Abstract
The unstable proteins Cdc6p and cdc18+ are essential and rate limiting for the initiation of DNA replication in Saccharomyces cerevisiae and Schizosaccharomyces pombe, respectively, and also participate in checkpoint controls that ensure DNA replication is completed before mitosis is initiated. We have identified Xenopus and human proteins closely related to Cdc6p/cdc18. The human protein, p62cdc6, is encoded on chromosome 17q21.3 and includes putative cyclin-dependent kinase phosphorylation sites, destruction boxes, a nucleotide binding/ATPase domain, and a potential leucine zipper. Expression of p62cdc6 mRNA and protein is suppressed in human diploid fibroblasts made quiescent by serum starvation, and peaks as cells reenter the cell cycle and replicate DNA following serum stimulation. Conservation of structure among proteins involved in initiation suggests that fundamental features of replication complexes are maintained in all eukaryotes.
DNA replication in yeasts is initiated from discrete chromosomal locations (replicators) based on the sequential assembly of a prereplication complex that includes the origin binding protein ORC (origin recognition complex) (1–10) and several associated factors. ORC consists of six essential protein subunits (4) that have homologues in other yeasts, plants, and metazoan species (11–16). Binding of ORC to replicator DNA sequences in vivo can be detected by footprinting techniques at all stages of the cell cycle, but different patterns of nucleotide contacts are evident prior to and following DNA replication (2, 3, 17–19), consistent with the viewpoint that additional factors interact with ORC to establish a competent complex for initiation (10).
In budding yeasts this function resides, at least in part, with the protein encoded by the CDC6 gene (Cdc6p) (5, 19–25). Fission yeasts contain a closely related protein, cdc18+, with similar properties (26–30). Yeast strains lacking CDC6 are nonviable, and temperature sensitive cdc6 mutants suffer growth arrest with partially unreplicated DNA at the restrictive temperature (5, 20–22). Even at temperatures permissive for viability, the frequency at which DNA replication is initiated from specific replicators is reduced in a strain with a cdc6 mutation (5, 25). Overexpression of cdc18+ results in repeated rounds of DNA replication, in the absence of mitosis, such that cells accummulate DNA at levels greater than that of normal diploid cells (28, 29, 31). In contrast, reduced activity of Cdc6p/cdc18+ causes underreplication of the genome and premature entry into mitosis (reductional anaphase) (5, 25–27, 30).
These data establish an important role for Cdc6p/cdc18+ in the initiation of DNA replication and in mechanisms by which cells delay mitosis until DNA replication is complete. We have determined that cells from vertebrate organisms contain a closely related protein, which we termed p62cdc6. Several protein structural motifs within human p62cdc6 predicted from the cDNA sequence have potential importance in the control of protein stability and function at replication origins. The chromosomal location and transcriptional start site of a gene encoding human p62cdc6 were mapped, and expression of the protein was characterized in relation to cell cycle progression and mitogenic stimulation in human cells.
MATERIALS AND METHODS
Cloning.
Degenerate oligonucleotide primers for the polymerase chain reaction (PCR) were designed using blocks of six or seven amino acids that were identical, or nearly so, in Cdc6p and cdc18+, but differed in two or more codons from sequences conserved among Orc1 proteins (4, 13). An abundant PCR product closely related to Cdc6p was amplified using cDNA prepared from Xenopus oocytes as template. 5′ and 3′ RACE (rapid amplification of cDNA ends) techniques were used to clone a Xenopus p62cdc6 partial cDNA. Based on the sequence of the Xenopus p62cdc6, new sets of nondegenerate oligonucleotide primers were synthesized, and further rounds of PCR were performed using human cDNA templates. A 687-nt fragment of human p62cdc6 cDNA was radiolabeled and used as the probe for screening a human cDNA library carried in λgt10. From 900,000 phage plaques, 5 positive clones were isolated following three rounds of plaque purification, and sequencing of inserts defined the complete nucleotide sequence of human p62cdc6 cDNA.
Generation of Antibodies and Analysis of mRNA and Protein Expression.
Segments of p62cdc6 cDNA were engineered into plasmid vectors for expression of recombinant protein in bacteria (32). Portions of human or Xenopus p62cdc6 were expressed as glutathione S-transferase fusion proteins in Escherichia coli and purified recombinant proteins were used to immunize rabbits to generate specific antibodies directed against p62cdc6. For detection of the human protein, the best results were obtained with an immunogen consisting of glutathione S-transferase fused to amino acids 51–172 of human p62cdc6. A purified IgG fraction prepared from serum of a rabbit inoculated with this antigen was used in the experiments reported here.
The specificity of this rabbit polyclonal antibody was verified in three ways: (i) in vitro translated p62cdc6 protein was specifically immunoprecipitated; (ii) in immunoblots probed at dilutions of 1/1000 or greater, this antibody recognizes the truncated recombinant protein used as the immunogen (amino acids 51–172), full-length recombinant human p62cdc6 in native form after cleavage from glutathione S-transferase, and an endogenous human nuclear protein of the predicted molecular size; (iii) recombinant p62cdc6 bearing a tag and expressed in HeLa cells was recognized both by anti-p62cdc6 and by anti-hemagglutination antigen antibodies.
Subconfluent cultures of human diploid Wi38 cells were serum starved for 48 hr (time 0) and protein or RNA extracts were prepared from sister cultures at selected times following addition of complete media (DMEM + 10% FBS). HeLa cells were cultured in suspension and cells proliferating in logarithmic phase were separated using centrifugal elutriation (33). DNA content of cells stained with propidium iodide was determined by flow cytometry to calculate the percentages of cells in G1, S, and G2/M phase at each sampling interval. Nuclear and cytoplasmic fractions were prepared as described (34), and proteins were separated by SDS/PAGE before transfer to nitrocellulose membranes, incubation with primary and secondary antibodies as described, and visualization of immune complexes using enhanced chemiluminescence (35). Total RNA was purified from HeLa or Wi38 cells as described (35), separated by agarose gel electrophoresis under denaturing conditions, transferred to nitrocellulose membranes, and probed under conditions of high stringency with human p62cdc6 cDNA radiolabeled with 32P.
Chromosomal Mapping and Determination of the Transcriptional Start Site.
Deoxyoligonucleotide primers based on sequences within human p62cdc6 cDNA were annealed to total human genomic DNA, or DNA extracted from chromosome-specific human-rodent hybrid cell lines (36), and products amplified by PCR. Fluorescence in situ hybridization was performed as described (37). Genomic DNA clones encoding human p62cdc6 were cloned from an arrayed cosmid library limited to segments of human chromosome 17, and restriction fragments were subcloned in plasmid vectors for PCR, Southern blot analysis, and sequencing. The position of the transcriptional start site was mapped by primer extension analysis using a synthetic deoxyoligonucleotide primer complementary to a segment from the 5′ untranslated region (UTR) of the cloned human p62cdc6 cDNA, and RNA isolated from asynchronously growing HeLa cells as the template.
RESULTS
cDNA Sequences Encoding Xenopus and Human p62.
Using PCR-based methods, we cloned a partial cDNA encoding a Xenopus protein closely related to Cdc6p. An identical Xenopus protein has been identified independently by Coleman et al. (9). Knowledge of the Xenopus p62cdc6 sequence was used to design new sets of deoxyoligonucleotide primers, which were successful for generation of specific PCR amplification products encoding segments of human p62cdc6. The largest (687 nt) fragment of human p62cdc6 cDNA obtained by PCR amplification was radiolabeled and used as the probe for screening a human cDNA library carried in bacteriophage λgt10. In the first round of screening of 900,000 phage plaques, 18 clones were positive in duplicate lifts. Of these 18 clones, 5 were positive in duplicate in a second round of screening. Each of these 5 clones was isolated following a third round of plaque purification after plating at low density. Phage DNA was purified and characterized by PCR and restriction digests. cDNA inserts were isolated and cloned into a plasmid vector for sequencing.
The nucleotide sequence of human p62cdc6 cDNA was determined, and the amino acid sequence predicted from the largest open reading frame of 1580 nt is shown in Fig. 1a. The cloned cDNA includes a 5′ UTR of 210 nt and a 3′ UTR of 763 nt. A stop codon in the same reading frame is present in the 5′ UTR, and multiple in-frame stop codons are present within the 3′ UTR downstream of the stop sequence that terminates the open reading frame. No poly(A) tail was detected in cloned cDNA sequences encoding p62cdc6, suggesting that native transcripts may include additional 3′ UTR sequences.
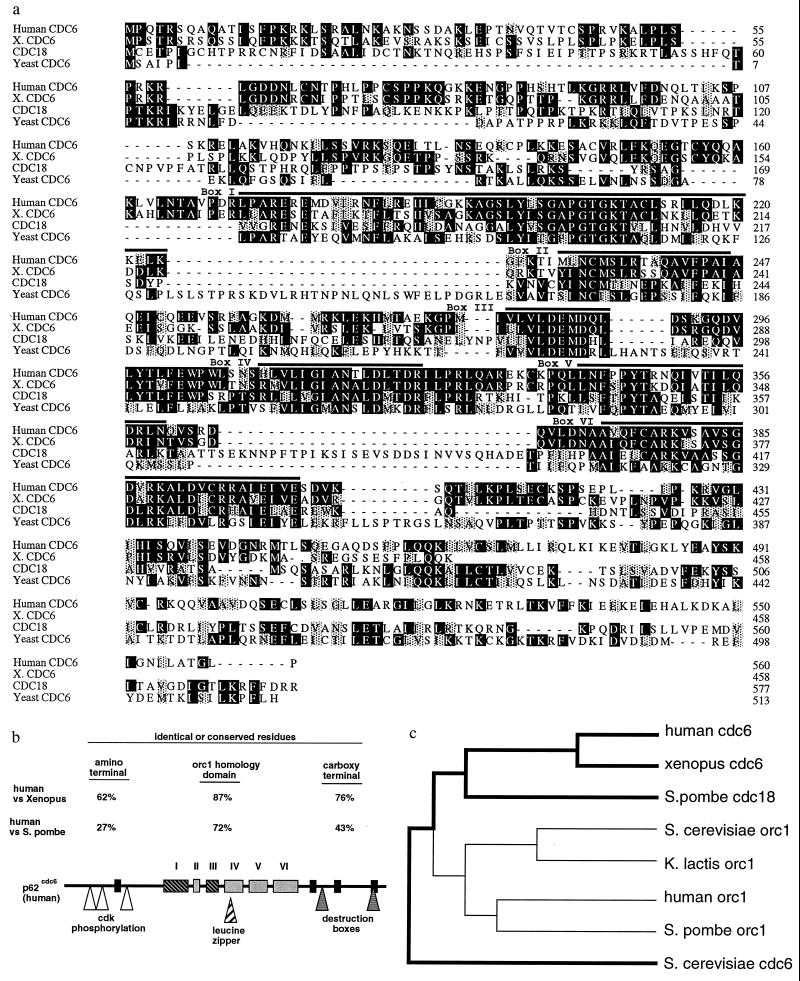
Figure 1.
(a) Multiple alignment of amino acid sequences of human p62cdc6 and related proteins from Xenopus laevis (partial sequence), Saccharomyces cerevisiae, and Schizosaccharomyces pombe. Amino acid residues that are identical are indicated by dark shading, and conservative substitutions are indicated by light shading. The six conserved sequence blocks (I–VI) previously found to be shared among the Orc1 family and fungal Cdc6p/cdc18+ proteins (4, 13), which include the nucleotide binding/ATPase domain (I and III), are found in the midportion of these proteins. Outside of this central domain, no regions of sequence similarity with Orc1-related proteins are evident. (b) Potential structural motifs and highly conserved regions of human p62cdc6. The regions similar to the Orc1 family of proteins are indicated (I–VI). Four other regions of near identity among the vertebrate and yeast Cdc6-related proteins (black rectangles) are present in the amino or carboxyl-terminal regions, and are not found in Orc1 proteins from any species. The positions of several additional motifs of potential biological significance also are indicated. (c) Dendrogram (Genetics Computer Group pileup program) indicating the relative similarities between Orc1- and Cdc6-related proteins from fungal and vertebrate species.
Sequence Conservation of Cdc6-Related Proteins and Potential Structural Motifs Within Human p62cdc6.
Alignment of the amino acid sequence of Cdc6p, cdc18+, Xenopus p62cdc6 (partial), and human p62cdc6 is shown in Fig. 1a, and the position of conserved sequence blocks and potential structural motifs within the human protein is illustrated schematically in Fig. 1b. The degree of sequence identity between human, Xenopus, and yeast cdc6-related proteins is highest in the midportion of the protein that contains six segments also conserved among Orc1-related proteins (4, 13) (Fig. 1 a and b). Other conserved sequence blocks of near identity among fungal and vertebrate cdc6-related proteins, but not found in Orc1 proteins, are also present, and conserved residues are found throughout the entire protein. Human p62cdc6 appears to be more closely related to cdc18+ and to Orc1p than to Cdc6p (Fig. 1c). Human p62cdc6 also includes consensus sites for phosphorylation by cyclin-dependent kinases (38) and destruction box motifs potentially mediating cell cycle-dependent degradation of this protein (39). A potential leucine zipper motif (40) overlaps with conserved sequence block IV.
Chromosomal Location of a Human Gene Encoding p62cdc6.
Two independent sets of primer pairs yielded abundant and specific amplification products in PCR reactions using total human genomic DNA as template. Both of these primer pairs produced identical PCR amplification products using DNA derived from human–hamster cell hybrids as the template, but only from those cells bearing human chromosome 17 (data not shown). No specific PCR amplification products were observed using these primers and hamster or murine genomic DNA as template, or DNA from hamster–human hybrid lines bearing any other human chromosome. The position of the gene encoding p62cdc6 on human chromosome 17 was defined at higher resolution by fluorescence in situ hybridization, which localized the gene to 17q21.3, near to the position of the BRCA1 gene (data not shown). Six overlapping cosmid clones encoding p62cdc6 were isolated from a chromosome 17-specific library. Partial sequencing and Southern blot analysis of these cosmid clones confirmed the presence of sequences identical to the cloned p62cdc6 cDNA.
Transcriptional Start Site and Proximal Promoter of the Human p62cdc6 Gene.
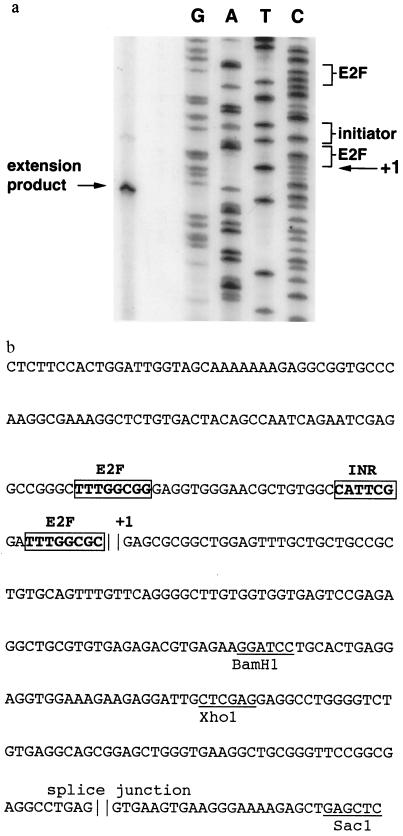
The 5′ end of native p62cdc6 gene transcripts was mapped by primer extension using RNA prepared from asynchronously growing HeLa cells as the template. The major extension product identified the transcriptional start site of the human gene at a position corresponding to the 5′ terminus of the cloned human p62cdc6 cDNA (Fig. 2a). Sequencing of the immediate 5′ flanking region revealed putative sites for transcriptional control, including an initiator element and potential binding sites for E2F proteins (Fig. 2b). No consensus TATA box is evident. This organization resembles that of other genes (cyclin E and ribonucleotide reductase) encoding proteins associated with the G1/S transition (41, 42). A splice junction is present at the point where the genomic sequence diverges from that of the cloned cDNA (Fig. 2b), defining exon 1 of this gene.
Figure 2.
(a) Primer extension analysis of HeLa cell RNA showing the transcriptional start site in relation to the sequence of the p62cdc6 gene cloned from human chromosome 17. The positions of putative transcriptional control elements (initiator element and potential E2F binding sites) are indicated. (b) Nucleotide sequence of proximal promoter region and exon 1 of human gene encoding p62cdc6. Putative transcriptional control elements are indicated in boldface type, and restriction sites are underlined. The first nucleotide (+1) in the cloned p62cdc6 cDNA is shown, and a putative splice junction is present at the point where the genomic sequence diverges from that of the cloned cDNA, defining exon 1.
Expression of Human p62cdc6 mRNA and Protein.
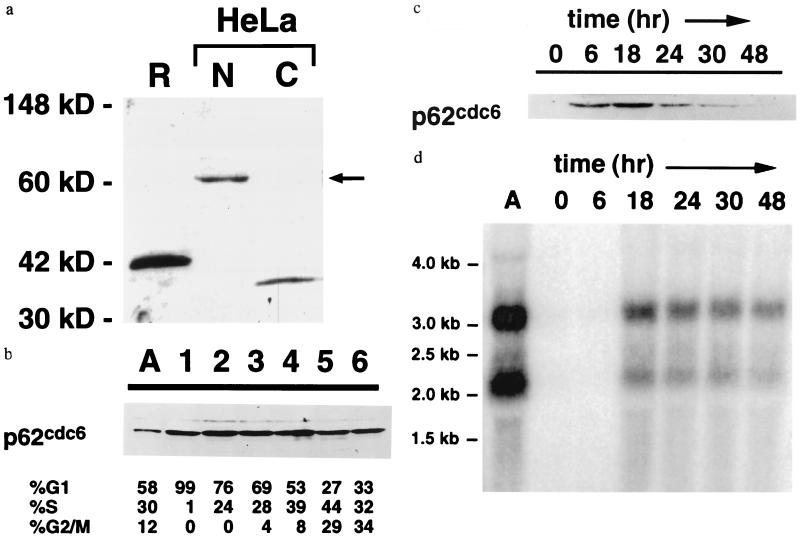
The p62cdc6 protein is localized to nuclei of human cells (Fig. 3a). Cytoplasmic protein extracts contain a cross-reacting antigen of approximately 35 kDa, the identity of which is unknown at this time. In HeLa cells separated by centrifugal elutriation, comparable levels of p62cdc6 were detected in all fractions, irrespective of the proportion of cells in specific stages of the cell cycle (Fig. 3b). The abundance of the 35-kDa cytoplasmic protein also was invariant among the cell populations separated by elutriation, and p62cdc6 was not detected in cytoplasmic extracts from any cell fraction.
Figure 3.
(a) Nuclear localization of p62cdc6 in HeLa cells. Specific antisera raised against a recombinant, truncated form of p62cdc6 (R) expressed in bacteria identified a nuclear protein of 62 kDa in nuclear extracts (N, 20 μg) but not cytoplasmic extracts (C, 50 μg) of asynchronously growing human HeLa cells. (b) Expression of p62cdc6 in asynchronous HeLa cells (A) and in fractions of HeLa cells separated by centrifugal elutriation. Flow cytometry results are indicated. (c) Expression of p62cdc6 in quiescent and proliferating human diploid fibroblasts (Wi38 cells). Cells were grown at low density in the absence of serum for 48 hr and harvested at the indicated times following the addition of serum. The timing of entry into S phase varied among individual experiments, but flow cytometry indicated that the majority of cells (58–70%) consistently were in S phase between 18 and 24 hr after the addition of serum, and this percentage declined to less than 27% as growth became asynchronous. In b and c, each lane was loaded with 32 μg of nuclear protein, and equivalent protein loading was confirmed by staining total proteins with Commassie brilliant blue. Elutriation and serum stimulation experiments were repeated three or more times with similar results, except that expression of p62cdc6 in Wi38 cells at the 6-hr time point following serum stimulation was variable and not detectable in some experiments. (d) Northern blot indicating the abundance of p62cdc6 mRNA transcripts in asynchronous HeLa cells (A) and in Wi38 cells subjected to serum starvation and refeeding, as in c. Lanes were loaded with 15 μg total RNA and equivalent loading was confirmed by staining of ribosomal RNA subunits with ethidium bromide.
In contrast, expression of p62cdc6 was strictly linked to cell proliferation in human diploid fibroblasts: the protein is not detected in quiescent Wi38 cells, but is up-regulated by serum stimulation coincident with resumption of DNA synthesis (Fig. 3c). The induction of p62cdc6 protein by serum in Wi38 cells is accompanied by an increase in p62cdc6 mRNA in these cells (Fig. 3d). Two sizes of human mRNA transcripts hybridize under conditions of high stringency to the cloned human p62cdc6 cDNA. A smaller transcript corresponds in size to the cloned cDNA, whereas a larger 3.2-kb transcript also is expressed in both HeLa and Wi38 cells. The relative abundance of the two transcripts is comparable in both cell types and remains constant during induction by serum in Wi38 cells. As cells recovering from serum starvation enter and complete mitosis, levels of p62cdc6 protein fall more rapidly than the decline in p62cdc6 mRNA (Fig. 3 c and d). This observation is consistent with more rapid degradation of p62cdc6 during G2/M phases of the cell cycle. Growth of these human diploid fibroblasts becomes asynchronous following the first cell cycle after serum stimulation, such that variations in p62cdc6 in subsequent cycles could not be assessed.
DISCUSSION
An extensive body of evidence now supports the replicon model for initiation of DNA replication in yeasts. Many of the proteins that interact physically at yeast replication origins have been identified, and features of the biochemical mechanisms that govern assembly of prereplication complexes and firing of origins are emerging from studies of these organisms. Our understanding of replication origins in metazoan species, including humans, is much less well developed. Progress in this field has been hampered by an inability to define discrete replicator regions at high resolution within the human genome, and by limited information regarding human initiator proteins.
Our current work demonstrates that vertebrates express a protein closely related to Cdc6p and cdc18+ from S. cerevisiae and Sch. pombe, respectively. These proteins are essential for DNA replication in yeasts, and are rate-limiting for firing of individual replication origins. The amino acid sequence of human p62cdc6 is remarkably conserved by comparison to its Xenopus and yeast counterparts. Recently, a Cdc6-related protein identified in Xenopus oocytes (identical to the protein we found) was shown to be required for initiation of DNA replication (9). The degree of sequence conservation across species among Cdc6-related proteins is similar to or greater than that observed previously for ORC protein subunits (13). Conservation of structure among proteins involved in the initiation of replication suggests that fundamental features of origin function are maintained in all eukaryotes.
The CDC6 and cdc18+ genes are expressed in yeast at specific stages of the cell cycle (25, 27, 30, 43). Expression of mRNA encoding Cdc6p peaks at the end of M phase in rapidly cycling S. cerevisiae cells, but a second peak of expression is evident if G1 is prolonged (25, 43). In contrast, the cdc18+ gene is expressed just prior to the G1 to S phase transition (27, 30). Both of these proteins are very unstable: the half-life of cdc18+ and Cdc6p has been estimated as 5 min or less (25, 29). The abundance of cdc18+ is down-regulated by the activity of mitotic cyclins and cyclin-dependent kinase activity, and up-regulated by cyclin-dependent kinase inhibitors (29). The decline in the concentration of Cdc6p/cdc18+ after the initiation of DNA replication appears to be necessary to release checkpoint controls and permit entry into mitosis (22). This observation, coupled with a requirement for renewed synthesis of Cdc6p/cdc18+ after each round of DNA replication, has suggested that Cdc6p/cdc18+ is an important component of the mechanism to ensure that each segment of chromosomal DNA is replicated once, and only once, in each cell cycle.
Our studies of the expression of human p62cdc6 are less complete, but demonstrate a strict association with cell proliferation and DNA synthesis in the transition of diploid fibroblasts from a quiescent, nonreplicating state to active cell cycling. The time course of the induction and subsequent decline in concentrations of p62cdc6 mRNA and protein are consistent with changes in both transcription and protein stability as these cells traverse the cell cycle. By contrast, we were unable to discern major variations in the abundance of p62cdc6 in fractions of asynchronously growing HeLa cells enriched for cells at specific cell cycle stages. The purity of fractions isolated by centrifugal elutriation may be inadequate, however, to exclude the possibility that nuclear concentrations of p62cdc6 may fluctuate within narrow segments of the cell cycle in HeLa cells. In addition, regulation of p62cdc6 expression across the cell cycle may differ in normal somatic cells by comparison to immortalized cell lines. It would not be surprising if mechanisms controlling the abundance and activity of initiatior proteins differ among species or cell types.
The amino acid sequence of human p62cdc6 includes a number of structural motifs predicted to have bearing on the regulation and function of the protein in DNA replication. We present no evidence at this time to demonstrate functions of these regions, but their presence within the human protein serves to generate several testable hypotheses. Phosphorylation by cyclin-dependent kinases may modify protein stability or influence interactions with other initiator factors. Destruction box motifs also may influence stability of human p62cdc6, perhaps by facilitating interactions with components of the ubiquitin–proteasome pathway of protein degradation (44). A potential leucine zipper motif provides a site that may be involved in protein–protein contracts important for origin function. Within the central region of the protein that is conserved among Cdc6- and Orc1-related proteins, conservation of a nucleotide binding domain predicts that human p62cdc6 has ATPase activity important for its function as a replication initiation protein. Amino acid substitutions within the nucleotide binding/ATPase domain of Cdc6p in S. cerevisiae impair DNA replication (M. Weinreich and B.S., unpublished data).
We identified and cloned a gene encoding p62cdc6 on human chromosome 17q21.3. Both HeLa cells and Wi38 human diploid fibroblasts express two mRNA transcripts of different sizes that hybridize to the cloned p62cdc6 cDNA under conditions of high stringency. This finding raises the possibility that a second gene encoding a closely related isoform of p62cdc6 is present in the human genome, but our data suggest that the two forms of p62cdc6 mRNA are more likely to arise by alternative splicing within the 3′ UTR of primary transcripts from a single gene. A putative promoter region from the human p62cdc6 gene includes potential binding sites for E2F transcription factors, suggesting parallels to the control of yeast initiator proteins by Swi/MBF proteins (25).
Cloning of human p62cdc6 provides new opportunities to increase our understanding of replicator elements within the human genome and of the molecular events that control the initiation of DNA replication in human cells. The overreplication phenotype produced in yeast by forced expression of cdc18+ suggests a potential role for p62cdc6 in human malignancies, either in initial stages of oncogenesis, or in later stages of tumor progression characterized by chromosomal instability and polyploidy. It should now be possible to determine whether inherited or somatic mutations at the p62cdc6 locus are present in human cancers. In addition, human p62cdc6 represents a novel target for development of drugs to limit cell proliferation in human diseases.
Acknowledgments
We are grateful for technical assistance from Miho Waga and Maggy Fina. Dr. Rong Li of Cold Spring Harbor Laboratory provided useful discussion and assistance with elutriation experiments. Fluorescense in situ hybridization was performed by Dr. Roger Schultz of the University of Texas Southwestern Medical Center. A panel of rodent/human hybrids was obtained from the Coriell Institute (Camden, NJ). Blots of an arrayed chromosome 17 cosmid library (from Larry Deaven at Los Alamos National Laboratory) were provided by Dr. Anne Bowcock of the University of Texas Southwestern Medical Center. This work was supported by National Institutes of Health Grants HL06296 and HL54794 (R.S.W.) and CA13106 (B.S.).
Footnotes
References
- 1.Bell S P, Stillman B. Nature (London) 1992;357:128–134. doi: 10.1038/357128a0. [DOI] [PubMed] [Google Scholar]
- 2.Diffley J F X, Cocker J H. Nature (London) 1992;357:169–172. doi: 10.1038/357169a0. [DOI] [PubMed] [Google Scholar]
- 3.Diffley J F X, Cocker J H, Dowell S J, Rowley A. Cell. 1994;78:303–316. doi: 10.1016/0092-8674(94)90299-2. [DOI] [PubMed] [Google Scholar]
- 4.Bell S P, Mitchell J, Leber J, Kobayashi R, Stillman B. Cell. 1995;83:563–568. doi: 10.1016/0092-8674(95)90096-9. [DOI] [PubMed] [Google Scholar]
- 5.Liang C, Weinreich M, Stillman B. Cell. 1995;81:667–676. doi: 10.1016/0092-8674(95)90528-6. [DOI] [PubMed] [Google Scholar]
- 6.Rowley A, Cocker J H, Harwood J, Diffley J F X. EMBO J. 1995;14:2631–2641. doi: 10.1002/j.1460-2075.1995.tb07261.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Loo S, Fox C A, Rine J, Kobayashi R, Stillman B, Bell S. Mol Biol Cell. 1995;6:741–756. doi: 10.1091/mbc.6.6.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rao H, Stillman B. Proc Natl Acad Sci USA. 1995;92:2224–2228. doi: 10.1073/pnas.92.6.2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coleman T R, Carpenter P B, Dunphy W G. Cell. 1996;87:53–63. doi: 10.1016/s0092-8674(00)81322-7. [DOI] [PubMed] [Google Scholar]
- 10.Stillman, B. (1996) Science, in press.
- 11.Ehrenhofer-Murray A E, Gossen M, Pak D T S, Botchan M R, Rine J. Science. 1995;270:1671–1674. doi: 10.1126/science.270.5242.1671. [DOI] [PubMed] [Google Scholar]
- 12.Rowles, A., Chong, J. P. J., Brown, L., Howell, M., Evan, G. I. & Blow, J. J. (1996) Cell, in press. [DOI] [PubMed]
- 13.Gavin K A, Hidaka M, Stillman B. Science. 1995;270:1667–1671. doi: 10.1126/science.270.5242.1667. [DOI] [PubMed] [Google Scholar]
- 14.Gossen M, Patk D T S, Hanse S K, Acharya J K, Botchan M R. Science. 1995;270:1674–1677. doi: 10.1126/science.270.5242.1674. [DOI] [PubMed] [Google Scholar]
- 15.Muzi-Falconi M, Kelly T J. Proc Natl Acad Sci USA. 1995;92:12475–12479. doi: 10.1073/pnas.92.26.12475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carpenter P B, Meuller P R, Dunphy W G. Nature (London) 1996;379:357–360. doi: 10.1038/379357a0. [DOI] [PubMed] [Google Scholar]
- 17.Dahmann C, Diffley J F X, Nasmyth K A. Curr Biol. 1995;5:1257–1269. doi: 10.1016/s0960-9822(95)00252-1. [DOI] [PubMed] [Google Scholar]
- 18.Diffley J F X. Yeast. 1995;11:1651–1670. doi: 10.1002/yea.320111608. [DOI] [PubMed] [Google Scholar]
- 19.Cocker J H, Piatti S, Santocanale C, Nasmyth K, Diffley J F X. Nature (London) 1996;379:180–182. doi: 10.1038/379180a0. [DOI] [PubMed] [Google Scholar]
- 20.Lisziewicz J, Godany A, Agoston D B, Küntzel H. Nucleic Acids Res. 1988;16:11507–11520. doi: 10.1093/nar/16.24.11507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou C, Huang S-H, Jong A. J Biol Chem. 1989;264:9022–9029. [PubMed] [Google Scholar]
- 22.Bueno A, Russell P. EMBO J. 1992;11:2167–2176. doi: 10.1002/j.1460-2075.1992.tb05276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hartwell L H. J Mol Biol. 1976;104:803–817. doi: 10.1016/0022-2836(76)90183-2. [DOI] [PubMed] [Google Scholar]
- 24.Hogan E, Koshland D. Proc Natl Acad Sci USA. 1992;89:3098–3102. doi: 10.1073/pnas.89.7.3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Piatti S, Lengauer C, Nasmyth K. EMBO J. 1995;14:3788–3799. doi: 10.1002/j.1460-2075.1995.tb00048.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kelly T J, Nurse P, Forsburg S L. Cold Spring Harbor Symp Quant Biol. 1993;58:637–644. doi: 10.1101/sqb.1993.058.01.071. [DOI] [PubMed] [Google Scholar]
- 27.Kelly T J, Martin G S, Forsburg S L, Stephen R J, Russo A, Nurse P. Cell. 1993;74:371–382. doi: 10.1016/0092-8674(93)90427-r. [DOI] [PubMed] [Google Scholar]
- 28.Nishitani H, Nurse P. Cell. 1995;83:397–405. doi: 10.1016/0092-8674(95)90117-5. [DOI] [PubMed] [Google Scholar]
- 29.Jallepalli P, Kelly T. Genes Dev. 1996;10:541–552. doi: 10.1101/gad.10.5.541. [DOI] [PubMed] [Google Scholar]
- 30.Muzi-Falconi M, Brown G W, Kelly T J. Proc Natl Acad Sci USA. 1996;93:1566–1570. doi: 10.1073/pnas.93.4.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leatherwood J, Lopez-Girona A, Russell P. Nature (London) 1996;379:360–363. doi: 10.1038/379360a0. [DOI] [PubMed] [Google Scholar]
- 32.Studier F W, Rosenberg A H, Dunn J J, Dubendorff J W. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 33.Buchkovich K, Duffy L A, Harlow E. Cell. 1989;58:1097–1105. doi: 10.1016/0092-8674(89)90508-4. [DOI] [PubMed] [Google Scholar]
- 34.Grayson J, Williams R S, Yu Y T, Bassel-Duby R. Mol Cell Biol. 1995;15:1870–1878. doi: 10.1128/mcb.15.4.1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bassel-Duby R, Hernandez M D, Yang Q, Rochelle J M, Seldin M F, Williams R S. Mol Cell Biol. 1994;14:4596–4605. doi: 10.1128/mcb.14.7.4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jackson C. In: Current Protocols In Human Genetics. Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. New York: Wiley; 1995. pp. 3.2.1–3.2.29. [Google Scholar]
- 37.Knoll J, Lichter P. In: Current Protocols in Human Genetics. Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. New York: Wiley; 1995. pp. 4.3.1–4.3.29. [Google Scholar]
- 38.Jans D A, Moll T, Nasmyth K, Jans P. J Biol Chem. 1995;270:17064–17067. doi: 10.1074/jbc.270.29.17064. [DOI] [PubMed] [Google Scholar]
- 39.Amon A, Irniger S, Nasmyth K. Cell. 1994;77:1037–1050. doi: 10.1016/0092-8674(94)90443-x. [DOI] [PubMed] [Google Scholar]
- 40.Landschultz W H, Johnson P F, McKnight S L. Science. 1988;240:1759–1764. doi: 10.1126/science.3289117. [DOI] [PubMed] [Google Scholar]
- 41.Ohtani K, DeGregori J, Nevins J R. Proc Natl Acad Sci USA. 1995;92:12146–12150. doi: 10.1073/pnas.92.26.12146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Johansson E, Skogman E, Thelander L. J Biol Chem. 1995;270:30162–30167. doi: 10.1074/jbc.270.50.30162. [DOI] [PubMed] [Google Scholar]
- 43.Zwerschke W, Rottjakob H-W, Küntzel H. J Biol Chem. 1994;269:23352–23356. [PubMed] [Google Scholar]
- 44.Kirschner, M. (1996) Science, in press.