruvate carboxylase from Brassica napus (rapeseed) suspension cell cultures. Eur. J. Biochem. 2000;267:4465–4476. doi: 10.1046/j.1432-1327.2000.01495.x. [DOI] [PubMed] [Google Scholar]
- 43.Toyota K., Koizumi N., Sato F. Transcriptional activation of phosphoenolpyruvate carboxylase by phosphorus deficiency in tobacco. J. Exp. Bot. 2003;54:961–969. doi: 10.1093/jxb/erg095. [DOI] [PubMed] [Google Scholar]
- 44.Sánchez R., Cejudo F. J. Identification and expression analysis of a gene encoding a bacterial-type phosphoenolpyruvate carboxylase from Arabidopsis and rice. Plant Physiol. 2003;132:949–957. doi: 10.1104/pp.102.019653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Walker A. R., Davison P. A., Bolognesi-Winfield A. C., James C. A., Srinivasan N., Blundell T. M., Esch J. J., Marks M. D., Gray J. C. The TRANSPARENT TESTA GLABRA1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 repeat protein. Plant Cell. 1999;11:1337–1349. doi: 10.1105/tpc.11.7.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nesi N., Debeaujon I., Jond C., Pelletier G., Caboche M., Lepiniec L. The TT8 gene encodes a basic helix-loop-helix domain protein required for expression of DFR and BAN genes in Arabidopsis siliques. Plant Cell. 2000;12:1863–1878. doi: 10.1105/tpc.12.10.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang F., Gonzalez A., Zhao M., Payne C. T., Lloyd A. A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis. Development. 2003;130:4859–4869. doi: 10.1242/dev.00681. [DOI] [PubMed] [Google Scholar]
- 48.Payne C. T., Zhang F., Lloyd A. M. GL3 encodes a bHLH protein that regulates trichome development in Arabidopsis through interaction with GL1 and TTG1. Genetics. 2000;156:1349–1362. doi: 10.1093/genetics/156.3.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Western T. L., Young D. S., Dean G. H., Tan W. L., Samuels L., Haughn G. W. MUCILAGE-MODIFIED4 encodes a putative pectin biosynthetic enzyme developmentally regulated by APETALA2, TRANSPARENT TESTA GLABRA1, and GLABRA2 in the Arabidopsis seed coat. Plant Physiol. 2004;134:296–306. doi: 10.1104/pp.103.035519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen M., Chory J., Fankhauser C. Light signal transduction in higher plants. Annu. Rev. Genet. 2004;38:87–117. doi: 10.1146/annurev.genet.38.072902.092259. [DOI] [PubMed] [Google Scholar]