Summary
Crystal structures of the 50S ribosomal subunit from H. marismortui complexed with two antibiotics have identified new sites at which antibiotics interact with the ribosome and inhibit protein synthesis. 13-deoxytedanolide binds to the E site of the 50S subunit at the same location as the CCA of tRNA, and thus appears to inhibit protein synthesis by competing with deacylated tRNAs for E site binding. Girodazole binds near the E-site region, but is somewhat buried and may inhibit tRNA binding by interfering with conformational changes that occur at the E site. The specificity of 13-deoxytedanolide for eukaryotic ribosomes is explained by its extensive interactions with protein L44e which is an E-site component of archaeal and eukaryotic ribosomes, but not of eubacterial ribosomes. In addition, protein L28, which is unique to the eubacterial E site, overlaps the site occupied by 13-deoxytedanolide, precluding its binding to eubacterial ribosomes. Girodazole is specific for eukarytes and archaea because it makes interactions with L15 that are not possible in eubacteria.
Introduction
Many clinically important antibiotics inhibit the growth of microorganisms by blocking ribosome function, and important insights into the mechanisms of action of these antibiotics have emerged recently from crystal structures of ribosome complexes with antibiotics (1–14). At the present, all of the anti-large ribosomal subunit antibiotics for which there are published crystal structures bind in or adjacent to its peptidyl transferase center (PTC). Here we present crystal structures of large subunit complexes with two less well known antibiotics, 13-deoxytedanolide and girodazole. Both bind to the E site of the large ribosomal subunit of Haloarcula marismortui (Hma), rather than the PTC.
13-deoxytedanolide is a protein synthesis inhibitor specific for eukaryotes (and presumably archaea) that binds to the large ribosomal subunit (15). It contains an 18-membered macrolide ring that lacks the sugar substituents characteristic of macrolides like erythromycin, which bind specifically near the PTC of eubacterial ribosomes (Figure 1c). Both 13-deoxytedanolide and tedanolide, the first compound of this class to be discovered, are found in marine sponges (16, 17), and both have anti-tumor activity.
Figure 1.
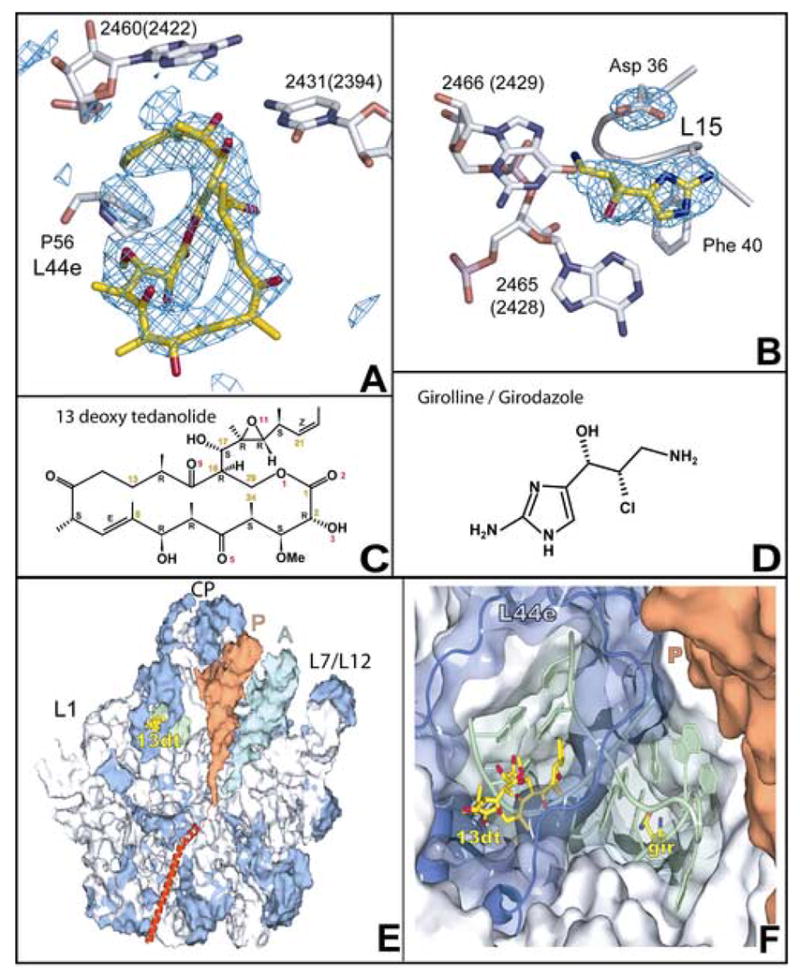
Location of the binding sites for 13-deoxytedanolide and girodazole. The regions in difference electron density maps calculated for each of the two complexes using observed amplitudes and calculated phases where drugs bind are shown with structural models for the drugs superimposed. (a) 13-deoxytedanolide, contoured at +3 sigma. (b) Girodazole, contoured at +4 sigma. The chemical structures of: (c) 13-deoxytedanolide, and (d) girodazole. (e) The location of the binding site for 13-deoxytedanolide (yellow), compared to the locations of th P-site (orange) tRNA and the A-site (cyan) tRNA. The large ribosomal subunit is oriented in the “crown” view but split through the tunnel and the part close to the viewer removed so that the trajectory of peptides down the exit tunnel can be seen (red helix). Ribosomal proteins are blue and rRNA is gray, except for the RNA segment, shown in green. that lies between the 13-deoxytedanolide and the girodazole. (f) A close up view of the E-site region showing the positions of the 13-deoxytednolide and girodazole and the intervening rRNA (green). Most of the macromolecular material in this view has been made transparent so that the bound girodazole molecule can be visualized. The coloring scheme for this panel is the same as in (e).
Like 13-deoxytedanolide, girodazole (Figure 1d), also known as girolline, is an antibiotic obtained from a marine sponge that has antitumor activity (18). Its cytotoxicity correlates with its activity as an inhibitor of eukaryotic protein synthesis (19). In vitro, it does not inhibit either the initiation or the elongation steps of protein synthesis, but rather it blocks the release of polypeptides from the ribosome (20).
Results
13-deoxytedanolide and girodazole bind to the E-site region of the 50S ribosomal subunit
Data sets were collected from crystals of the Hma large ribosomal subunit that had been soaked in solutions containing either 13-deoxytedanolide or girodazole, and difference electron density maps were computed using experimental amplitudes for both the liganded and apo-subunit and phases calculated from the coordinates of the subunit (Materials and Methods). The difference maps for both drug complexes exhibited only a single positive feature that was large enough to accommodate the drug. 13-deoxytedanolide binds in the E-site of the large ribosomal subunit, far from both the PTC and the polypeptide exit tunnel (Figure 1e). The girodazole binding site is close to the 13-deoxytedanolide binding site, but is more deeply buried in the interior of the particle (Figure 1f).
Both drugs can be modeled into the corresponding difference electron density map
The difference electron density maps for 13-deoxytedanolide and girodazole are shown in Figures 1a and 1b, respectively, with the structures of the drugs superimposed. (Table 1 summarizes the crystallographic and refinement statistics for these structures.) While it is clear that the difference electron density for each drug is appropriate in size and shape, the fitting of the structures of these drugs into their respective electron densities was not straightforward.
Table 1.
Crystallographic statistics
| Data Collection | |||||
|---|---|---|---|---|---|
| Antibiotic | Res (Å)a | I/σb | Rmergec | Completed | Rede |
| 13-deoxytedanolide | 50.0-2.92 (3.00-2.92) | 11.0 (1.8) | 0.163 (.98) | 99.9 (100) | 6.9 |
| girodazole | 50.0-2.83 (2.91-2.83) | 15.7 (2.2) | 0.106 (.88) | 99.8 (100) | 6.7 |
| Model Refinement | |||||
| Antibiotic | Res (Å) | Rcryst f | Rfree g | Bond (Å) h | Ang.i |
|
| |||||
| 13-deoxytedanolide | 50.0-2.92 | 0.194 | 0.238 | .005 | 1.02 |
| girodazole | 50.0-2.7 | 0.204 | 0.249 | .005 | 1.05 |
The resolution range of the X-ray diffraction data. The resolution range of the highest-resolution data bin included in the data is indicated in parentheses. Statistics provided in parentheses elsewhere in this table are those for that highest resolution bin.
Average intensity/average standard error of the intensity.
Rmerge = {∑hkl∑i|Ii(hkl) − i(hkl)|}/{∑hkl∑i|Ii(hkl)|}, where i(hkl) is the average value of the intensity of reflection (hkl) in the data set, and Ii(hkl) is the intensity of the ith observation of that reflection.
Completeness, percentage of the possible number of reflections that was actually observed.
Average redundancy of the data set.
Rcryst = {∑hkl|Fc(hkl) − Fo(hkl)|}/{∑hkl|Fo(hkl)|}, where Fc(hkl) is the structure factor for the (hkl) reflection (hkl) computed from the structural model, and Fo(hkl) is the structure factor measured for that reflection.
Rfree is computed the same way as Rcryst using a small, but randomly chosen subset of reflections measured that is not included in the data set used for refinement.
The root mean squared difference of bond lengths from expectation.
The root mean squared difference of bond angles from expectation in degrees.
No independent small molecule crystal structure exists for girodazole, and at the resolution of the electron density maps obtained in this study (Figure 1b) it was not possible to determine its conformation with high accuracy. 13-deoxytedanolide, on the other hand, is a much bigger molecule, and there is clear electron density for both its macrolide ring and its “tail” (Figure 1a). However, the crystal structure of tedanolide (16), which differs from 13-deoxytedanolide by only a single hydroxyl group, fits the difference electron density for 13-deoxytedanolide so poorly that its coordinates could not be refined into the map. A model for the drug that does fit the difference density was obtained by opening the macrolide ring of a model for 13-deoxytedanolide derived from the crystal structure of tedanolide, fitting the two halves of the ring manually into the electron density, and then reforming the broken bond. The conformation of the resulting model was optimized (in the gas phase) using the program BOSS 4.6 (21) and the OPELS/CM1A force field (22). The optimized model differs only slightly from the starting model and fits the corresponding difference electron density well. The computed conformational energy for the refined model is only slightly higher than that of the small-molecule crystal of tedanolide
13-deoxytedanolide is likely to be a competitive inhibitor of E-site function
The only ribosomal base with which 13-deoxytendanolide forms hydrogen bonds is C2431(2394) (Figure 2a), which is a functionally critical base in the ribosome that is >99% conserved in all three kingdoms (23). (Bases in 23S rRNA are identified as follows: (base type, Hma number)(base type, if different, E. coli number).) When tRNA binds to the E site of the ribosome, its 3′ terminal nucleotide, A76, forms hydrogen bonds with the N4 and N3 of C2431(2394) (24;25) (Figures 2b and 2d). In addition to blocking access to C2431(2394), the drug fills most of the rest of the space occupied by the CCA end of deacylated tRNAs when it is bound to the E site in Hma. Since the CCA end of tRNAs and 13-deoxytendanolide cannot simultaneously bind to the large ribosomal subunit part of the E site, it is highly likely that the physiological effects of 13-deoxytedanolide derive from its acting as a competitive inhibitor of tRNA interaction with the E site.
Figure 2.
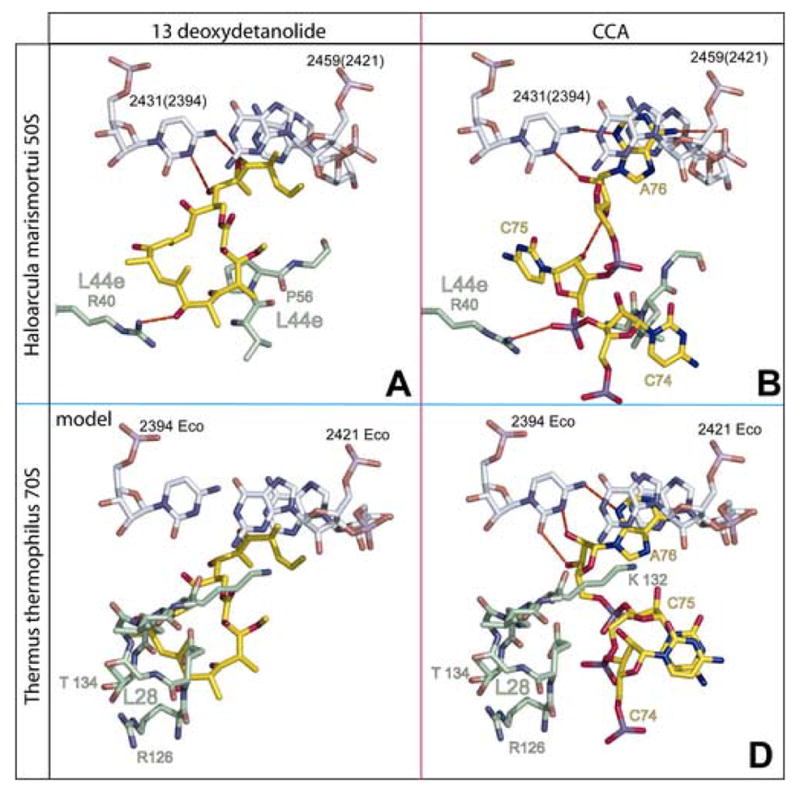
The interactions of 13-deoxytedanolide and the 3′ terminal CCA of deacylated tRNAs with the E sites of the large ribosomal subunit from Hma and Thermus thermophilus compared (a) The interactions of 13-deoxytedanolide (yellow) with the E site of the large ribosomal subunit of Hma. rRNA is shown in gray and protein L44a in green. (b) The interaction of the CCA end of deacylated tRNA with the E site of the large ribosomal subunit of Hma showing C75 and C74 unstacked (24). The CCA sequence is yellow. The RNA and protein are colored as in (a). (c) 13-deoxytedanolide has been homology modeled into the E site of the large ribosomal subunit from T. thermophilus. Phosphorus atoms in the E site region of the large ribosomal subunit of the T. thermophilus 70S ribosome (25) were superimposed on the corresponding atoms of the complex of 13-deoxytedanolide with the Hma large ribosomal subunit and the 13-deoxytedanolide part of the Hma structure is shown in yellow, and the rest of the atoms shown are T. thermophilus atoms. The E-site atoms shown are those of T. thermophilus with the rRNA in gray and protein L28 (which overlaps the drug) in green. (d) The stacked conformation of the 3′-terminal CCA sequence observed when deacylated tRNAs interact with the E site of the large ribosomal subunit of T. thermophilus (25). The CCA sequence is yellow, and the RNA and protein L28 are colored as in (a).
Hydrophobic interactions stabilize the binding of 13-deoxytedanolide to the ribosome
In addition to forming functionally critical hydrogen bonds with C2341(2394), 13-deoxytedanolide makes extensive hydrophobic interactions with both 23S rRNA and ribosomal protein L44e (Figure 2a, supplementary material). The drug has a surface area of 818 Å2, and 65% of its surface becomes inaccessible to solvent when the drug binds to the ribosome. 43% of the buried surface is buried by virtue of the drug’s interactions with L44e, primarily Pro56 and Gly 57. Insertion of its hydrophobic tail into the gap between G2459(2421) and A2460(2422) accounts for most of the rest of the buried surface area.
Girodazole binds near the E site, but not in it
Girodazole binds to the large ribosomal subunit in the densely packed region in domain 5 of 23S rRNA where helices 80, 82, 88 and 74 come together. C2431(2394), the base that hydrogen bonds to A76 of E site- bound tRNA, lies at the bottom of helix 88, about 5 base-pairs removed from the girodazole binding site (Figure 3b). Girodazole forms hydrogen bonds with A2465(G2428), G2466(2429) and Asp36 of ribosomal protein L15 (Figure 3a). As is evident in Figure 1b, the side chain of Asp 36 reorients significantly in the presence of the drug; in the absence of the drug its side chain extends away from the girodazole binding site.
Figure 3.
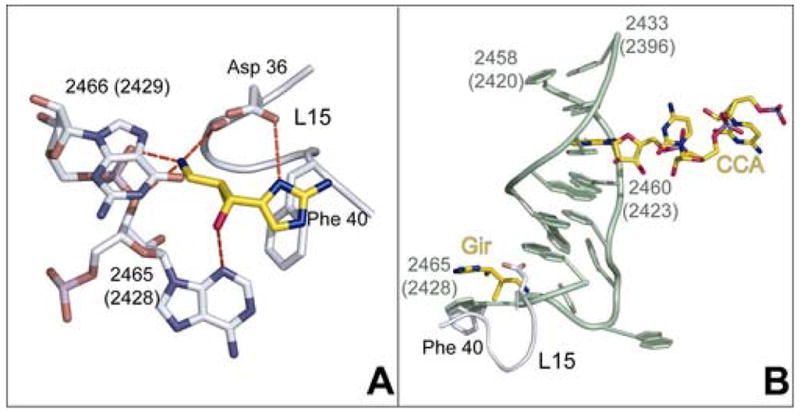
Interactions of girodazole with the large ribosomal subunit from Hma. (a) Specific interactions observed between girodazole (yellow) and the ribosomal RNA and protein L15. (b) The relationship between the binding site for girodazole and the E site bound CCA. The drug is shown in yellow in the lower left of this panel. The structure of the segment of 23S rRNA that lies between the girodazole and CCA binding sites is green. The CCA sequence of a deacylated tRNA bound to the E site of the large ribosomal subunit of Hma is shown in yellow in the upper right part of this panel (24).
Discussion
Both 13-deoxytedanolde and girodazole inhibit protein synthesis in eukaryotic cells, and presumably also archaea, but not in eubacterial cells, and the structures of the complexes they form with the Hma 50S subunit explain why. The sites where they interact with the large ribosomal subunit, which have not previously been identified as antibiotic binding sites, are located at or near the place where the CCA ends of E-site bound tRNAs interact with the large subunit. The large subunit component of the E site is formed, in part, of proteins, and the E-site proteins of the eubacterial large ribosomal subunit are not the same as the E-site proteins of archaeal and eukaryotic large ribosomal subunits, which are similar. These protein differences account, in part, for the kingdom specificity of the two antibiotics as well as for the differences between the conformations the CCA ends of tRNAs adopt when bound to the E sites of eubacterial and archaeal large ribosomal subunits.
The structure of 13-deoxytedanolide bound to the ribosome predicts that it should block protein synthesis by competitively inhibiting the binding of deacylated tRNAs to the E site. The source of its specificity for eukaryotes is also clear. 13-deoxytedanolide interacts extensively with L44e, which is the dominant protein found in the E sites of both archaea and eukaryotes (24). The protein found in the same region in the eubacterial ribosome is L28, but its structure is unrelated to that of L44e (25). In the eubacterial ribosome, residues from L28 fill much of the space that is occupied by 13-deoxytedanolide when it binds to the large ribosomal subunit of Hma and thus would preclude its binding to the eubacterial ribosome in the same manner (Fig. 2c).
The same structural differences in the E site that explain the species specificity of 13-deoxytedanolide, also are responsible for the differences in the conformations that the 3′ terminal CCA sequences of tRNAs adapt when bound to the E sites of eubacteria (25, 26) and archaea (24). When that sequence is bound to the Hma E site, the base of C75 splays out filling an area that is occupied by L28 in eubacteria (Figures 2b and 2d). Similarly, L44e in Hma sterically prevents E-site bound CCA sequences from adapting the stacked conformation that is observed (25, 26) in the eubacterial E site.
If the structure of 13-deoxytedanolide bound to the Hma 50S subunit described here is physiologically relevant it should provide structural explainations for the relative activities of chemical derivatives of 13-deoxytendanolide as protein synthesis inhibitors. Nishimura and coworkers have prepared 11 chemical derivatives of 13-deoxytedanolide and measured their activities as inhibitors of protein synthesis using a poly U-dependent, phenylalanine incorporating system derived from Saccharomyces cerevisiae (27). They observed, for example, that reduction of either of the molecule’s two carbon-carbon double bonds has little effect on activity. One of those double bonds, (C21-C22), is part of the molecule’s tail that inserts into the gap between G2459 and A2460. Its reduction should not interfere with the hydrophobic interaction that part of the tail normally makes with those bases. Reduction of the other double bond (C8-C9) should have little effect because that part of the drug does not interact with the ribosome. Reduction of the carbonyl oxygen at C11 also has little effect on activity, as expected because that oxygen does not interact with the ribosome, but removal of the entire “tail” of 13-deoxytendanolide results in a large reduction in drug activity. The “tail-less” derivative Nishimura and colleagues prepared cannot interact hydrophobically with G2459 and A2460, and also lacks the groups that normally hydrogen bond with C2431(2394)l. Its affinity for the ribosome may be further compromised by conformational alterations in the macrolide ring near C16 caused by the chemistry done to remove the tail.
The only derivative Nishimura and colleagues prepared whose properties are hard to explain structurally is the one they obtained by acetylating both O3 and O6 with pentenoyl chloride. When either O3 and O6 are acetylated separately this way there is no effect on activity as expected because O3 does not contact the ribosome, and acetylation of O6 should not prevent it from accepting a hydrogen bond from R40 of L44e, which is what it does when the unmodified drug binds. The surprise is that the di-acetylated derivative is inactive, as is the compound obtained when O3, O6, and O10 are all acetylated. However, the inactivity of the tri-acetylated derivative can be explained structurally entirely in terms of O10. Acetylation of 13-deoxytendanolide at O10 ought to prevent the O10 hydroxyl group from making a hydrogen bond with the N3 of C2431 (2394). It is also hard to explain why the removal of the last two carbon atoms from the “tail” of 13-deoxytendanolide and their replacement by an aldehyde group (at C21) should reduce the activity of the drug as dramatically as it does, but it may adversely affect the hydrophobic interactions the tail of the drug normally makes with the ribosome. While the capacity of the structure of the 13-deoxytedanolide-ribosome complex described here to rationalize almost all the structure-activity data just discussed does not prove that it is physiologically relevant, if it did not, it would be decisive proof that it is irrelevant, and it is unlikely that the structure would rationalize much of that data at all if it were physiologically irrelevant.
13-deoxytedanolide is the first antibiotic that we have found whose activity results from binding to the E site of the large ribosomal subunit, but it is unlikely to be unique. Fusetani and coworkers have identified three antibiotics of unrelated chemical structure that compete with 13-deoxytedanolide for binding to the eukaryotic large ribosomal subunit: pederin, theopederin and onnamide A (15). Cycloheximide may be another eukaryote-specific antibiotic that targets the E site since mutation of the unstructured loop in the yeast homolog of L44e (RPL41) can confer cycloheximide resistance on that organism (28, see also 29). Nishimura and colleagues looked for, but did not detect competition between the binding of 13-deoxytedanolide and the binding of cycloheximide, but it is possible that the measurements they did would not have done so. The dissociation constant for cycloheximide binding to the ribosome is in the micromolar range (30), but the dissociation constant for 13-deoxytedanolide/ribosome complexes is ~2 nM (15). The concentration of cycloheximide used in the competition experiment reported by Nishimura and colleagues (3.9 μM) may not have been high enough to displace from the ribosome the trace amounts of radioactive 13-deoxytedanolide used to detect competition.
Most antibiotics that target the large subunit for which structural information is available exert their effects either by competitively blocking substrate binding or sterically blocking polypeptide product egress, whereas most of the small subunit antibiotics that have been studied crystallographically interfere with conformational changes (2). It is possible, but far from proven at this point, that girodazole inhibits protein synthesis by preventing conformational changes in the large subunit. Girodazole binds to the large subunit close to the E site, but its binding site does not overlap with the E-site bound deacylated tRNAs, and thus it cannot be a direct, competitive inhibitor of that interaction. The structures of CCA or of tRNA acceptor stem/loop mimics bound to the Hma large ribosomal subunit show no indication that the conformation of the ribosome changes in the region of the girodazole binding site when the E site is occupied (24), nor does the structure of girodazole bound to the Hma 50S subunit indicate that drug binding causes a conformational change in the E site. Thus if the drug inhibits E-site activity at all, it must block conformational changes that occur in that region during protein synthesis.
Although the structural basis of girodazole’s inhibition remains unclear, its binding site is consistent with the drug’s known specificity for eukaryotes. The two RNA bases that interact with girodazole are components of the single-stranded region that connects helices 88 and 74 of 23S rRNA. In this region the sequences of eubacterial 23S rRNAs diverge significantly from those of both archaeal and eukaryotic large ribosomal subunit RNAs, which are similar. The connecting sequence is 12 nucleotides long in eubacteria, but only 9 nucleotides long in archaea and eukaryotes (23), and thus, expectedly, their conformations are not the same either. When the structures of the large ribosomal subunit from E. coli (31) and Hma are superimposed using 10 backbone phosphorus atoms from this region, the rmsd is 1.95 Å, but when a similar superposition is done using 180 phosphorus atoms distributed over the entire PTC, the rmsd is just over 1.0 Å. Nonetheless, it is clear that the nucleotide that interacts most extensively with the drug, A2465 in Hma, corresponds to G2428 in E. coli. The two bases occupy the same space in the two molecules, and are oriented the same way. There are three reasons why girodazole ought not to bind to the eubacterial version of its site with high affinity. First, girodazole cannot interact with G2428 in the E. coli ribosome the way it interacts with A2465 in the Hma large ribosomal subunit because the N2 amino group of G2428 would interfere sterically. Second, insertion of the drug into this site would be interfered with by the O6 of G2466(2429), which is about 2 Å closer to A2465(G2428) in E. coli than it is in Hma. Third, in the E. coli large ribosomal subunit, the prototypical Gram-negative bacterium, the adjacent part of ribosomal protein L15 has no residues that can hydrogen bond with the drug the way Asp 36 does in the Hma subunit. Thus girodazole should bind to this site in the eubacterial large ribosomal subunit with an affinity far lower than its affinity for the corresponding site in archaeal and eukaryotic ribosomes.
Materials and Methods
The 50S ribosomal subunits were prepared from Hma and crystallized as previously described (32, 33). Crystals were equilibrated at least twice with solutions containing antibiotic dissolved in buffer B (12 % w/v PEG 6000, 20% v/v ethylene glycol, 1.7 M NaCl, 0.5 M NH4Cl, 1mM CdCl2, 100 mM potassium acetate, 6.5 mM acetic acid, pH 6.0, 30mM MgCl2 or 100mM SrCl2, 5mM β-mercaptoethanol) at 4°C over the course of 1–4 hours when SrCl2 was included in soak solutions (33), and 16h to 24 h when MgCl2 was included in soak solutions. The antibiotic concentrations used were 1 mM for 13-deoxytedanolide, and 40 mM for girodazole.
13-deoxytendanolide was provided by Drs. Nobuhiro Fusetani and Shigeki Matsunaga (Laboratory of Aquatic Natural Products Chemistry, University of Tokyo). Girodazole was given to us by Dr. Christiane Poupat of the Institut de Chimie des Substances Naturelles du CNRS, Gif-sur-Yvette, France. The samples of 13-deoxytedanolide and girodazole used were both isolated from natural sources, and only a few milligrams of each were available. There are few experiments that could have been done with samples so small that would have yielded as much information as was obtained for these two antibiotics from the crystallographic experiments reported above.
Data were collected as previously described (32) using the following synchrotron beamlines: ALS 8.2.2, APS 19ID, and NSLS X29. Following merging of the diffraction data, pairwise cross-R factors were computed between each data set for the drug complex with the 50S subunit and every data set in the collection of native data sets available in this laboratory in order to identify the native data set that is most similar to that of the drug complex data set. Difference electron density maps were computed for each complex of a drug with the ribosome using as amplitudes (|Fo(hkl)|drug/ribosome − |Fo(hkl)|best native) and phases obtained by rigid body refinement of the native structure (pdb # 1S72; 34) into the drug complex data set. Refinements done using the program CNS (34) included rigid body refinement, energy minimization, and B factor refinement. In the final rounds of refinement only those atoms within a 25–35 Å sphere centered on each antibiotic binding site were allowed to vary in position.
Because crystal structures have not been reported for girodazole, the starting model used for it was an energy-minimized structure created using either CS Chem3D Pro (Cambridge Soft Corp.) or PRODRG2 (http://davapc1.bioch.dundee.ac.uk/programs/prodrg) (36). The procedure used to obtain a satisfactory initial structure for 13-deoxytedanolide is described above. Modeling was done using O (37), and figures were generated using PyMol (38) (http://www.pymol.org).
The structures described in the paper are available from the Protein Data Bank. The PDB number of the 13-deoxytedanolide structure is XXXX, and the PDB number of the girodazole structure is YYYY.
Supplementary Material
Acknowledgments
We are indebted to Drs. Shigeki Matsunaga, and Christiane Poupat for providing us with the antibiotic samples that made this study possible. This work was supported by grants from the National Institutes of Health (PO1-GM022778, to TAS and PBM; F32-GM067354 to S.J.S; GM32136 to J. T.-R.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Brodersen DE, Clemons WM, Carter AP, Morgan-Warren RJ, Wimberly BT, Ramakrishnan V. The structural basis for the action of the antibiotics tetracycline, pactamycin and hygromycin B on the 30S subunit. Cell. 2000;103:1143–1154. doi: 10.1016/s0092-8674(00)00216-6. [DOI] [PubMed] [Google Scholar]
- 2.Carter AP, Clemons WM, Brodersen DE, Morgan-Warren RJ, Wimberly BT, Ramakrishnan V. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature. 2000;407:340–348. doi: 10.1038/35030019. [DOI] [PubMed] [Google Scholar]
- 3.Pioletti M, Schlunzen F, Harms J, Zarivach R, Glumann M, Avila H, Bashan A, Bartels H, Auerbach T, Jacobi C, Hartsch T, Yonath A, Franceschi F. Crystal structures of complexes of the small ribosomal subunit with tetracycline, edeine and IF3. EMBO J. 2001;20:1829–1839. doi: 10.1093/emboj/20.8.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schluenzen F, Zarivach R, Harms J, Bashan A, Tocilj A, Albrecht R, Yonath A, Franceschi F. Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria. Nature. 2001;413:814–821. doi: 10.1038/35101544. [DOI] [PubMed] [Google Scholar]
- 5.Hansen JL, Ban N, Nissen P, Moore PB, Steitz TA. The structures of four macrolide antibiotics bound to the large ribosomal subunit. Molec Cell. 2002;10:117–126. doi: 10.1016/s1097-2765(02)00570-1. [DOI] [PubMed] [Google Scholar]
- 6.Auerbach T, Bashan A, Harms J, Schluenzen F, Zarivach R, Bartels H, Agmon I, Kessler M, Pioletti M, Franceschi F, Yonath A. Antibiotics targeting ribosomes: crystallographic studies. Curr Drug Targ Infect Disord. 2002;2:169–186. doi: 10.2174/1568005023342506. [DOI] [PubMed] [Google Scholar]
- 7.Berisio R, Harms J, Schluenzen F, Zarivach R, Hansen HAS, Fucini P, Yonath A. Structural insight into the antibiotic action of telithromycin against resistant mutants. J Bacteriol. 2003;185:4276–4279. doi: 10.1128/JB.185.14.4276-4279.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hansen JL, Moore PB, Steitz TA. Structure of five antibiotics bound at the peptidyl transferase center of the large ribosomal subunit. J Mol Biol. 2003;330:1061–1075. doi: 10.1016/s0022-2836(03)00668-5. [DOI] [PubMed] [Google Scholar]
- 9.Harms JM, Bartels H, Schluenzen F, Yonath A. Antibiotics acting on the translational machinery. J Cell Sci. 2003;116:1391–1393. doi: 10.1242/jcs.00365. [DOI] [PubMed] [Google Scholar]
- 10.Schluenzen F, Harms JM, Franceschi F, Hansen HAS, Bartels H, Zarivach R, Yonath A. Structural basis for the antibiotic activity of ketolides and azalides. Structure. 2003;11:329–338. doi: 10.1016/s0969-2126(03)00022-4. [DOI] [PubMed] [Google Scholar]
- 11.Harms JM, Schluenzen F, Fucini P, Bartels H, Yonath A. Alterations at the peptidyl transferase centre of the ribosome induced by the synergistic action of the streptogramins dalfopristin and quinupristinh. BMC Biology. 2004;2:4. doi: 10.1186/1741-7007-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schluenzen F, Pyetan E, Fucini P, Yonath A, Harms JM. Inhibition of peptide bond formation by pleuromutilins: the structure of the 50S ribosomal subunit from Deinococcus radiodurans in complex with tiamulin. Molec Microbiol. 2004;54:1287–1295. doi: 10.1111/j.1365-2958.2004.04346.x. [DOI] [PubMed] [Google Scholar]
- 13.Tu D, Blaha G, Moore PB, Steitz TA. Structures of MLSBK antibiotics bound to mutated large ribosomal subunits provide a structural explanation for resistance. Cell. 2005;121:257–270. doi: 10.1016/j.cell.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 14.Wilson DN, Harms JM, Nierhaus KH, Schlunzen F, Fucini P. Species-specific antibiotic-ribosome interactions: implications for drug development. Biol Chem. 2005;386:1239–1252. doi: 10.1515/BC.2005.141. [DOI] [PubMed] [Google Scholar]
- 15.Nishimura S, Matsunaga S, Yoshida M, Hirota H, Yokoyama S, Fusetani N. 13-deoxytedanolide, a marine sponge-derived antitumor macrolide, binds to the 60S large ribosomal subunit. Bioorgan Med Chem. 2005;13:449–454. doi: 10.1016/j.bmc.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 16.Schmitz FJ, Gunasekera SP, Yalamanchili G, Hossain MB, van der Helm D. Tedanolide: a potent cytotoxic macrolide from the Caribbean sponge Tedania ignis. J Am Chem Soc. 1984;106:7251–7252. [Google Scholar]
- 17.Fusetani N, Sugawara T, Matsunaga S, Hirota H. Cytotoxic metabolites of the marine sponge Mycale adhaerans Lambe. J Org Chem. 1991;56:4971–4974. [Google Scholar]
- 18.Ahond A, Bedoya ZM, Colin M, Fizames C, Laboute P, Lavelle F, Laurent D, Poupat C, Pusset J, Pusset M, Thoison O, Potier P. La giroline, nouvelle substance antitumorale extraite de l’eponge Pseudaxinyssa cantharella. CR Acad Sci Paris. 1988;307:145–148. [Google Scholar]
- 19.Lavelle F, Zerial A, Dizames C, Rabault B, Curaudeau A. Antitumor activity and mechanism of action of the marine compound girodazole. Investigat New Drugs. 1991;9:233–244. doi: 10.1007/BF00176976. [DOI] [PubMed] [Google Scholar]
- 20.Colson G, Rabault B, Lavelle F, Zerial A. Mode of action of the antitumor compound girodazole (RP 49532A, NSC 627434) Biochem Pharmacol. 1992;43:1717–1723. doi: 10.1016/0006-2952(92)90701-j. [DOI] [PubMed] [Google Scholar]
- 21.Jorgensen WL, Tirado-Rives J. Molecular modeling of organic and biomolecular systems using BOSS and MCPRO. J Comput Chem. 2005;26:1689–1700. doi: 10.1002/jcc.20297. [DOI] [PubMed] [Google Scholar]
- 22.Jorgensen WL, Tirado-Rives J. Chemical theory and computation special feature: potential energy function for atomic-level simulation of water and organic and biomolecular systems. Proc Natl Acad Sci USA. 2005;102:6665–6670. doi: 10.1073/pnas.0408037102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cannone JJ, Subramanian S, Schnare MN, Collett JR, D’Souza LM, Du Y, Feng B, Lin N, Madabusi LV, Muller KM, Pande N, Shang Z, Yu N, Gutell RR. The comparative RNA web (CRW) site: an outline database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BioMed Centr Bioinform. 2002;3:2. doi: 10.1186/1471-2105-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schmeing TM, Moore PB, Steitz TA. Structure of deacylated tRNA mimics bound to the E site of the large ribosomal subunit. RNA. 2003;9:1345–1352. doi: 10.1261/rna.5120503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Selmer M, Dunham CM, Murphy FVI, Weixibaumer A, Petry S, Kelley AC, Weir JR, Ramakrishnan V. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 26.Korostelev A, Trakhanov S, Laurberg M, Noller HF. Crystal structure of a 70S ribosome-tRNA complex reveals functional interations and rearrangements. Cell. 2006;126:1065–1077. doi: 10.1016/j.cell.2006.08.032. [DOI] [PubMed] [Google Scholar]
- 27.Nishimura S, Matsunaga S, Yoshida S, Nakao Y, Hirota H, Fusetani N. Structure-activity relationship study on 13-deoxytedanolide, a highly antitumor macrolide from the marine sponge Mycale adhaerens. Bioorgan Med Chem. 2005;13:455–462. doi: 10.1016/j.bmc.2004.10.014. [DOI] [PubMed] [Google Scholar]
- 28.Kawai S, Mirato S, Mochizuki M, Shibaya I, Yano K, Tagaki M. Drastic alteration of cycloheximide sensitivity by substitution of one amino acid in the L41 ribosomal protein of yeast. J Bacteriol. 1992;174:254–262. doi: 10.1128/jb.174.1.254-262.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pestova TV, Hellen CUT. Translation elongation after assembly of ribosomes on the cricket paralysis virus internal ribosome entry site without initiation factor or initiator tRNA. Gen & Dev. 2003;17:181–186. doi: 10.1101/gad.1040803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cooper D, Banthorpe DV, Wilkie D. Modified ribosomes conferring resistance to cylcloheximide in mutants of Saccharomyces cerevisiae. J MolBiol. 1967;26:347–350. doi: 10.1016/0022-2836(67)90302-6. [DOI] [PubMed] [Google Scholar]
- 31.Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, VCila-Sanjuro A, Holton JH, Doudna Cate JH. Structure of the bacterial ribosome at 3.5 A resolution. Science. 2005;310:827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- 32.Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- 33.Schmeing TM, Huang KS, Strobel SA, Steitz TA. An induced-fit mechanism to promote peptide bond formation and exclude hydrolysis of peptidyl-tRNA. Nature. 2005;438:520–524. doi: 10.1038/nature04152. [DOI] [PubMed] [Google Scholar]
- 34.Klein DJ, Moore PB, Steitz TA. The roles of ribosomal proteins in the structure, assemby and evolution of the large ribosomal subunit. J Mol Biol. 2004;340:141–177. doi: 10.1016/j.jmb.2004.03.076. [DOI] [PubMed] [Google Scholar]
- 35.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography and NMR system (CNS): a new software suite for macromolecular structure determination. Acta Cryst D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 36.Schuettelkopf AW, van Aalten DMF. PRODRG: a tool for high-throughput crystallography of protein0ligand complexes. Acta Cryst. 2004;D60:1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- 37.Jones TA, Cowan S, Zou J-Y, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. 1991;A46:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 38.DeLano WL. The PyMol molecular gaphics system. DeLano Scientific; San Carlos, CA, USA: 2002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


