Abstract
The insufficient selectivity of drugs is a bane of present-day therapies. This problem is significant for antibacterial drugs, difficult for antivirals, and utterly unsolved for anticancer drugs, which remain ineffective against major cancers, and in addition cause severe side effects. The problem may be solved if a therapeutic agent could have a multitarget, combinatorial selectivity, killing, or otherwise modifying, a cell if and only if it contains a predetermined set of molecular targets and lacks another predetermined set of targets. An earlier design of multitarget drugs [Varshavsky, A. (1995) Proc. Natl. Acad. Sci. USA 92, 3663–3667] was confined to macromolecular reagents such as proteins, with the attendant difficulties of intracellular delivery and immunogenicity. I now propose a solution to the problem of drug selectivity that is applicable to small (≤1 kDa) drugs. Two ideas, codominant interference and antieffectors, should allow a therapeutic regimen to possess combinatorial selectivity, in which the number of positively and negatively sensed macromolecular targets can be two, three, or more. The nature of the effector and interference moieties in a multitarget drug determines its use: selective killing of cancer cells or, for example, the inhibition of a neurotransmitter-inactivating enzyme in a specific subset of the enzyme-containing cells. The in vivo effects of such drugs would be analogous to the outcomes of the Boolean operations “and,” “or,” and combinations thereof. I discuss the logic and applications of the antieffector and interference/codominance concepts, and the attendant problem of pharmacokinetics.
Keywords: pharmacology, cancer, codominance, selectivity
The many successes of pharmacology (1, 2) do not include the problem of cancer. Major human cancers are incurable once they have metastasized. A few relatively rare cancers, such as testicular carcinoma in men, Wilms’ kidney tumor, and some leukemias in children, can often be cured through chemotherapy but require cytotoxic treatments of a kind that cause severe side effects and are themselves carcinogenic (2).
The main reason for the failure of cytotoxic therapies is their insufficient selectivity for tumors. For example, treatments with radiation or alkylating agents perturb many functions that are common to all cells. The more selective cytotoxic drugs, for instance, methotrexate, taxol, and etoposide, perturb the functions of specific macromolecular targets (dihydrofolate reductase, microtubules, and topoisomerase II), but these targets are present in both normal and malignant cells (1, 2). Hence the low therapeutic index of anticancer drugs and their systemic toxicity at clinically relevant doses. Because the mitotic activity of cells in a tumor is often lower than the mitotic activity of normal cells in self-renewing tissues such as the bone marrow (3), one might not have expected these drugs to work at all—to have any preference for the killing of cancer cells. That such preference actually exists stems in part from the fact that tumor cells are often perturbed by their mutations into stress-hypersensitive states. Consequently, these tumor cells die an apoptotic death at the level of a drug-imposed metabolic stress that induces apoptosis in some but not in most of the organism’s normal cells (3).
With some cancers, cytotoxic therapies are ineffective from the beginning. In other cases, these therapies yield a partial, sometimes clinically complete, but almost invariably transient remission of a cancer, in part because these treatments select for tumor cell variants that retain tumorigenicity but are more resistant to either apoptosis per se or a drug that induces apoptosis. Because a significant increase in the drug or radiation dosage is precluded by their low therapeutic index, these therapies become ineffective when resistant clones of malignant cells, selected by a drug treatment, present themselves as a cancer recurrence.
The failure of small cytotoxic drugs to produce a cure for cancer has given rise to other strategies, in particular the insightful suggestion that solid tumors can be targeted by selectively inhibiting neovascularization, a process that these tumors depend on for growing to a clinically significant size (4). Another approach, immunotoxins, involves the linking of a toxin to a ligand such as an antibody or a growth factor that binds to a target on the surface of tumor cells (5). Among the limitations of present-day immunotoxins is their incapacity, on entering a cell, to adjust their toxicity in response to the intracellular protein composition. Yet another approach is to enhance the ability of the immune system to identify and selectively destroy tumor cells. The current revival of this strategy holds the promise of a rational and curative treatment (6). Given the complicated regimens and unsolved problems of immunotherapies, it is clear that this and other recent approaches (7, 8) are motivated in part by the perception that small-drug pharmacology, so successful against bacterial infections, is unlikely to prove effective against cancer. In contrast to this view, the premise of the strategy described in the present work is that small anticancer drugs may become curative and free of severe side effects if a way is found to confer on these compounds a multitarget, combinatorial selectivity.
Most cancers are monoclonal: cell lineages of both the primary tumor and the metastases originate from a single founder cell. This cell is a breakthrough descendant of a cell lineage that has been accumulating mutations for some time, often in proximity to other neoplastic but still nonmetastatic cell lineages within an indolent proliferative lesion such as a benign tumor (9, 10). Given the monoclonality of a cancer, cells of both the primary tumor and the metastases share the initial mutations that yielded the founder cell, even if these cells differ at other loci that accumulated mutations in the course of the later tumor progression. Some of the early mutations are in genes that encode tumor suppressors (9, 11, 12). In most cancers, both alleles of a tumor suppressor gene are inactivated, sometimes through deletions that encompass the gene on the two homologous chromosomes. Thus, a monoclonal cancer, although heterogeneous genetically, always contains a set of founder mutations that is shared by all of its cells.
A drug that kills a cell only if it lacks a specific macromolecular target would distinguish tumor cells from many other cells of an organism, provided that the target is a product of a gene that had been deleted or inactivated in this cancer at the stage of its founder cell. Such a drug may be especially selective against cancers that lack a gene for a ubiquitously expressed tumor suppressor, for example, the retinoblastoma (Rb) protein (11, 12). An example of the negative-target approach is the use of a mutant adenovirus that replicates selectively in human cancer cells lacking the tumor suppressor p53 and has been shown to kill these cells in a model setting (8).
However, other tumor suppressors may not be expressed at comparable levels in most cells. A drug that kills a cell if it lacks a nonubiquitous tumor suppressor would be toxic to a subset of normal cells as well. This problem could be reduced through the use of a drug that is toxic only if a cell lacks two specific macromolecules, termed negative targets. Two judiciously chosen negative targets may, together, suffice to distinguish all of the cancer cells from all of the organism’s normal cells. If they do not, a third negative target that had been deleted or rendered defective in a given cancer can be employed as well. This strategy requires a drug that possesses the ability to kill a cell if it lacks two or more of the predetermined targets, but would spare a cell containing either one of these targets.
Other changes in a founder cell may involve a missense mutation, an amplification and overexpression, an ectopic expression, or a translocation/fusion of a specific protooncogene such as, for example, Ras or Myc (9, 10, 13). A single oncoprotein may not be a unique enough target by itself, for reasons similar to those described above in the context of negative targets. However, a combination of two or more distinct oncoproteins that were either mutated or inappropriately expressed in the founder cell can be employed to formulate the unique multiprotein signature of a specific cancer that comprises both positive and negative targets.
These considerations suggest that a conditionally cytotoxic therapeutic regimen that is exquisitely specific for a given cancer, and therefore would eliminate it without significant side effects, must possess, in most cases, a multitarget, combinatorial (positive/negative) selectivity of the kind defined above. Conversely, even an informed choice of the molecular target for a single-target drug may not suffice to define unambiguously the cell type to be eliminated. Note that simply combining two single-target drugs against two different targets in a multidrug regimen would not yield a multitarget selectivity, because the two drugs together would perturb not only cells containing both targets but also cells containing either one of the targets.
Although the problem of insufficient selectivity is not as acute with noncytotoxic drugs, it is relevant to them as well. Among the multitude of examples are side effects of therapies with antipsychotic agents. The side effects are caused in part by the insufficient molecular specificity of drugs, which is exemplified by the ability of antidepressants that inhibit monoamine oxidase to perturb other proteins as well (2). This difficulty will continue to abate with the development of more specific single-target inhibitors. But an entirely distinct, major, and unsolved problem with inhibitors as drugs is the current impossibility of restricting their action to a specific subset of cells among those that contain the inhibitor’s target. For example, even an exquisitely specific inhibitor of a clinically relevant enzyme is likely to have side effects, because the target enzyme is present, in most cases, not only in the cells where its inhibition is clinically beneficial but also in the cells where its inhibition is physiologically inappropriate. The present work describes a possible solution of this problem.
A previously proposed approach to designing multitarget drugs utilized degradation signals (degrons) and analogous signals that exhibit the property of codominance (14, 15). As a result, this strategy was confined to macromolecular reagents such as proteins, with the attendant problems of immunogenicity, extravasation, and intracellular delivery. The latter difficulty is especially significant, because either gene-therapy or direct-delivery methods for introducing large molecules into cells work reasonably well with cells in culture but are still inefficient with cells in an intact organism. The challenge, then, is to attain a multitarget, combinatorial selectivity in the setting of small (≤1 kDa) drugs, where the immunogenicity and delivery problems are less severe. A solution, described below, invokes a modification of the earlier idea of codominant interference (14) in conjunction with the new concept of antieffectors. This solution is applicable to either cytotoxic or noncytotoxic therapies.
RESULTS AND DISCUSSION
Multitarget Compounds Specific for Negative Targets: The Concept of Codominant Interference.
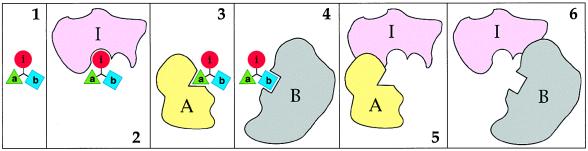
Previous work (14) suggested that the property of codominance, characteristic of degradation signals (degrons) and many other signals in biopolymers, can be employed to design protein-based reagents that possess multitarget, combinatorial selectivity of the kind defined above. Codominance refers to the ability of two or more signals in the same molecule to function independently and not to interfere with each other. It is shown below that a distinct version of the interference/codominance (IC) concept (14) is applicable to small (≤1 kDa) compounds. Consider a reagent containing three small moieties a, b, and i, which can bind, respectively, to three macromolecular targets A, B, and I. Because the moieties a, b, and i are much smaller than the macromolecules A, B, and I, it should be possible to arrange these moieties in the compound abi in such a way that the binding of A or B to a or b would preclude, through steric hindrance, the binding of moiety i to I (Fig. 1).
Figure 1.
The interference/codominance concept. (1–4) A small (<1 kDa) moiety i is linked to two other small moieties, a and b. The moieties a, b, and i are ligands of the macromolecules A, B, and I, respectively. The distances between a, b, and i, and their mutual arrangement in the tripartite compound abi are such that the interaction i-I is mutually exclusive with either the interaction a-A or the interaction b-B. Specifically, the macromolecule I in its complex with the small moiety i would sterically clash with the macromolecules A or B if either A or B is positioned to bind a or b of abi (5 and 6). In the diagram, the interactions a-A and b-B are also mutually exclusive, but this constraint is not essential. (Note that if the interactions a-A and b-B were mutually nonexclusive, the compound abi would promote the binding of A to B.) The codominance aspect of the IC concept allows this design to accommodate more than two of the a, b-like competition modules (not shown). In Figs. 2–4, the i moiety is an inhibitor of an essential enzyme I. In fact, the only constraint on the identities of i and I is the requirement for an i-I interaction to alter the functional activity of a macromolecule I. In other words, the choice of I is determined by the intended effect of the (unsequestered) compound abi (see the main text).
That the interactions of a small bipartite compound with its two macromolecular ligands can be made mutually exclusive is expected from basic physicochemical considerations. This has also been demonstrated directly, in a context unrelated to the present discussion. When lisinopril, an inhibitor of the angiotensin-converting enzyme (ACE), was connected, via an 11-atom linker, to the biotin moiety, the resulting bivalent compound could bind to and inhibit ACE in the absence but not in the presence of the biotin-binding protein streptavidin (16). Small compounds comprising two linker-connected moieties such as cyclosporin and FK506, which are specific for two macromolecular targets, have previously been employed as in vivo dimerization devices, making it possible to bring together two otherwise noninteracting proteins (17). However, the linker moiety of these bipartite compounds was chosen to allow simultaneous interactions with the targets, in contrast to the mutual exclusivity of interactions in the IC approach (Fig. 1).
If the moiety i is an inhibitor of an essential cellular enzyme I, the presence of the macromolecular targets A or B in a cell would reduce the inhibition of enzyme I by abi, because the complexes abi-A and abi-B would be mutually exclusive with the complex abi-I (Fig. 1). Note that A and B are codominant in their ability to reduce the inhibition of I by abi. Therefore, there is, formally, no limit on the number of a, b-like competition modules that can be used to construct an abi-like compound whose activity is sensitive to the presence of several distinct macromolecules, called negative targets. The fractional occupancy of the macromolecular targets A, B, and I by the a, b, and i moieties of abi would be determined in part by the targets’ intracellular concentrations. There are also specific pharmacokinetic constraints on the selectivity of abi, an issue discussed below.
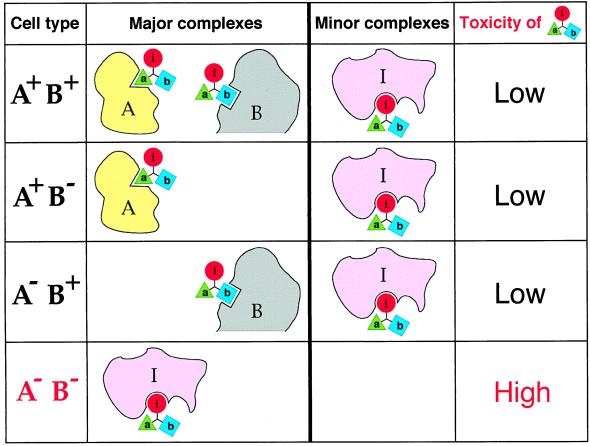
A tabulation of the relative toxicities of abi for cells that either lack or contain the negative targets A and B is shown in Fig. 2. It can be seen that abi would be relatively nontoxic to three of the four cell types and toxic exclusively to the cells that lack both A and B (Fig. 2). Thus, the IC concept allows the construction of small compounds that exhibit multitarget selectivity for negative targets. One more idea is required to accomplish the same for positive targets and to link the two strategies.
Figure 2.
Multitarget selectivity of a compound that utilizes interference/codominance. This diagram tabulates the relative toxicities of the compound abi for cells that either lack or contain macromolecular targets A and B. The i moiety of the compound abi (see the legend to Fig. 1) inhibits an essential enzyme I. The interaction i-I is mutually exclusive with the interaction a-A and the interaction b-B, the macromolecules A and B being negative targets of abi. It is assumed that concentrations of the targets A and B in cells that contain at least one of them significantly exceed the concentration of I (see the main text). In A+ B+, A+ B−, and A− B+ cells, the enzyme I would be at most partially inhibited by the i moiety of abi, because of the competing interactions of abi with A and/or B. By contrast, in A− B− cells, the bulk of abi molecules would be available for interaction with I, resulting in the selective toxicity of abi to these cells. The selectivity pattern of abi requires that certain pharmacokinetic conditions are met as well (see the main text). Note that the physiological effects and the uses of abi-type compounds are not confined to cytotoxic regimens.
Multitarget Compounds Specific for Positive Targets: The Concept of Antieffectors.
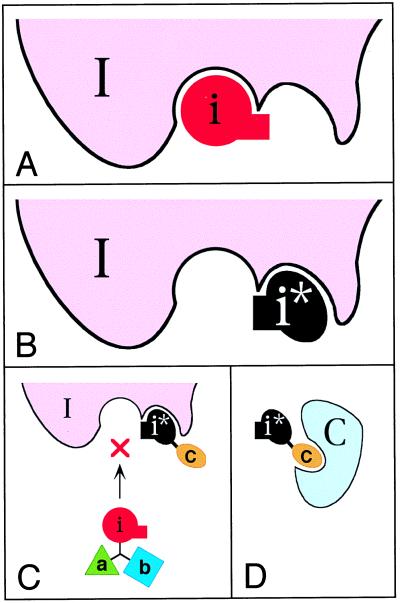
Consider a small compound i* that binds to enzyme I in the vicinity of its active site, but does not perturb the catalytic activity of I toward its physiological substrates (Fig. 3B). Suppose further that the compound i*, termed an antiinhibitor, was designed to interfere, sterically, with the binding of an inhibitor i to the enzyme’s active site while at the same time allowing the binding of physiological substrates. One way to achieve this would be to endow either i, or i*, or both of them with a set of chemical groups, termed a “bump,” whose function is to produce steric hindrance that makes the interactions i-I and i*-I mutually exclusive (Fig. 3 A and B). A moiety that functions as a bump may also be designed to enhance specific binding of either the inhibitor i or the antiinhibitor i* to enzyme I, but this consideration is secondary to the bump’s essential purpose.
Figure 3.
The antieffector concept. The particular case illustrated here is that of an antiinhibitor i*, defined as a compound whose binding to an enzyme I does not inhibit the activity of I but does preclude the inhibition of I by an inhibitor i. In this example, the antiinhibitor i* has the following properties. First, it binds to I in the vicinity of the I’s active site, but does not perturb the catalytic activity of I toward its physiological substrates. Second, i*, in its bound state, sterically interferes with the interaction between I and its inhibitor i. To implement the second condition, either i, or i*, or both of them bear additional moiety, a “bump,” denoted by the rectangular protrusions in i and i*. The function of the bump is to produce steric hindrance that makes the interactions i-I and i*-I mutually exclusive. The inhibitor i described here and in the main text is a competitive inhibitor, but i could be a noncompetitive inhibitor as well. An allosteric antiinhibitor, which functions through binding to a remote site of enzyme I, is yet another possibility. (A) A complex of the enzyme I with its inhibitor i. (B) A complex of I with its antiinhibitor i*. Note that the bumps of the bound i and i* spatially overlap. (C) The antiinhibitor i* is linked to c, a small moiety that can bind to a macromolecular target C. The design of ci* is analogous to abi (Figs. 1 and 2), in that the interactions of ci* with I and C are mutually exclusive. When ci* is bound to I, the inhibitor i, shown here as a part of the compound abi (Figs. 1 and 2), is unable to bind to and inhibit the enzyme I. (D) A complex between ci* and its macromolecular target C. This complex, being mutually exclusive with the ci*-I complex, reduces the ability of ci* to protect the enzyme I from inhibition by abi.
In one application of the antiinhibitor i*, it is linked to c, a small moiety that can bind to a macromolecular target C. The mutual arrangement of i* and c in ci* is such that the interactions of ci* with I and C are mutually exclusive. In the absence of C, ci* would compete with abi for the binding to enzyme I, thereby partially protecting I from inhibition by abi (Fig. 3C). This protective effect of ci* would be suppressed in the presence of its macromolecular target C (Fig. 3D). In the logic of codominance, discussed above in the context of negative targets, a compound bearing an antiinhibitor moiety i* could contain more than one c-like moiety. For example, a compound cdi*, whose moieties c and d can bind, respectively, to the macromolecules C and D, would reduce the inhibition of enzyme I by abi only in the absence of both C and D. Yet another pattern of multitarget selectivity can be produced, in this context, by separating the competition moieties c and d. The resulting ci* and di*, if administered together with abi, would reduce the inhibition of enzyme I by abi if just one of the targets, C or D, is absent. As shown below, the key merit of the antiinhibitor idea is that it allows the effect of a single inhibitor i to be modulated by both negative and positive macromolecular targets.
On the Difference Between Antieffectors and Antagonists.
The distinctions between substrates, inhibitors, and antiinhibitors were described above. The concept of antieffectors is also relevant to ligand-binding biopolymers other than enzymes. For example, an agonist binds to its receptor and evokes a physiological response. An antagonist binds to a site of the receptor that overlaps with the agonist-binding site, does not activate the receptor, and in addition precludes the binding of agonist (1, 2). By contrast, an antieffector, which would be called, in this setting, an antiantagonist, binds to the receptor in such a way that the receptor can still bind, and respond to, the agonist, but cannot bind the antagonist. To this end, either an antagonist, or an antiantagonist, or both must possess a bump, an additional moiety described above in the context of enzymes and antiinhibitors (Fig. 3). The idea of antieffectors is thus distinct from that of antagonists or inhibitors and is new, to the best of my knowledge. I am also not aware of a naturally occurring pair of compounds that satisfy the definition of effectors/antieffectors in a physiologically relevant setting.
Interference/Codominance and Antieffector in a Regimen That Possesses Combinatorial Selectivity.
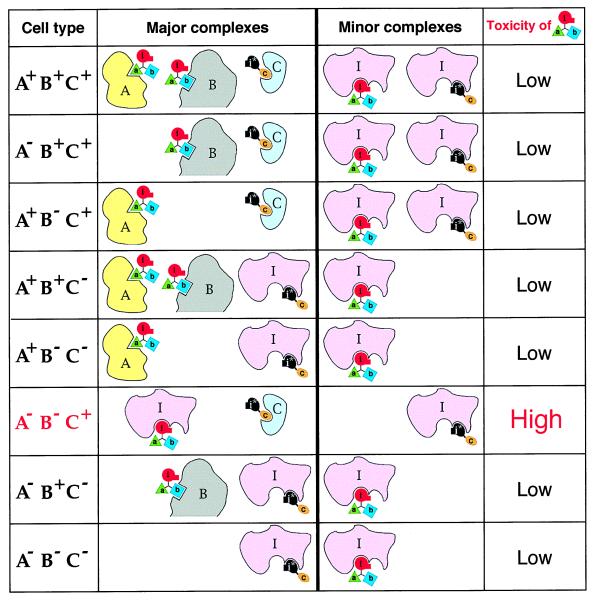
Applying the IC and antieffector concepts together yields regimens that possess true combinatorial selectivity, i.e., sensitivity to both negative and positive targets. Consider a population of cells that either contain or lack the macromolecular targets A, B, and C. Our aim is to devise a treatment that would be toxic to cells that lack A and B but contain C (A− B− C+ cells) and relatively nontoxic to the other cell types (Fig. 4). A regimen of two compounds, abi (Fig. 3C) and ci* (Fig. 3D), has the requisite selectivity, as shown in Fig. 4, which tabulates the outcomes of this treatment for different cell types. Specifically, in the absence of C (four cell types out of eight), the antiinhibitor-containing ci* would compete with the inhibitor-containing abi for binding to the essential enzyme I, thereby reducing the inhibition of I by abi, and hence reducing the toxicity of abi. In three other cell types, whose common property is the presence of C and at least one of the other two targets, A or B, the antiinhibitor-containing ci* would be largely sequestered by C, and hence inactive, but the inhibitor-containing abi would be sequestered as well, by either A or B. In only one type of cells, those that lack A and B but contain C (A− B− C+ cells), is the inhibitor abi fully available for interaction with I, resulting in higher toxicity (Fig. 4). The differences in the toxicity of abi to different cell types would be determined by the relative stoichiometries and absolute concentrations of the cellular targets involved (A, B, C, and I), by the affinities of the moieties a, b, c, i, and i* for these targets, and by the pharmacokinetic properties of abi and ci*.
Figure 4.
Combinatorial (positive/negative) selectivity of a regimen that utilizes interference/codominance and antiinhibitor. The diagram tabulates the relative toxicities of the compound abi in the setting of a two-compound treatment of cells that either lack or contain the macromolecular targets A, B, and C. The inhibitor-containing compound abi is described in the main text and Figs. 1, 2, and 3C. The antiinhibitor-containing compound ci* is described in the main text and Fig. 3. A and B are negative targets, in that they reduce, through the binding to the moieties a and b of abi, the inhibition of an essential enzyme I by abi. C is a positive target, in that it reduces, through the binding to the c moiety of ci*, the binding of ci* to enzyme I. This results in a larger fraction of enzyme I available for the inhibition by abi. It is assumed that the concentrations of A, B, and C in cells that contain them significantly exceed the concentration of I (see the main text). In all of the cell types except A− B− C+, the enzyme I would be at most partially inhibited by the i moiety of abi, because of the competing interactions of abi with A and/or B, and also because in C− cells a fraction of enzyme I would be protected from the inhibition by abi through the interaction of I with the antiinhibitor moiety i* of ci*. By contrast, in A− B− C+ cells, the antiinhibitor-containing ci* would be sequestered by C, whereas abi would not be sequestered by A or B, which are absent from these cells. As a result, a larger fraction of the inhibitor-containing abi molecules would be available for the interaction with I, resulting in the selective toxicity of abi to A− B− C+ cells. The selectivity pattern of abi requires that certain pharmacokinetic conditions are met as well (see the main text). Note that the physiological effects and the uses of abi-type compounds are not confined to cytotoxic regimens.
Straightforward variations of the abi and ci* designs that utilize IC and the properties of antiinhibitors would allow selective targeting of any one of the eight cell types that differ by the presence or absence of three macromolecular targets. Moreover, there is no formal limit on the total number of negative and/or positive targets that can be simultaneously sensed by regimens that employ abi- and ci*-type compounds bearing multiple interference moieties. Note that the cell type selectivity of regimens such as abi + ci* (Fig. 4) is analogous to the outcomes of the Boolean operations “and,” “or,” and combinations thereof.
Stoichiometries, Affinities, and Pharmacokinetics.
The selectivity of the proposed compounds results from mutually exclusive, competing interactions between individual moieties of these compounds and their macromolecular targets (Figs. 2–4); hence, the importance of the targets’ intracellular concentrations, relative to each other and the enzyme I, which is inhibited by an effector moiety of these drugs. The choice of I is not confined to essential enzymes. The target I could be, for instance, a DNA-binding repressor of terminal differentiation, a repressor of apoptosis, or, in the example of a noncytotoxic therapy, a neurotransmitter-inactivating enzyme. In other words, the choice of I is determined by the intended effect of the (unsequestered) compound abi.
The sequestration of abi- and ci*-type compounds by their macromolecular ligands A, B, and C serves to prevent their binding to the enzyme I (Figs. 3 and 4). Therefore, in schemes of the type considered above, the molar concentration of I should be significantly (if possible, considerably) lower than the molar concentrations of A, B, and C. In addition, the concentration of ci* in a ci* + abi regimen should significantly exceed that of abi, because ci* is the sole obstacle to the inhibition of enzyme I by abi in the A− B− C− cells (Fig. 4). It is assumed, furthermore, that the total intracellular concentrations of abi and ci*, bound and unbound, would remain significantly below the concentrations of the interference targets A, B, and C. The affinities of A, B, C, and I for the respective moieties of the compounds abi and ci*, and also the targets’ intracellular locations are among the independent parameters that can be varied in designing these compounds.
Yet another, and major, constraint on the nature and pharmacokinetics of multitarget drugs stems from the fact that the selectivity patterns described above (Figs. 2 and 4) may not be observed under equilibrium conditions, where the influx of a drug into cells equals its outflux. To illustrate one clear difficulty, let us oversimplify and suppose that an abi-type compound is metabolically inert, in addition to being capable of crossing the plasma membranes and other lipid bilayers. If abi is initially outside the cells, and if the extracellular pool of abi (e.g., in the blood plasma) is large enough, it can be shown that the subsequently reached equilibrium state would be characterized by equal concentrations of the free abi in both the A+ B+ and A− B− cells, thereby resulting in the equal occupancies of the enzyme I by abi in these cells, contrary to the pattern illustrated in Fig. 2.
By contrast, the selectivity patterns of Figs. 2 and 4 would be observed during the initial influx of drugs into cells. Thus, one requirement for the multitarget selectivity of abi- and ci*-type regimens is the avoidance of equilibrium states such as the one described above. This and related considerations indicate that despite the logical simplicity of the proposed designs, their implementation will have to address pharmacokinetic problems that do not necessarily arise with single-target drugs.
Selection of Targets and Construction of Multitarget Drugs.
Appropriate macromolecular targets of the A–C class (Figs. 3 and 4) are suggested by the protein composition of the tumor cells to be eliminated. The choice of an essential intracellular enzyme I (Figs. 2–4) is determined by the presence of I at least in tumor cells, its physiological concentration, and the feasibility of an efficacious inhibitor of I. Among potentially suitable enzymes for which cell-penetrating inhibitors already exist is dihydrofolate reductase. Its high-affinity inhibitors include methotrexate, which enters cells through carrier-mediated pathways, and the more lipophilic trimetrexate, which can enter cells by diffusing through lipid bilayers (18).
Each of the modules in the abi- and ci*-type compounds (Figs. 1–4) would bind its macromolecular target in the absence of the other modules. Therefore, the cytotoxic i-type modules of IC-based compounds would be similar to the stand-alone cytotoxic drugs of today. By contrast, the interference modules of these compounds, i.e., their a-, b-, and c-type moieties (Figs. 1–3), are supposed to bind to their macromolecular targets but preferably not impair them functionally. This specification of a competition module simplifies its design in comparison to that of inhibitors, because many sites on the target’s surface, and not just the active site, would be acceptable.
Antiinhibitors (Fig. 3) are a new class of physiologically active compounds. The opportunities and problems of their design are similar to those for the interference moieties a–c (Figs. 1–3), but there are two other difficulties as well. First, the antiinhibitor moiety i* must bind in the vicinity of, but not at, the active site of enzyme I. Second, the moiety i* must also bear chemical groups (a bump) whose function is to preclude, through steric hindrance, the binding of the inhibitor moiety i to the enzyme I (Fig. 3).
A substrate-binding cleft is not the only indentation in a folded protein molecule. The other clefts tend to be smaller, but they are present as well (19), and some of them may be located next to the enzyme’s active site (Fig. 3). In addition, even relatively flat molecular surfaces can be, in principle, the sites of high-affinity interactions with small ligands (20). These optimistic comments notwithstanding, the development of antiinhibitors is certain to be a complex undertaking. As discussed above, the pharmacokinetic aspects of the proposed designs are also complex. Yet simplicity is good only if it works. Single-target anticancer drugs remain unsatisfactory, in spite of decades of immense effort. It may therefore be wise to attempt a more complex but also more effective solution.
A recent advance in drug design, termed SAR by NMR (structure–activity relationships by nuclear magnetic resonance), provides an especially promising route to constructing ligands for specific regions of a protein molecule (21). In this approach, a library of small molecules is screened for binding to an 15N-labeled protein by using NMR, which can detect weak interactions, and in addition assigns them to specific nitrogens of a protein, thereby identifying the site of binding. Finding two small compounds that bind to adjacent patches of the target protein molecule and covalently linking these compounds produces a higher-affinity ligand. This powerful strategy (21), which already yielded tightly binding ligands of specific proteins, may prove sufficient for constructing the a–c competition modules and the i* antiinhibitor modules of the proposed designs (Figs. 1–4).
Noncytotoxic Multitarget Drugs.
Many useful drugs are the inhibitors of intracellular enzymes that are not essential for cell viability (2). The problem of insufficient selectivity is relevant to these drugs as well. For example, even an exquisitely specific inhibitor of a clinically relevant enzyme is likely to have significant side effects, because the target enzyme is present, in most cases, not only in the cells where its inhibition is clinically beneficial but also in the cells where its inhibition is physiologically inappropriate. The logic of abi-type inhibitors (Figs. 1 and 2) and ci*-type antiinhibitors (Figs. 3 and 4) is applicable in these settings, because an informed choice of the competition moieties a, b, and c would sharpen up the cell selectivity of the inhibitor moiety i in the way described above for cytotoxic drugs (Fig. 4), resulting in the inhibition of the (nonessential) enzyme I in a predetermined subset of the enzyme-containing cells. Note that the same considerations apply to extracellular settings as well. The examples above are but a glimpse of the drug-engineering vistas that are opened up by the IC and antieffector concepts. At the same time, there are significant pharmacokinetic constraints on the properties of the proposed drugs, as discussed above. These constraints are likely to complicate the implementation of the IC/antieffector strategies.
The Problem of Drug Resistance.
With small anticancer drugs that are in use today, the macromolecular target of a drug serves two distinct functions. First, the target is a cell-selectivity determinant that may bias the treatment against tumor cells. Second, the target is also a device whose inhibition by the drug brings about the desired effect, e.g., cell death. Consequently, when drug-resistant tumor cells, selected by a drug treatment, present themselves as a cancer recurrence, the necessity of employing another therapeutic agent (if such an option exists) robs the physician of whatever cell-selectivity advantage there was with the earlier drug.
The situation is qualitatively different with IC-based compounds. Suppose that a treatment that included the drug abi (Figs. 1 and 2) results in the appearance of abi-resistant tumor cells that contain, for example, an altered or overproduced enzyme I. If so, replacement of the i moiety by another small cytotoxic moiety, specific for another essential enzyme, would retain the cell selectivity of the new ab-containing drug. Thus, one advantage of modularity inherent in the designs of IC/antiinhibitor-based compounds (Figs. 1–4) lies in the separation of the effector aspect of a drug from its selectivity aspect. As a result, once an efficacious arrangement of the selectivity modules in abi- or ci*-type compounds has been identified, it can be reutilized in drugs bearing effector moieties other than i and i*.
Concluding Remarks.
The above considerations are based on the existing understanding of single-target drugs and on the notion of steric hindrance. By introducing the new concept of antieffectors and a modification of the previously proposed idea of codominant interference (14), we can now attempt the construction of small modular compounds that possess a multitarget, combinatorial selectivity (Fig. 4). The IC/antieffector strategies are not confined to cytotoxic therapies and are relevant, in principle, to all pharmacological settings. As indicated above, one expected difficulty in implementing these strategies stems from significant pharmacokinetic constraints that do not necessarily arise with single-target drugs.
This work was motivated by the premise that the confinement of anticancer drug research and development to single-target compounds will prove insufficient for the task at hand, because even the informed choices of targets for such drugs may not define unambiguously enough the cell type to be eliminated. The remedy, described above, is to aim for drugs that possess qualitatively different selectivity—multitarget and combinatorial. If this view is correct, the future ascent of multitarget drugs may transform not only the treatment of cancer but also approaches in other settings where the killing or modification of undesirable cells or organelles is carried out in the presence of nearly identical cells or organelles that must be spared. These applications of multitarget drugs encompass more discriminating antiviral and antifungal therapies, as well as the selective killing of activated lymphocytes in autoimmune diseases and the selective elimination of damaged mitochondria in aging cells (14, 15). In yet another class of applications, a noncytotoxic multitarget drug would be used to inhibit a clinically relevant nonessential enzyme in a specific subset of the enzyme-containing cells, thereby retaining the benefits of inhibition while reducing its side effects.
Acknowledgments
I thank L. Peck, G. Turner, D. Anderson, A. Rich, S. Mayo, E. Berezutskaya, and A. Kashina for comments on the manuscript. Studies in my laboratory are supported by grants from the National Institutes of Health.
ABBREVIATION
- IC
interference/codominance
References
- 1.Gilman A G, Rall T W, Nies A S, Taylor P. The Pharmacological Basis of Therapeutics. New York: Pergamon; 1990. [Google Scholar]
- 2.Munson P L, Mueller R A, Breese G R. Principles of Pharmacology. New York: Chapman & Hall; 1996. [Google Scholar]
- 3.Waldman T, Zhang Y, Dillehay L, Yu J K, Vogelstein B, Williams J. Nat Med. 1997;3:1034–1036. doi: 10.1038/nm0997-1034. [DOI] [PubMed] [Google Scholar]
- 4.Folkman J. Adv Cancer Res. 1985;43:175–203. doi: 10.1016/s0065-230x(08)60946-x. [DOI] [PubMed] [Google Scholar]
- 5.Thrush G R, Lark L R, Clinchy B C, Vitetta E S. Annu Rev Immunol. 1996;14:49–71. doi: 10.1146/annurev.immunol.14.1.49. [DOI] [PubMed] [Google Scholar]
- 6.Rosenberg S A. Annu Rev Med. 1996;47:481–491. doi: 10.1146/annurev.med.47.1.481. [DOI] [PubMed] [Google Scholar]
- 7.da Costa L T, Jen J, He T-C, Chan T A, Kinzler K W, Vogelstein B. Proc Natl Acad Sci USA. 1996;93:4192–4196. doi: 10.1073/pnas.93.9.4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bischoff J R, Kirn D H, Williams A, Heise C, Horn S, Muna M, Ng L, Nye J A, Sampson-Johannes A, Fattaey A, McCormick F. Science. 1996;274:373–376. doi: 10.1126/science.274.5286.373. [DOI] [PubMed] [Google Scholar]
- 9.Bishop J M. Genes Dev. 1995;9:1309–1315. doi: 10.1101/gad.9.11.1309. [DOI] [PubMed] [Google Scholar]
- 10.Kinzler K W, Vogelstein B. Cell. 1996;87:159–170. doi: 10.1016/s0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- 11.Weinberg R A. Cell. 1995;81:323–330. doi: 10.1016/0092-8674(95)90385-2. [DOI] [PubMed] [Google Scholar]
- 12.Knudson A G. Proc Natl Acad Sci USA. 1993;90:10914–10921. doi: 10.1073/pnas.90.23.10914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hunter T. Cell. 1997;88:333–346. doi: 10.1016/s0092-8674(00)81872-3. [DOI] [PubMed] [Google Scholar]
- 14.Varshavsky A. Proc Natl Acad Sci USA. 1995;92:3663–3667. doi: 10.1073/pnas.92.9.3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Varshavsky A. Cold Spring Harbor Symp Quant Biol. 1996;60:461–478. doi: 10.1101/sqb.1995.060.01.051. [DOI] [PubMed] [Google Scholar]
- 16.Bernstein K E, Welsh S L, Inman J K. Biochem Biophys Res Commun. 1990;167:310–316. doi: 10.1016/0006-291x(90)91766-l. [DOI] [PubMed] [Google Scholar]
- 17.Crabtree G R, Schreiber S L. Trends Biochem Sci. 1996;21:418–422. doi: 10.1016/s0968-0004(96)20027-1. [DOI] [PubMed] [Google Scholar]
- 18.Takimoto C H, Allegra C J. Oncology. 1995;9:649–659. [PubMed] [Google Scholar]
- 19.Laskowski R A, Luscombe N M, Swindells M B, Thornton J M. Protein Sci. 1996;5:2438–2452. doi: 10.1002/pro.5560051206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mattos C, Ringe D. Nat Biotech. 1996;14:595–599. doi: 10.1038/nbt0596-595. [DOI] [PubMed] [Google Scholar]
- 21.Hajduk P J, Meadows R P, Fesik S W. Science. 1997;278:497–499. doi: 10.1126/science.278.5337.497. [DOI] [PubMed] [Google Scholar]