Abstract
Peptide nucleic acid (PNA) is a DNA mimic that has shown considerable promise as a lead compound for developing gene therapeutic drugs. We report that PNAs targeted to functional and accessible sites in ribosomal RNA can inhibit translation in an Escherichia coli cell-free transcription/translation system, with 50% reductions caused by nanomolar PNA concentrations. The effect in vitro is quantitatively similar to that of the known translation inhibitor and antibiotic tetracycline. Also, the targeted PNAs inhibited bacterial growth on agar plates and in liquid culture. A strain of E. coli (AS19) that is more permeable to antibiotics was approximately 10-fold more sensitive to the active PNAs, suggesting that the effect on growth indeed was caused by PNAs that entered cells. Inhibition was not observed when using control PNAs of similar composition but with an unrelated or mismatched sequence. The results demonstrate that ribosomal RNA is a possible target for sequence-designed novel antibiotics based on DNA analogues or mimics.
The RNA component of ribosomes (rRNA) is essential for protein synthesis (1) and therefore is an attractive target for antimicrobial drugs. Indeed, many natural antibiotics disrupt protein synthesis and most of these appear to act by binding rRNA (2). Previous studies have indicated that nucleic acid oligomers also can inhibit translation in vitro by binding to rRNA (3–7) and that short methylphosphonate oligonucleotides targeted at the Shine–Delgarno sequence have some growth inhibitory potential in permeable Escherichia coli cells (4). We considered that the superior hybridization and stability properties of the DNA mimic peptide nucleic acid (PNA) (8, 9) (Fig. 1A) should enhance such effects. This is demonstrated by its ability to block DNA and RNA polymerases and ribosome progression when bound to DNA or RNA templates (10–13) as well as its high potency to inhibit the activity of telomerase by binding to its RNA component (14).
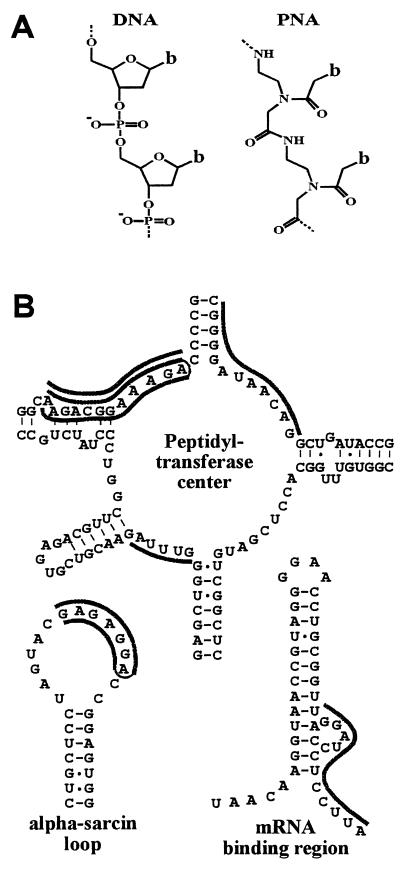
Figure 1.
(A) Chemical structure of a PNA oligomer compared with that of DNA. b indicates the nucleobases adenine, cytosine, guanine, thymine, or pseudo-isocytosine(20). (B) Target sites for antiribosomal PNAs. The binding sites for PNAs are shown as dark lines adjacent to the sequence. Triplex forming bis-PNAs (18) are shown connected by an ethylene glycol linker (thin line) (see also Table 1).
MATERIALS AND METHODS
Strains, Plasmid, and PNA.
E. coli strains K-12 (wild type) and D10 (rna-10) were from the E. coli genetic stock center (Yale University, New Haven, CT). The permeable strain AS19 (15) was obtained from Steen Pedersen (University of Copenhagen). A derivative of D10 (D10–1) containing the lacIq gene for repressor overproduction was constructed by transfer of the F′ episome from strain JM105 as described (16). The plasmid pMAS2 (17), which carries the E. coli gene for β-galactosidase was obtained from Michael Sørensen (University of Copenhagen). The peptide nucleic acids used in this study (Table 1) were synthesized as previously described (18, 19).
Table 1.
PNAs and inhibitory concentrations
| PNA | Target region | Inhibitory concentrations (IC50)
|
|
|---|---|---|---|
| Cell-free translation, nM | Cell growth, μM | ||
| Duplex-forming | |||
| 1176 H-GCAAGCGACTGTGGA-Lys-NH2 | Control | >500 | >20 |
| 1284 H-GGTCATAGCTGTTTC-Lys-NH2 | Control | >500 | >20 |
| 111 H-CCCCTATTGTCC-Lys-NH2 | P.T. center | >500 | >20 |
| 112 H-TTCTGCCTTTCT-Lys-NH2 | P.T. center | >500 | >20 |
| 1140 H-TAAAC-NH2 | P.T. center | >500 | >20 |
| 1142 H-AAGGAGGTGA-Lys-NH2 | mRNA binding domain | >500 | >20 |
| Triplex-forming | |||
| 977 H-TTJTTJTTTT-(eg1)3-TTTTCTTCTT-Lys-NH2 | Control | >500 | >20 |
| 1197 H-TJJJTTJ-(eg1)3-CTTCCCT-Lys-NH2 | Control | >500 | >20 |
| 1440 H-JTTTJJT-(eg1)3-TCCTTTC-Lys-NH2 | Control | >500 | >20 |
| 647 H-Lys-TTJTJJJTTTJT-(eg1)3-TCTTTCCGTCTT-Lys-NH2 | P.T. center | 80 | 5 |
| 1143 H-JTJTJJT-(eg1)3-TCCTCTC-Lys-NH2 | α-sarcin loop | 100 | 2 |
| Tetracycline | 70 | 0.1 | |
The PNAs are written from their amino to carboxy termini, with the carboxy termini containing an amide or lysine amide. PNA 647 contains a G residue to help accommodate a nonpurine C residue in the target (Fig. 1). The IC50 values are the levels that caused a 50% inhibition of cell-free translation or cell growth, relative to control reactions and cultures that lacked inhibitors. The values for inhibition of cell growth are for strain AS19 grown in 0.1× LB broth. Control PNAs are of unrelated or mismatched sequence. P.T. indicates peptidyl transferase. The J bases indicate pseudo-isocytosine (20).
Cell-Free Transcription and Translation.
Strain D10–1 was grown to mid-log phase in Luria–Bertani (LB) media supplemented with 4 g/liter of glucose. The preparation of S-30 cell extracts and coupled transcription/translation reactions was carried out as described (20) by using plasmid pMAS2. The reaction components were aliquoted into microfuge tubes on ice to a total of 30 μl, vortexed briefly and incubated at 37°C for 30 min. β-galactosidase activity was measured by using the substrate o-nitrophenyl-β-galactoside and absorbance measurements at 420 nm as described (16).
Transcription in the cell-free system was assayed by using 30-μl reactions that contained 34 μM unlabeled UTP and 1 μCi 32P-UTP. After 30 min, the reactions were stopped with 10 vol of 5% trichloroacetic acid (TCA) and incubated on ice for 30 min. Translation in the cell-free system was assayed by using reactions that contained 1 mCi [35S]methionine and 1 μg of chloramphenicol acetyltransferase gene mRNA, which was generated by T3 RNA polymerase transcription as described (12). After 30 min, the reactions were stopped with an equal volume of 1 M NaOH and placed at 37°C for 15 min. The incorporated radioactivity was precipitated with 1 vol of ice-cold 50% TCA containing 2% casamino acids, collected by vacuum filtration using Watman GF/A filters, and rinsed five times with 5% TCA. Cerénkov and scintillation counting were used to determine 32P-UTP and [35S]methionine incorporation, respectively.
Growth Assays.
LB media at one-tenth normal strength was inoculated with 1% (vol/vol) of an overnight E. coli culture. For solid media cultures, 4 ml of molten LB/agar media was inoculated and spread onto prewarmed LB/agar plates, and the excess molten media was poured off. PNA or antibiotic solutions were pipetted (2 μl) directly onto the solidified overlay, and the plates were incubated overnight at 37°C. For liquid cultures, 100 μl of inoculated LB media was aliquoted into microtiter plate wells containing PNAs or antibiotic. The plates were incubated overnight at 37°C, and growth was measured by absorbance at 550 nm.
RESULTS AND DISCUSSION
Duplex- and triplex-forming antiribosomal PNAs were designed to target sites within the peptidyl transferase center, the α-sarcin loop, and the mRNA binding domain at the 3′ end of 16S rRNA (Fig. 1B). As well as being functionally active, these sites are accessible for interaction with certain antibiotics, translation factors, structure probing agents, other RNA molecules, and short oligonucleotides (21–23).
The potential for using PNAs to inhibit protein synthesis was evaluated initially by using an in vitro assay, in which plasmid DNA encoding β-galactosidase was added to a template-depleted E. coli S-30 cell extract along with the reagents necessary for coupled transcription and translation (20). The production of β-galactosidase was measured colorimetrically by using the substrate o-nitrophenyl-β-galactoside (16). The assay is sensitive to inhibitors that prevent complete or accurate translation. Two of the antiribosomal PNAs (647 and 1143) inhibited β-galactosidase production in a dose-dependent manner, with 50% reductions caused by nanomolar PNA concentrations (Fig. 2A). The inhibitory PNAs both have a bis construction (18) and target purine regions at the peptidyl transferase center or the α-sarcin loop. The bis construction stabilizes triplex formation, which may be needed for efficient ribosome inhibition because none of the duplex-forming PNAs are effective. However, further studies are required to characterize the limits of PNA design and target site selection. PNAs of similar composition but unrelated sequence (977 and 1197) and a PNA containing a one-base mismatch (1440) were not inhibitory. The PNAs used in this study and their effects on bacterial cell-free protein synthesis are summarized in Table 1. The inhibiting concentrations found for the active PNAs are comparable with those obtained for the known translation inhibitor and antibiotic tetracycline.
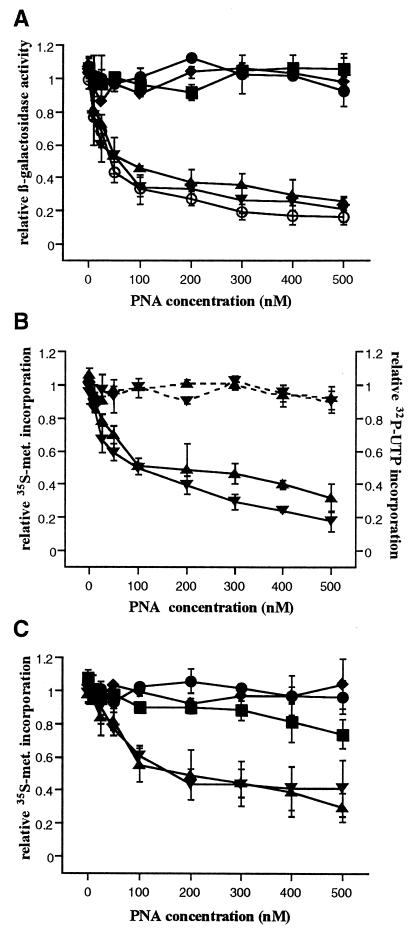
Figure 2.
Inhibition of cell-free translation. (A) Relative β-galactosidase activity after coupled transcription/translation in the presence of control PNAs 997 (⧫), 1197 (•), 1440 (▪); antiribosomal PNAs 647 (▴) and 1143 (▾) and the antibiotic tetracycline (○). (B) Transcription (broken lines) and translation (solid lines) in the cell-free system were assayed by measuring 32P-UTP and [35S]methionine incorporation in independent reactions in the presence of the antiribosomal PNAs. (C) Translation of a synthetic mRNA, assayed by measuring [35S]methionine incorporation in the presence of control and antiribosomal PNAs. The values represent the average of duplicate measurements with the SD shown as error bars.
To ascertain that the reduction in β-galactosidase production observed in the coupled transcription/translation assays was caused by inhibition of ribosome activity, the incorporation of 32P-UTP and [35S]methionine was assayed as indicators of transcription and translation activities, respectively. With increasing concentrations of targeted PNA, the level of radioactive methionine incorporation was reduced, whereas radioactive UTP incorporation was unaffected (Fig. 2B). Also, a synthetic mRNA was generated and used to assay the effect of the antisense PNAs on [35S]methionine incorporation. Again, increasing concentrations of targeted PNA reduced the level of incorporation and the control PNAs were not inhibitory (Fig. 2C). Therefore, the PNA-mediated inhibition of β-galactosidase production appears to occur at the translation level.
To determine whether antiribosomal PNAs could be used to limit bacterial growth, inhibition assays were performed by using E. coli grown on solid media. LB/agar plates were prepared with a thin overlay of media containing an inoculum of E. coli strain K-12. The LB media was used at one-tenth its normal strength to overcome solubility limitations with the PNAs. Solutions containing PNA were applied by pipetting 2-μl aliquots directly onto the solidified overlay. After overnight incubation at 37°C, a lawn of bacterial cells was established and growth inhibition was evident as zones of clearing in the lawn at sites of PNA application (Fig. 3). Consistent with the results from the cell-free assay, the two antiribosomal PNAs that showed strong inhibitory effects in vitro were found to inhibit cell growth when applied directly onto solid media cultures (647 and 1143). Again, control PNAs of similar composition but of unrelated (977 and 1197) or mismatched (1440) sequence did not inhibit growth, and the antiribosomal PNAs that showed no effects on translation in vitro also were inactive against cell growth.
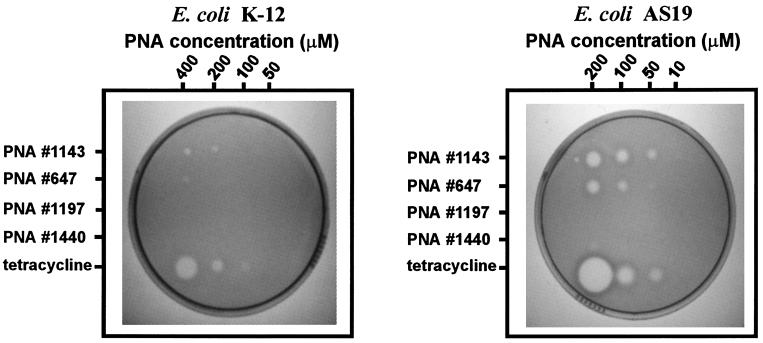
Figure 3.
Inhibition of cell growth on solid media for wild-type E. coli strain K-12 and the permeable strain AS19 (15). Antiribosomal PNAs and tetracycline were aliquoted by direct pipetting onto overlay cultures before overnight growth. Zones of inhibition are visible at sites of application as clear regions in the lawn of cells. Contrast between zones of clearing and the bacterial lawn was improved by using 5-bromo-4-chloro-3-indoyl β-d-galactoside (X-Gal) media and a red filter for photography.
If the antiribosomal PNAs used in this study inhibit growth by entering cells and binding to rRNA, their activity should be limited by the integrity of the E. coli cell wall or outer membrane, which generally limits the action of antibiotics (24). To address the extent to which the E. coli cell barrier limits the action of the antiribosomal PNAs, the growth inhibition assay was repeated by using a strain of E. coli (AS19) that is more permeable to many antibiotics (15). Inhibition occurred with PNA concentrations approximately one-tenth of the level required for similar inhibition of the standard E. coli strain K-12. (Fig. 3). This increase in sensitivity parallels that observed for known antibiotics (15) and further suggests that the PNAs entered cells and inhibited growth via their effect on protein synthesis. To determine the PNA concentrations that are inhibitory to strain AS19, growth assays in liquid media were performed in 100-μl cultures. Two of the antiribosomal PNAs inhibited growth in liquid culture when present in the low micromolar range (Table 1).
The inhibitory effects we have observed for PNAs targeted to rRNA are consistent with reports of in vitro translation inhibition by modified and unmodified antiribosomal oligonucleotides (3–7). Also, structural probing experiments have revealed regions of the ribosome that are accessible for hybridization (6). Therefore, it appears that sequence-specific targeting of rRNA is possible and can be used to effectively inhibit translation. Previously, short methylphosphonate oligomers were reported to inhibit E. coli growth, however, the effect was temporary, limited to a permeable strain, and not shown to be sequence dependent (4). Thus, PNAs appear to offer greatly improved opportunities for targeting accessible sequences within rRNA, not the least of which for in vivo use.
Our results showing that PNA molecules are able to inhibit E. coli growth is somewhat unexpected. PNA and other nucleobase oligomers are inefficiently taken up by cultured eukaryotic cells (25) and are slow to move across phospholipid vesicle (liposome) membranes (26). Furthermore, the molecular weight of typical oligonucleotides exceeds the expected cut-off for efficient passive diffusion through the nonspecific porin channels that span the E. coli cell wall (27). However, uptake of larger molecules through the porin openings may be more efficient for long or flexible structures (24), such as PNA, which is furthermore uncharged and largely hydrophilic.
The experiments show that sequence-specific nucleic acid binding molecules can be designed to inhibit bacterial growth, opening possibilities for developing sequence-designed antiribosomal agents to limit bacterial growth in research or medical applications. Thus, “designer antibiotics” may be developed by this principle. An important feature of the approach is the potential to define the spectrum of target species by exploiting RNA sequence differences between species, which occurs at several functional sites within rRNA, including the peptidyl transferase center, but not the highly conserved α-sarcin loop. Also, if resistance arose because of rRNA sequence changes in the target organism, it should be possible to restore the inhibitory effect by redesigning the PNA. Finally, it may be possible to extend these studies to other RNA targets, such as the spliceosome and other ribonucleoprotein complexes.
Acknowledgments
We thank Steen Pedersen and Michael Sørensen for strain AS19, plasmid pMAS2, and helpful discussions. This research was supported by the Danish National Research Foundation and a fellowship to L.G. from the Natural Sciences and Engineering Research Council of Canada.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: PNA, peptide nucleic acid; rRNA, ribosomal RNA; LB, Luria–Bertani.
References
- 1.Noller H F, Hoffarth V, Zimniak L. Science. 1992;256:1416–1419. doi: 10.1126/science.1604315. [DOI] [PubMed] [Google Scholar]
- 2.Cundliffe E. In: The Ribosome. Hill W E, Dahlberg A, Garrett R A, Moore P B, Schlessinger D, Warner J R, editors. Washington, D.C.: Am. Soc. Microbiol.; 1990. pp. 479–490. [Google Scholar]
- 3.Taniguchi T, Weissmann C. Nature (London) 1978;275:770–772. doi: 10.1038/275770a0. [DOI] [PubMed] [Google Scholar]
- 4.Jayaraman K, McParland K, Miller P, Ts’o P O. Proc Natl Acad Sci USA. 1981;78:1537–1541. doi: 10.1073/pnas.78.3.1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Walker K, Abou Elela S, Nazar R N. J Biol Chem. 1989;265:2428–2430. [PubMed] [Google Scholar]
- 6.Saxena S K, Ackerman E J. J Biol Chem. 1990;265:3263–3269. [PubMed] [Google Scholar]
- 7.Meyer H-A, Triana-Alonso F, Spahn C M T, Twardowski T, Sobkiewicz A, Nierhaus K H. Nucleic Acids Res. 1996;24:3996–4002. doi: 10.1093/nar/24.20.3996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nielsen P E, Egholm M, Berg R H, Buchardt O. Science. 1991;254:1497–1500. doi: 10.1126/science.1962210. [DOI] [PubMed] [Google Scholar]
- 9.Egholm M, Buchardt O, Christensen L, Behrens C, Freier S M, Driver D A, Berg R H, Kim S K, Nordén B, Nielsen P E. Nature (London) 1993;365:566–568. doi: 10.1038/365566a0. [DOI] [PubMed] [Google Scholar]
- 10.Hanvey J C, Peffer N J, Bisi J E, Thomson S A, Cadilla R, Josey J A, Ricca D J, Hassman C F, Bonham M A, Au K G, et al. Science. 1992;258:1481–1485. doi: 10.1126/science.1279811. [DOI] [PubMed] [Google Scholar]
- 11.Nielsen P E, Egholm M, Buchardt O. Gene. 1994;149:139–145. doi: 10.1016/0378-1119(94)90422-7. [DOI] [PubMed] [Google Scholar]
- 12.Knudsen H, Nielsen P E. Nucleic Acids Res. 1996;24:494–500. doi: 10.1093/nar/24.3.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taylor R W, Chinnery P F, Turnbull D M, Lightowlers R N. Nat Genet. 1997;15:212–215. doi: 10.1038/ng0297-212. [DOI] [PubMed] [Google Scholar]
- 14.Norton J C, Piatyczek J A, Wright W E, Shay J W, Corey D R. Nat Biotechnol. 1996;14:615–619. doi: 10.1038/nbt0596-615. [DOI] [PubMed] [Google Scholar]
- 15.Sekiguchi M, Iida S. Proc Natl Acad Sci USA. 1967;58:2315–2320. doi: 10.1073/pnas.58.6.2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miller J H. Experiments in Molecular Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1972. [Google Scholar]
- 17.Sørensen M A, Kurland C G, Pedersen S J. J Mol Biol. 1989;207:365–377. doi: 10.1016/0022-2836(89)90260-x. [DOI] [PubMed] [Google Scholar]
- 18.Egholm M, Christensen L, Dueholm K, Buchardt O, Coull J, Nielsen P E. Nucleic Acids Res. 1995;23:217–222. doi: 10.1093/nar/23.2.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Christensen L, Fitzpatrick R, Gildea B, Petersen K H, Hansen H F, Koch T, Egholm M, Buchardt O, Nielsen P E. J Peptide Sci. 1995;3:175–183. doi: 10.1002/psc.310010304. [DOI] [PubMed] [Google Scholar]
- 20.Thorson, J. S., Cornish, V. W., Barrett, J. E., Cload, S. T., Yano, T. & Schultz, P. G. (1998) Methods Mol. Biol., in press. [DOI] [PubMed]
- 21.Steitz J A, Jakes K. Proc Natl Acad Sci USA. 1975;72:4634–4638. doi: 10.1073/pnas.72.12.4734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Egebjerg J, Larsen N, Garrett R A. In: The Ribosome. Hill W E, Dahlberg A, Garrett R A, Moore P B, Schlessinger D, Warner T R, editors. Washington, DC: Am. Soc. Microbiol.; 1990. pp. 168–179. [Google Scholar]
- 23.Hill W E, Camp D G, Tapprich W E, Tassanakajohn A. Methods Enzymol. 1988;164:401–419. doi: 10.1016/s0076-6879(88)64057-2. [DOI] [PubMed] [Google Scholar]
- 24.Nikaido H, Vaara M. Microbiol Rev. 1985;49:1–32. doi: 10.1128/mr.49.1.1-32.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bonham M A, Brown S, Boyd A L, Brown P H, Bruckenstein D A, Hanvey J C, Thomson S A, Pipe A, Hassman F, Bisi J E, et al. Nucleic Acids Res. 1995;23:1197–1203. doi: 10.1093/nar/23.7.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wittung P, Kajanus J, Edwards K, Nielsen P, Nordén B, Malmström B G. FEBS Lett. 1995;365:27–29. doi: 10.1016/0014-5793(95)00409-3. [DOI] [PubMed] [Google Scholar]
- 27.Decad G M, Nikaido H. J Bacteriol. 1976;128:325–336. doi: 10.1128/jb.128.1.325-336.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]