Abstract
Assembly of the mammalian spliceosome is known to proceed in an ordered fashion through several discrete complexes, but the mechanism of this assembly process may not be universal. In an early step, pre-mRNAs are committed to the splicing pathway through association with U1 small nuclear ribonucleoprotein (snRNP) and non-snRNP splicing factors, including U2AF and members of the SR protein family. As a means of studying the steps of spliceosome assembly, we have prepared HeLa nuclear extracts specifically depleted of the splicing factor U2AF. Surprisingly, the SR protein SC35 can functionally substitute for U2AF65 in the reconstitution of pre-mRNA splicing in U2AF-depleted extracts. This reconstitution is substrate-specific and is reminiscent of the SC35-mediated reconstitution of splicing in extracts depleted of U1 snRNP. However, SC35 reconstitution of splicing in U2AF-depleted extracts is dependent on the presence of functional U1 snRNP. These observations suggest that there are at least three distinguishable mechanisms for the binding of U2 snRNP to the pre-mRNA, including U2AF-dependent and -independent pathways.
Pre-mRNA splicing occurs via two sequential transesterification reactions in a 60S complex known as the spliceosome, which assembles on the pre-mRNA substrate in an ordered fashion through several discrete complexes (E, A, B, C; ref. 1). The spliceosome includes the small nuclear ribonucleoprotein (snRNP) particles U1, U2, U4/6, and U5, as well as associated splicing factors (2–4). Commitment of a pre-mRNA substrate to assembly of the spliceosome involves the ATP-independent formation of the E (early) or commitment complex (5–8). This complex, in the mammalian system, contains U1 snRNP as well as non-snRNP protein factors, including the U2 auxiliary factor, U2AF, and members of the SR protein family (8).
SR proteins contain extensive serine/arginine (SR) repeats and a subset contain RNA recognition motifs; the predominant members of the family are conserved from Drosophila to humans. Many SR proteins are important splicing factors that function in both constitutive and alternative RNA splicing (9). SR proteins containing RNA recognition motifs display modest affinity and sequence specificity in their association with RNA and probably bind cooperatively in association with other factors, including other SR proteins (9). SR family members associate with pre-mRNAs early in spliceosomal assembly (10, 11) and this association may persist through the chemistry of splicing (12). It has been suggested that SR proteins are required for specific transitions during the course of spliceosomal assembly such as the progression from A to B complex (13). The 35-kDa SR protein SC35 has been reported to stimulate E complex formation (8) and has been shown to be associated with a complex formed at the 3′ end of the intron at early stages in spliceosome assembly (14).
Conserved sequence elements at the 5′ and 3′ splice sites and in the branch sequence and pyrimidine tract of the pre-mRNA direct formation of the spliceosome (3, 4). In particular, recognition of the pyrimidine tract is required early in the formation of commitment complexes. A number of polypeptides have been reported to bind specifically to the pyrimidine tract including the U2 auxiliary factor, U2AF (15–18).
The splicing factor U2AF was first identified as an activity required for the stable association of U2 snRNP with the pre-mRNA branch site in the formation of the A complex (15). U2AF is a heterodimer consisting of both a large (65 kDa) and a small (35 kDa) subunit (18). The large subunit, U2AF65, is an essential splicing factor containing an N-terminal SR domain and three C-terminal RNA recognition motif domains. U2AF65 binds with avidity to the pyrimidine tract, while U2AF35 is in turn tightly associated with the larger subunit through protein–protein interactions (17, 18).
The interactions between the various components of the commitment complex and their precise role in progression through spliceosomal assembly remain to be determined. U2AF has been detected in affinity-selected commitment (E) complexes isolated by gel filtration (8). Although U2AF is clearly important for the transition from E to A complex, it has not been found to be stably associated with either the A or B/C complexes (19). In the commitment complex, U2AF is probably bound to the pyrimidine tract, while U1 snRNP is associated with the 5′ splice site. Far Western analysis has suggested that the SR protein SC35 is associated with both the U1 snRNP-associated factor U1-70K and U2AF through interactions with the small subunit U2AF35 (20). Thus, SC35 may function as a bridge across the 5′ and 3′ splice sites.
To study the mechanism of spliceosomal assembly, we have prepared HeLa nuclear extracts depleted of the splicing factor U2AF and reconstituted splicing of pre-mRNA substrates by the combination of both column fractions and purified recombinant splicing proteins. Interestingly, the SR protein SC35 can functionally substitute for U2AF65 in this reconstitution in a manner that is both substrate-specific and dependent on the presence of functional U1 snRNP.
MATERIALS AND METHODS
RNA Transcription.
The PIP85.A RNA pre-mRNA substrate was transcribed from plasmid pPIP85.A (21). The PIPβG pre-mRNA substrate is a chimera consisting of the 5′ portion of PIP85.A and the 3′ portion of β-globin (22). It was transcribed from a template constructed by ligating the PCR product representing β-globin sequences between +252 and +386 (which include the last 92 nt of the intron and the complete 3′ exon) to pPIP85.A digested with XbaI and HindIII.
Nuclear Extracts.
Nuclear extracts were prepared from HeLa cells as described by Dignam et al. (23). Extracts depleted of poly(U)-binding proteins including U2AF65 (P.S.M. and P.A.S., unpublished work) were prepared by dialyzing nuclear extract directly into 1 M KCl/buffer D (20 mM Hepes, pH 7.9/20% glycerol/0.2 mM EDTA/0.05% Nonidet P-40/0.5 mM DTT). The resulting extract was passed over a poly(U)-Sepharose 4B column (Pharmacia) at 0.1 ml/min and subsequently dialyzed against 0.1 M KCl/buffer D. After washing this column with 1 M KCl/buffer D, the column was eluted with buffer D containing 2 M KCl and the eluate was dialyzed against 0.1 M KCl. Combination of the poly(U)-depleted nuclear extract with the 2 M KCl eluate gave an extract, ΔU2AF NE, specifically depleted of U2AF activity. The extent of U2AF depletion in this extract was assayed by Western analysis with α-U2AF65 antibody.
U2AF and SC35 Preparation.
Recombinant His6-tagged U2AF65 was prepared from a 60% ammonium sulfate precipitate of a soluble Escherichia coli lysate (P.S.M. and P.A.S., unpublished work). This was loaded onto a Ni2+-NTA-agarose column (Qiagen), eluted with 250 mM imidazole/buffer D, and dialyzed into 0.1 M KCl/buffer D.
SC35 was purified as described elsewhere (10) from baculovirus-infected Hi5 insect cells. The protein recovered from phenyl-Sepharose chromatography was treated with micrococcal nuclease, concentrated by precipitation with 20 mM MgCl2, and then resuspended in buffer D.
Splicing Assays.
Splicing reactions (25 μl) were performed under standard conditions (24) using 20% HeLa nuclear extract, incubated at 30°C, and resolved on 20% denaturing polyacrylamide gels. Reactions containing U2AF-depleted extract, ΔU2AF NE, were supplemented with 500 ng of recombinant U2AF65 and/or 300 ng of recombinant SC35. For the U1 blocking experiments, mock or U2AF-depleted nuclear extract was incubated in the presence of 7 μM (≈15-fold excess over U1 snRNP; ref. 25) α-U1 2′-OMe oligonucleotide (26) for 15 min at 30°C, followed by addition of substrate RNA and SR protein and incubation under splicing conditions for 60 min.
RESULTS
U2AF65 and SC35-Mediated Reconstitution of Splicing in U2AF-Depleted Extracts.
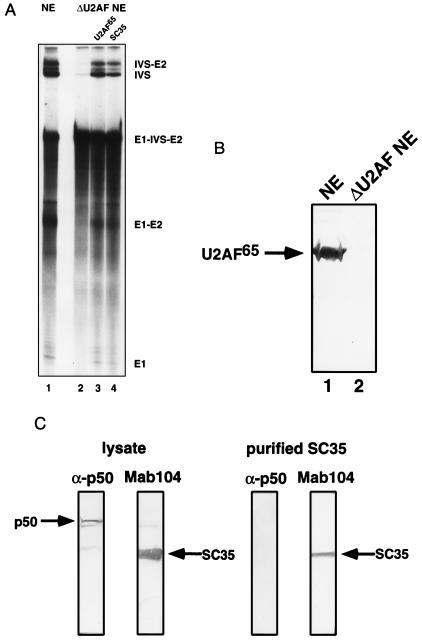
To examine the mechanism of action of the splicing factor U2AF, HeLa nuclear extracts were depleted of this activity. HeLa nuclear extracts depleted of poly(U) binding factors and supplemented with a 2 M KCl column fraction (P.S.M. and P.A.S., unpublished work) were efficiently depleted of the splicing factor U2AF65 (400-fold by Western analysis; Fig. 1B and data not shown). Splicing of pre-mRNA substrates including PIPβG could be restored to the U2AF-depleted extract by either the addition of a 3 M KCl column fraction containing U2AF65 or recombinant U2AF65 (Fig. 1A and data not shown).
Figure 1.
SC35 functionally substitutes for U2AF65 in the reconstitution of pre-mRNA splicing in U2AF-depleted extracts. (A) Splicing of PIPβG pre-mRNA in: mock depleted extract (lane 1); U2AF-depleted extract (lane 2); and U2AF65-reconstituted (lane 3) and SC35-reconstituted (lane 4) U2AF-depleted extract. (B) Western analysis with anti-U2AF65 antibody of mock depleted (lane 1) and U2AF-depleted (lane 2) extract. (C) Western analysis of (Left) crude lysate of SC35 overexpression with α-p50 (anti-Drosophila U2AF50 antibody) and anti-SC35 antibody (mAb104) and (Right) purified SC35 with α-p50 and anti-SC35 antibody (mAb104).
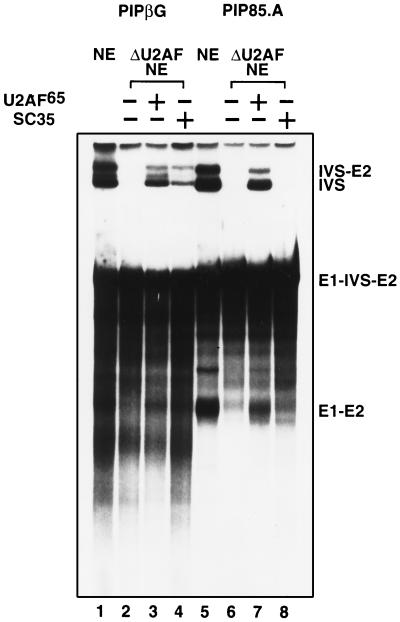
Combinations of U2AF-depleted extract, purified SR proteins, and recombinant U2AF65 were tested for reconstitution of splicing. Surprisingly, addition of SR proteins to nuclear extracts depleted of U2AF65 reconstituted splicing of the PIPβG pre-mRNA (data not shown). To examine this effect more closely, reconstitution reactions were carried out with specific SR proteins purified from baculovirus-infected insect cells. Addition of recombinant SC35, purified from insect cells, restored splicing of the PIPβG pre-mRNA in U2AF-depleted extracts (Fig. 1A). This effect was substrate-specific, since splicing of the PIP85.A pre-mRNA in depleted extract could not be reconstituted with the addition of SC35, although addition of recombinant U2AF65 resulted in reconstitution of PIP85.A splicing at the levels observed with the PIPβG pre-mRNA substrate (Fig. 2). The SC35 reconstitution activity was not due to the presence of the insect homologue of U2AF; insect U2AF50 was detected by Western analysis in crude insect cell lysates but not in samples of purified SC35 (Fig. 1C).
Figure 2.
SC35 reconstitution of splicing in U2AF-depleted reactions is substrate-specific. Splicing of PIPβG pre-mRNA in mock (lane 1), U2AF-depleted (lane 2), U2AF65-reconstituted (lane 3), and SC35-reconstituted (lane 4) extracts. Splicing of PIP85.A in mock (lane 5), U2AF-depleted (lane 6), U2AF65 reconstituted (lane 7) and SC35 reconstituted (lane 8) extracts.
Factor-Dependent Splicing in U1-Blocked Extracts.
SR proteins, specifically SC35, have been shown to reconstitute splicing in a substrate-specific manner in reactions in which U1 snRNP has been either depleted by affinity selection (22, 27) or blocked by the preincubation of extract with an antisense-U1 oligonucleotide (25). Because of this observation and because U1 snRNP is a component of the commitment complex, we examined the role of U1 snRNP in the reconstitution of U2AF-depleted extracts.
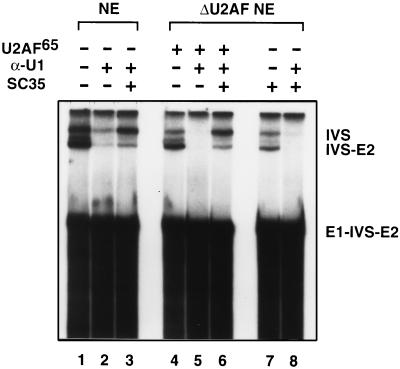
U1 snRNP was inactivated in both mock and U2AF-depleted extracts by preincubation of the extracts with a 15-fold excess (over endogenous U1 snRNP; ref. 25) of an antisense-U1 2′-OMe oligonucleotide. This blocking of U1 snRNP severely decreased the splicing of PIPβG pre-mRNA (Fig. 3; lanes 2 and 5). In accordance with previous observations (25), an excess of SC35 restored splicing activity in U1-blocked reactions in the presence of U2AF (Fig. 3; lanes 3 and 6). More interestingly, SC35 could only restore splicing activity in U2AF-depleted reactions in the presence of functional U1 snRNP (Fig. 3; compare lanes 7 and 8).
Figure 3.
SC35 reconstitutes pre-mRNA splicing in U2AF-depleted extracts dependent on the presence of functional U1 snRNP. Splicing of PIPβG pre-mRNA in: nuclear extract (lane 1); nuclear extract preblocked with α-U1 oligonucleotide (lane 2); nuclear extract preblocked with α-U1 oligonucleotide and supplemented with SC35 (lane 3); U2AF-depleted extract supplemented with U2AF65 (lane 4); U2AF-depleted extract supplemented with U2AF65 and preblocked with α-U1 oligonucleotide (lane 5); U2AF-depleted extract supplemented with U2AF65, preblocked with α-U1 oligonucleotide, and supplemented with SC35 (lane 6); U2AF-depleted extract supplemented with SC35 (lane 7);and U2AF-depleted extract blocked with α-U1 oligonucleotide and supplemented with SC35 (lane 8).
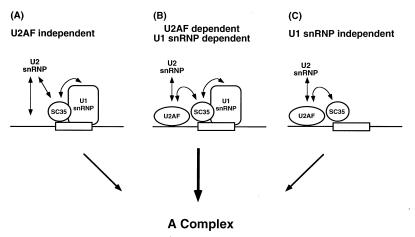
We have shown that SC35 functionally substitutes for U2AF65 in a U2AF-depleted extract (Fig. 1; Fig. 3, lane 7). However, when U2AF-depleted extract was preincubated with an antisense-U1 oligonucleotide in the absence of added U2AF65, addition of recombinant SC35 alone did not restore splicing of the PIPβG pre-mRNA (Fig. 3, lane 8). Thus, SC35 can only functionally substitute for U2AF in the presence of U1 snRNP. These results distinguish a U2AF-independent pathway from a U1 snRNP-independent pathway (Fig. 4; ref. 28).
Figure 4.
Three distinct pathways resulting in spliceosome assembly. Under typical splicing conditions, both U2AF and U1 snRNP are required for spliceosome assembly with a network of stabilizing interactions between U2AF, U1 snRNP, and SR proteins (B; ref. 20). In a substrate-specific manner, excess SC35 reconstitutes splicing in U2AF-depleted reactions in a U1 snRNP-dependent pathway (A) and in U1 snRNP-depleted reactions in a U2AF-dependent pathway (C).
DISCUSSION
HeLa nuclear extracts depleted of the splicing factor U2AF were reconstituted for splicing of pre-mRNA substrates by the addition of purified recombinant SC35. Furthermore, the SR protein SC35 restored splicing in a U2AF-depleted extract in a manner that was both substrate-specific and U1 snRNP-dependent. This suggests that U2AF65 is not an essential factor in either spliceosome assembly or RNA splicing in the presence of high concentrations of SC35; it is likely that the enhanced concentrations of SR domains provided by the addition of SC35 complement the U2AF deficiency.
The pyrimidine tract binding protein U2AF65 has been shown previously to be required for the formation of the first stable complex formed between the pre-mRNA and U2 snRNP, the A complex (15). However, addition of purified SR proteins and, more particularly, recombinant SC35 can functionally substitute for U2AF65 in the reconstitution of splicing in U2AF-depleted HeLa nuclear extracts. This complementation required addition of SR protein to approximately a 10-fold excess over levels present in a typical extract. Complementation by SC35 was substrate-specific: the PIPβG pre-mRNA was efficiently spliced in an SC35-reconstituted extract, while the PIP85.A pre-mRNA was not. The basis for this specificity is unclear, but it is intriguing in that it mirrors the specificity of the SC35-mediated reconstitution of splicing in extracts depleted of U1 snRNP (27, 28). There are no obvious specific SC35 binding sites (29) in the PIPβG substrate, but the specific interaction of SR proteins with the pre-mRNA substrate may only occur in the context of complex protein–RNA and RNA–RNA interactions (9).
Several lines of evidence suggest that the requirement of U2AF for in vitro splicing may not be stringent. First, it does not appear that U2AF is absolutely required for spliceosome assembly. Green and coworkers (30, 31) have shown that either the SR domain of U2AF65 fused to a heterologous RNA binding domain or heterologous SR domains fused to the U2AF65 RNA binding domains can restore U2AF function in a depleted extract. These results suggest that the function of U2AF is to position an SR domain in the vicinity of the branch region/pyrimidine tract. Second, U2AF does not appear to be present in catalytically active spliceosomes isolated by gel filtration, and thus the chemistry of splicing probably occurs in the absence of U2AF (19). Finally, the reported Saccharomyces cerevisiae homologue of U2AF, MUD2, is not a required splicing factor indicating that its function in U2 snRNP pre-mRNA association is either not essential or redundant (32). Thus, although in the mammalian system U2AF is highly conserved, it is possible that there are a variety of mechanisms or factors responsible for directing U2 snRNP to the branch region of the pre-mRNA.
The mechanism of the SC35 complementation for U2AF65 deficiency is not clear. Members of the SR protein family have been proposed to recruit U2AF to the branch site via exon enhancers (33), and it is possible that trace amounts of U2AF in the depleted extract were recruited to a commitment complex by excess SC35. However, this seems unlikely for several reasons. First, U2AF cannot be detected in either the depleted extract or in the purified SC35, which complements the reactions—the upper limit of contamination is 1/400th of endogenous levels of U2AF (Fig. 1 and data not shown). The U2AF-depleted extracts contained at least 10-fold less U2AF than that required for restoration of splicing (as determined by adding mock depleted extract to depleted extract; data not shown). Second, a recruitment mechanism would suggest that excess SC35 should reconstitute a U2AF-depleted extract, even when U1 activity was blocked in accordance with the observed SC35 reconstitution of U1 snRNP blocked (25) or depleted extracts (27, 28). This was not the case: preincubation of U2AF-depleted extract with antisense-U1 oligonucleotide effectively blocked the SC35 reconstitution (Fig. 3; see below).
It is likely that SC35 plays several roles in the reconstitution reactions, including substitution for the activity of U2AF (Fig. 4). The mechanism of U2AF activity in recruitment of U2 snRNP to the branch region is not well understood but clearly involves recognition of the N-terminal SR region (18). While most SR domains are believed to be involved in protein–protein interactions, it has been suggested that the SR domain of U2AF functions as an RNA annealing activity (34). Most probably, SC35 complements the U2AF deficiency in depleted extracts by providing a surrogate SR domain required for critical interactions in the course of spliceosome assembly.
High levels of SR proteins, including SC35, promote the splicing of the PIPβG substrate in the absence of U1 snRNP (22). Under these conditions, the SR proteins facilitate the formation of the U2 snRNP containing A complex independent of U1 snRNA and the association of U6 snRNA with the substrate RNA can become rate-limiting (28). The U1 snRNP-bypass reaction does not occur with PIP85.A pre-mRNA. The observed substrate specificity is identical to that in the SC35-mediated U2AF bypass reaction. This intriguing observation might have reflected a common pathway in which the U1 snRNP-independent reaction was also U2AF-independent. However, this was not the case: interference with the activity of U1 snRNP inactivated splicing in U2AF-depleted extracts, even in the presence of high concentrations of SC35. The common substrate specificities in the two reconstitutions perhaps reflect the sequence specificity of SC35–pre-mRNA interaction and not common splicing pathways. Thus, there are at least three different pathways to the formation of an active spliceosome: the conventional pathway requiring both U1 snRNP and U2AF65; a second pathway, which is independent of U1 snRNP but dependent on U2AF; and a third pathway, which is independent of U2AF but dependent upon U1 snRNP (Fig. 4). Any one of these mechanisms could be operative for a particular intron under specific conditions in vivo. However, since the splicing of a typical intron requires both U1 snRNP and U2AF, both of these entities probably act at a common step in stabilizing the interaction of U2 snRNP with the pre-mRNA.
Acknowledgments
We thank M. Green for the generous gift of U2AF65 antibody; R. Kanaar and D. Rio for the generous gift of Drosophila U2AF50 antibody; and B. Blencowe for the generous gift of anti-U1 oligonucleotide. We also thank L. Lim, J. Pomerantz, and C. Query for their critical reading of the manuscript; M. Siafaca for her ever-present assistance; and M. Beddall and R. Issner for indispensable technical support. This work was supported by United States Public Health Service MERIT Award R37-GM34277 and Grant RO1-AI32486 from the National Institutes of Health to P.A.S., and partially by a Cancer Center Support (Core) Grant P30-CA14051 from the National Cancer Institute. A.M.M. was supported by the Medical Research Council of Canada.
Footnotes
Abbreviation: snRNP, small nuclear ribonucleoprotein.
References
- 1.Konarska M M, Sharp P A. Cell. 1986;46:845–855. doi: 10.1016/0092-8674(86)90066-8. [DOI] [PubMed] [Google Scholar]
- 2.Guthrie C. Science. 1991;253:157–163. doi: 10.1126/science.1853200. [DOI] [PubMed] [Google Scholar]
- 3.Moore M J, Query C C, Sharp P A. In: The RNA World. Gesteland R F, Atkins J F, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1993. pp. 303–357. [Google Scholar]
- 4.Krämer A. In: Pre-mRNA Processing. Lamond A I, editor. Austin, TX: Landes; 1995. pp. 35–64. [Google Scholar]
- 5.Ruby S R, Abelson J. Science. 1988;242:1028–1035. doi: 10.1126/science.2973660. [DOI] [PubMed] [Google Scholar]
- 6.Séraphin B, Rosbash M. Cell. 1989;59:349–358. doi: 10.1016/0092-8674(89)90296-1. [DOI] [PubMed] [Google Scholar]
- 7.Michaud S, Reed R. Genes Dev. 1991;5:2534–2546. doi: 10.1101/gad.5.12b.2534. [DOI] [PubMed] [Google Scholar]
- 8.Staknis D, Reed R. Mol Cell Biol. 1994;14:7670–7682. doi: 10.1128/mcb.14.11.7670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fu X-D. RNA. 1996;1:663–680. [PMC free article] [PubMed] [Google Scholar]
- 10.Fu X-D. Nature (London) 1993;365:82–85. doi: 10.1038/365082a0. [DOI] [PubMed] [Google Scholar]
- 11.MacMillan A M, Query C C, Allerson C R, Chen S, Verdine G L, Sharp P A. Genes Dev. 1994;8:3008–3020. doi: 10.1101/gad.8.24.3008. [DOI] [PubMed] [Google Scholar]
- 12.Blencowe B J, Nickerson J A, Issner R, Penman S, Sharp P A. J Cell Biol. 1994;127:583–607. doi: 10.1083/jcb.127.3.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roscigno R F, Garcia-Blanco M A. RNA. 1995;1:692–706. [PMC free article] [PubMed] [Google Scholar]
- 14.Fu X-D, Maniatis T. Proc Natl Acad Sci USA. 1992;89:1725–1729. doi: 10.1073/pnas.89.5.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ruskin B, Zamore P D, Green M R. Cell. 1988;52:207–219. doi: 10.1016/0092-8674(88)90509-0. [DOI] [PubMed] [Google Scholar]
- 16.Zamore P D, Green M R. Proc Natl Acad Sci USA. 1989;86:9243–9247. doi: 10.1073/pnas.86.23.9243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zamore P D, Green M R. EMBO J. 1991;10:207–214. doi: 10.1002/j.1460-2075.1991.tb07937.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zamore P D, Patton J G, Green M R. Nature (London) 1992;355:609–614. doi: 10.1038/355609a0. [DOI] [PubMed] [Google Scholar]
- 19.Bennett M, Michaud S, Kingston J, Reed R. Genes Dev. 1992;6:1986–2000. doi: 10.1101/gad.6.10.1986. [DOI] [PubMed] [Google Scholar]
- 20.Wu J Y, Maniatis T. Cell. 1993;75:1061–1070. doi: 10.1016/0092-8674(93)90316-i. [DOI] [PubMed] [Google Scholar]
- 21.Moore M J, Sharp P A. Science. 1992;256:992–997. doi: 10.1126/science.1589782. [DOI] [PubMed] [Google Scholar]
- 22.Crispino J D, Mermoud J E, Lamond A I, Sharp P A. RNA. 1996;2:664–673. [PMC free article] [PubMed] [Google Scholar]
- 23.Dignam J D, Lebovitz R M, Roeder R D. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grabowski P J, Padgett R A, Sharp P A. Cell. 1984;37:415–427. doi: 10.1016/0092-8674(84)90372-6. [DOI] [PubMed] [Google Scholar]
- 25.Tarn W-Y, Steitz J A. Genes Dev. 1994;8:2704–2717. doi: 10.1101/gad.8.22.2704. [DOI] [PubMed] [Google Scholar]
- 26.Barabino S M L, Blencowe B J, Ryder U, Sproat B S, Lamond A I. Cell. 1990;63:293–302. doi: 10.1016/0092-8674(90)90162-8. [DOI] [PubMed] [Google Scholar]
- 27.Crispino J D, Blencowe B J, Sharp P A. Science. 1994;265:1866–1869. doi: 10.1126/science.8091213. [DOI] [PubMed] [Google Scholar]
- 28.Crispino J D, Sharp P A. Genes Dev. 1995;9:2314–2323. doi: 10.1101/gad.9.18.2314. [DOI] [PubMed] [Google Scholar]
- 29.Tacke R, Manley J L. EMBO J. 1995;14:3540–3551. doi: 10.1002/j.1460-2075.1995.tb07360.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Valcárcel J, Singh R, Zamore P D, Green M R. Nature (London) 1993;362:171–175. doi: 10.1038/362171a0. [DOI] [PubMed] [Google Scholar]
- 31.Valcárcel J, Gaur R J, Singh R, Green M R. Science. 1996;273:1706–1709. doi: 10.1126/science.273.5282.1706. [DOI] [PubMed] [Google Scholar]
- 32.Abovich N, Liao X C, Rosbash M. Genes Dev. 1994;8:843–854. doi: 10.1101/gad.8.7.843. [DOI] [PubMed] [Google Scholar]
- 33.Wang Z, Hoffmann H M, Grabowski P J. RNA. 1995;1:21–35. [PMC free article] [PubMed] [Google Scholar]
- 34.Lee C G, Zamore P D, Green M R, Hurwitz J. J Biol Chem. 1993;268:13472–13478. [PubMed] [Google Scholar]