Abstract
For many years it was accepted that isopentenyl diphosphate, the common precursor of all isoprenoids, was synthesized through the well known acetate/mevalonate pathway. However, recent studies have shown that some bacteria, including Escherichia coli, use a mevalonate-independent pathway for the synthesis of isopentenyl diphosphate. The occurrence of this alternative pathway has also been reported in green algae and higher plants. The first reaction of this pathway consists of the condensation of (hydroxyethyl)thiamin derived from pyruvate with the C1 aldehyde group of d-glyceraldehyde 3-phosphate to yield d-1-deoxyxylulose 5-phosphate. In E. coli, d-1-deoxyxylulose 5-phosphate is also a precursor for the biosynthesis of thiamin and pyridoxol. Here we report the molecular cloning and characterization of a gene from E. coli, designated dxs, that encodes d-1-deoxyxylulose-5-phosphate synthase. The dxs gene was identified as part of an operon that also contains ispA, the gene that encodes farnesyl-diphosphate synthase. d-1-Deoxyxylulose-5-phosphate synthase belongs to a family of transketolase-like proteins that are highly conserved in evolution.
Keywords: isopentenyl diphosphate, pyruvate, glyceraldehyde 3-phosphate, synthase
Isoprenoids are ubiquitous compounds found in all living organisms. Some isoprenoids play essential roles in particular cell functions such as sterols, contributing to eukaryotic membrane architecture, acyclic polyprenoids found in the side chain of ubiquinone, plastoquinone, and chlorophylls, sugar carriers for polysaccharide biosynthesis, or carotenoids in photosynthetic organisms. Although the physiological role of other isoprenoids is less evident, like that of the vast array of plant secondary metabolites, some are known to play key roles in the adaptative responses to different environmental challenges. In spite of the remarkable diversity of structure and function, all isoprenoids originate from a single metabolic precursor, isopentenyl diphosphate (1, 2).
For many years, it was accepted that isopentenyl diphosphate was synthesized through the well known acetate/mevalonate pathway. However, recent studies have demonstrated that the mevalonate-dependent pathway does not operate in all living organisms (3, 4). An alternative mevalonate-independent pathway for isopentenyl diphosphate biosynthesis was initially characterized in bacteria (4, 5) and later also in green algae (6) and higher plants (7–11). The first reaction of the novel mevalonate-independent pathway involves the condensation of (hydroxyethyl)thiamin derived from pyruvate with the C1 aldehyde group of d-glyceraldehyde 3-phosphate to yield d-1-deoxyxylulose 5-phosphate (5, 12). In Escherichia coli, d-1-deoxyxylulose (most likely in the form of d-1-deoxyxylulose 5-phosphate) is efficiently incorporated into the prenyl side chain of menaquinone and ubiquinone (12, 13). In plants, the incorporation of d-1-deoxyxylulose into isoprenoids has also been reported (11, 14). In addition, d-1-deoxyxylulose has also been described as a precursor for the biosynthesis of thiamin and pyridoxol. d-1-Deoxyxylulose is the precursor molecule of the contiguous 5-carbon unit (C4′–C4–C5–C5′–C5") of the thiazole ring of thiamin in E. coli (15, 16) and in higher plant chloroplasts (17). The role of d-1-deoxyxylulose in the biosynthesis of pyridoxol in E. coli is also well documented (18–20).
In spite of the extensive genetic analysis in E. coli, no genes involved in isopentenyl diphosphate biosynthesis have been reported to date. The only genes identified in E. coli related to isoprenoid biosynthesis are ispA and ispB, which encode farnesyl-diphosphate synthase (21) and octaprenyl-diphosphate synthase (22), respectively. In this paper we report the cloning and characterization of a gene from E. coli, designated dxs, that encodes d-1-deoxyxylulose-5-phosphate synthase. The dxs gene was identified as part of an operon that also contains the ispA gene.
MATERIALS AND METHODS
Bacterial Strains, Bacteriophages, and Plasmids.
E. coli strains XL1-Blue and DH5α were used for the cloning, maintenance, and propagation of plasmids. Wild-type E. coli strain E15 (23) was used for RNA isolation. Bacteriophage λ19F6, from the E. coli aligned genomic library of Kohara et al. (24), was propagated on E. coli strain LE392. Phage R408 was used as a helper for the isolation of single-stranded DNA from plasmid pBluescript (Stratagene) derivatives, with E. coli strain RZ1032 as a host. Plasmid pTACTAC (25) was used as expression vector with strain XL1-Blue as a host.
Plasmid Construction.
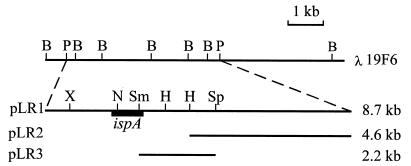
Plasmid pLR1 was constructed by cloning the 8.7-kb PstI–PstI fragment of bacteriophage λ19F6 into the PstI site of plasmid pBluescript (Fig. 1). Plasmid pLR2 was constructed by subcloning the 4.6-kb HindIII–PstI fragment generated by digestion of the 8.7-kb PstI–PstI fragment with HindIII into plasmid pBluescript (Fig. 1). Plasmid pLR3 was constructed by subcloning the 2-kb SmaI–SphI fragment excised from plasmid pLR1 into plasmid pBluescript (Fig. 1). An NdeI site was created at the ATG translation start codon of ORF2 (Fig. 2) in plasmid pLR3 by site-directed mutagenesis (26). The resulting plasmid was designated pLR4. Plasmid pTAC-ORF2 was constructed by cloning the 2-kb NdeI–SalI fragment of pLR4 into the corresponding sites of plasmid pTACTAC.
Figure 1.
Restriction map of the genomic region containing the ispA gene from E. coli. The genomic region cloned in λ19F6 (21) is shown on top. The fragments subcloned into plasmids pLR1, pLR2, and pLR3 are also indicated. The position of the ispA gene is represented by a solid box. Restriction sites are as follows: B, BglII; H, HindIII; N, NdeI; P, PstI; Sm, SmaI; Sp, SphI; X, XhoI.
Figure 2.
Analysis of a transcription unit from E. coli containing ORF1, ispA, ORF2 (dxs), and ORF3. (A) The previously reported genes ispA and pgpA and the short ORF cotranscribed with ispA (ORF1) are represented by open boxes. ORF2 and ORF3 are represented by shaded boxes. ATG and TAA indicate translation start and termination codons, respectively. The length (in bp) of the coding and intergenic regions is indicated in the line below. (B) A transcript containing ispA, ORF2, and ORF3 was detected by reverse transcription–PCR (RT-PCR) analysis with primers P1, P2, P3, and P4. Primers P2 and P4 were used for the synthesis of first strand cDNA by using reverse transcriptase. The PCR products obtained with the sets of primers P1/P2 and P3/P4 are shown below the mRNA. The enzymes used for the restriction enzyme mapping of the amplification products are also indicated.
DNA Sequencing and Databases.
Appropriate restriction fragments were subcloned into plasmid pBluescript. Both strands of DNA were sequenced by the dideoxynucleotide chain termination method (27) with the T7 sequencing kit (Pharmacia). Computer alignment and nucleotide and amino acid sequence analysis were performed with the blast Program of the National Center of Biotechnology Information (Bethesda, MD). The Wisconsin Package (version 9.0) of the Genetics Computer Group (GCG) (Madison, WI) was used for the editing of sequences.
Reverse Transcription–PCR Analysis.
DNA-free RNA was obtained from E. coli strain E15 according to the method described by Garrido et al. (28). First strand cDNA was obtained as described by Dumas Milne Edwards et al. (29) by using reverse transcriptase (Moloney murine leukemia virus, Promega) and the gene-specific primers P2 and P4 (see below). PCR reactions were performed as described by Ausubel et al. (30) by using two sets of primers. Primers P1 (5′-CCGTTTTATCGCCCCACTG-3′) and P2 (5′-GCAGCAACCGCAATACC-3′) were designed to amplify a region extending from nucleotide +56 of ispA to nucleotide +410 of ORF2. Primers P3 (5′-CCCGTGCTGAACATTGG-3′) and P4 (5′-ATACTGACAAACTGCGCCC-3′) were designed to amplify a region extending from nucleotide +1740 of ORF2 to nucleotide +545 of ORF3. In all cases, numbering refers to the ATG translation start codon (position +1) of the corresponding coding regions. The PCR products were analyzed by agarose gel electrophoresis and further characterized by restriction enzyme mapping. The PCR product obtained with primers P1 and P2 was digested with NheI and NdeI, and that obtained with primers P3 and P4 was digested with SphI and EcoRV (Fig. 2).
Expression of Plasmid pTAC-ORF2 in E. coli.
E. coli XL1-Blue cells harboring plasmid pTAC-ORF2 were grown at 37°C in 2× TY medium (30) supplemented with ampicillin (100 μg/ml) to an OD600 of 0.6 and then induced with isopropyl β-d-thiogalactoside (1 mM). E. coli strain XL1-Blue harboring plasmid pTACTAC was used as a control. Aliquots of 1 ml were withdrawn at several time intervals after induction, and the cells were harvested by centrifugation (8,000 × g for 1 min). The cell pellet was resuspended in 100 μl of extraction buffer (60 mM Tris⋅HCl/1% 2-mercaptoethanol/1% SDS/10% glycerol/0.01% bromophenol blue, pH 6.8) and heated in boiling water for 5 min. Samples (20 μl) were subjected to SDS/PAGE (10% acrylamide) (30), and the separated proteins were stained with Coomassie brilliant blue R250.
Assay of d-1-Deoxyxylulose-5-Phosphate Synthase Activity.
Bacterial cells were grown in M9 medium (30) supplemented with glucose (0.4%) and thiamin (15 nM) to an OD600 of 0.6, induced by the addition of isopropyl β-d-thiogalactoside (0.5 mM), and harvested by centrifugation. The cell pellet was washed first in 0.85% NaCl solution and subsequently in buffer A (40 mM Tris⋅HCl/2.5 mM MgCl2/1 mM thiamin diphosphate/0.1 mM phenylmethanesulfonyl fluoride/5 mM 2-mercaptoethanol, pH 8.0). Cells were resuspended in five times their wet weight of buffer A containing lysozyme (1 mg/ml), incubated for 20 min at 37°C, and then disrupted by sonication (30 W, 5 bursts of 1 min and cooling in an ice bath for 2 min between each burst). After centrifugation at 13,000 × g for 30 min at 4°C the supernatant was recovered and supplemented with protamine sulfate (1.25 mg/ml), incubated at room temperature for 15 min, and then centrifuged at 13,000 × g for 20 min at 4°C. The supernatant was used as enzyme sample. Protein concentration was determined with the Bio-Rad protein assay.
The enzyme reaction mixture consisted of 40 mM Tris⋅HCl/2.5 mM MgCl2/1 mM thiamin diphosphate/0.1 mM phenylmethanesulfonyl fluoride/5 mM 2-mercaptoethanol/0.2 mM [2-14C]pyruvate (15.9 mCi/mmol, DuPont/NEN)/50 mM pyruvate/100 mM dl-glyceraldehyde 3-phosphate (or 50 mM d-glyceraldehyde)/enzyme sample in a final volume of 50 μl. After incubation for 1–2 h at 37°C, the reactions were stopped by heating at 80°C for 3 min. After centrifugation at 13,000 × g for 5 min, aliquots of the supernatant (2–5 μl) were loaded onto a TLC plate (silica gel 60, Merck). Labeled d-1-deoxyxylulose 5-phosphate (or d-1-deoxyxylulose) was separated from [2-14C]pyruvate by using n-propyl alcohol/ethyl acetate/H2O (6:1:3) as solvent and quantified by autoradiography (Molecular Imager, Bio-Rad). The radioactivity incorporated into d-1-deoxyxylulose 5-phosphate (or d-1-deoxyxylulose) was referred to the radioactivity of [2-14C]pyruvate present at the beginning of the reaction (time 0). When appropriate amounts of enzyme sample were used, the reaction was linear for up to 2 h. Enzyme activity is expressed as micromoles of d-1-deoxyxylulose 5-phosphate (or d-1-deoxyxylulose) synthesized per min per mg of protein in the conditions described above. For qualitative assays, [2-14C]pyruvate was omitted from the reaction mixture, and the d-1-deoxyxylulose or d-1-deoxyxylulose 5-phosphate synthesized was identified by staining with p-anisaldehyde/sulfuric acid (31). d-1-Deoxyxylulose (Rf 0.63) and d-1-deoxyxylulose 5-phosphate (Rf 0.35) stained blue and purple, respectively. Pyruvate (Rf 0.59) stained pale yellow, and d-glyceraldehyde (Rf 0.65) and dl-glyceraldehyde 3-phosphate (Rf 0.40) stained orange. Control reactions lacked pyruvate or dl-glyceraldehyde 3-phosphate (or d-glyceraldehyde).
Enzymatic Synthesis of d-1-Deoxyxylulose in Vitro and NMR Analysis.
For bulk production of the enzyme, E. coli XL1-Blue cells harboring plasmid pTAC-ORF2 were grown under aerobic conditions at 37°C in 1 liter of medium containing meat peptone (0.5%), meat extract (0.3%), and ampicillin (50 μg/ml). When cultures reached an OD600 between 0.5 and 0.8, expression of the ORF2-encoded product was induced by addition of isopropyl β-d-thiogalactoside (0.2 mM) and incubation at 37°C. Cells were harvested by centrifugation (6,000 × g) for 30 min at 4°C and first washed in 100 ml of 0.85% NaCl solution and then resuspended in buffer B (80 mM Tris⋅HCl/5 mM MgCl2/2 mM EDTA/0.1 mM phenylmethanesulfonyl fluoride, pH 8.0) and freeze-dried. Freeze-dried cells (0.3 g) were resuspended in buffer B (20 ml) containing lysozyme (1 mg/ml) and incubated at 37°C for 30 min. Cells were disrupted by sonication at 4°C (40 W, five bursts of 1 min and cooling on an ice bath for 2 min between each burst). The cell-free extract was obtained by centrifugation at 18,000 × g for 30 min at 4°C. The cell-free extract, containing about 20 mg of protein, was adjusted to 20 ml with buffer B, supplemented with sodium pyruvate (0.9 mmol), d-glyceraldehyde (0.9 mmol), and thiamin diphosphate (0.01 mmol), and incubated in an orbital shaker at 37°C for 20 h. The reaction was stopped by heating at 80°C for 5 min. Denatured proteins were removed by centrifugation at 18,000 × g for 30 min, and the supernatant was lyophilized. The freeze-dried sample (300 mg) was dissolved in a small amount of methanol and separated on a chromatographic column of silica gel 60 (Merck) with chloroform/methanol (100:20) as eluent. d-1-Deoxyxylulose (Rf 0.37) was detected by examination of all fractions by TLC on silica gel plates (Merck, F254, 0.25 mm) with the same eluent.
d-1-Deoxyxylulose was extracted from the TLC plates by using chloroform/methanol (8:2) and subjected to 1H and 13C NMR structural analysis. Both open form (75%) and two closed hemiketal α- and β-furanose forms (25%, in a nearly 1:1 ratio, as deduced from the relative signal intensities of the C-1 methyl groups in the 1H NMR spectrum) were observed. The NMR spectra were: 1H NMR (200 MHz, DMSO-d6). Open form: 4.95 (d, J = 6.6, OH); 4.70 (d, J = 6.6, OH); 4.67 (t, J = 5.4, OH); 4.00 (dd, J1 = 6.4, J2 = 2.2, H-3); 3.75 (dq, J1 = 6.6, J2 = 2.2, H-4); 3.44 (ddd, J1 = 10.0, J2 = 6.6, J3 = 5.5, H-5); 3.33 (ddd, J1 = 10.1, J2 = 6.6, J3 = 5.5, H-5); 2.13 (s, methyl). C-1 methyl groups of α- and β-furanose forms: 1.27 (s, 3H); 1.20 (s, 3H); 13C NMR (50 MHz, DMSO-d6). Open form: 211.70 (s, C-2); 76.91 (d, C-3∗); 72.48 (d, C-4∗); 61.80 (t, C-5); 26.86 (q, C-1). α- and β-furanose forms: 105.59; 102.02; 82.02; 81.66; 76.74; 75.37; 70.35; 69.83; 24.99; 22.36. Assignments of signals bearing a superscript may be interchanged.
Alternatively, d-3,4,5-triacetyl-1-deoxyxylulose was identified after acetylation of the enzymatic reaction product. The crude material obtained after freeze-drying of the whole reaction mixture (300 mg) was stirred in equal amounts of pyridine and acetic anhydride (1:1) overnight at room temperature. After removing the excess of reagents under vacuum, the acetylated compound was isolated by preparative TLC on silica gel plates (Merck, F254) (toluene/ethyl acetate, 80:20, Rf 0.22). Only the open form of d-3,4,5-acetyl-1-deoxyxylulose was detected. The NMR spectra were: 1H NMR (200 MHz, CDCl3); 5.58 (ddd, J1 = 6.7, J2 = 5.9, J3 = 3.0, H-4); 5.24 (d, J = 3.0, H-3); 4.28 (dd, J1 = 11.6, J2 = 5.8, H-5); 4.12 (dd, J1 = 11.6, J2 = 6.7, H-5); 2.198 (s, methyl); 2.195 (s, methyl); 2.062 (s, methyl); 2.047 (s, methyl); 13C NMR (50 MHz, CDCl3); 201.38 (s, C-2); 170.31 (s, acetate); 170.00 (s, acetate); 169.73 (s, acetate); 76.33 (d, C-3∗); 68.77 (d, C-4∗); 61.50 (t, C-5); 26.86 (q, methyl); 20.60 and 20.46 (q, 3 methyls). Assignments of signals bearing a superscript may be interchanged. GC–MS (chemical ionization with isobutane as reactant gas): m/z (%) = 261 ([M + H]+, 28%), 201 ([M + H − AcOH]+, 99%), 141 ([M + H − 2AcOH]+, 100%).
RESULTS
The 3′-Flanking Region of the E. coli ispA Gene Contains Two Open Reading Frames Encoding Proteins Showing Similarity to Transketolases and Members of the Aldo–Keto Reductase Superfamily.
In bacteria, the genes encoding enzymes corresponding to specific metabolic pathways are usually organized in operons. Thus, we hypothesized that ispA, the gene encoding farnesyl-diphosphate synthase in E. coli (21), could be part of an operon containing other isoprenoid biosynthetic genes. To test this hypothesis, we cloned and characterized the genomic region flanking ispA. Because the ispA coding region is preceded by functional promoter sequences (21) our work was focused on the characterization of its 3′-flanking region. By using clone λ19F6 from the E. coli aligned genomic library of Kohara et al. (24) we first subcloned the 8.7-kb PstI–PstI fragment known to contain the ispA gene (21) to create plasmid pLR1 (Fig. 1). This plasmid was further used to obtain two overlapping subclones (plasmids pLR2 and pLR3) containing the ispA 3′-flanking region (Fig. 1). The nucleotide sequence analysis of the inserts cloned in plasmids pLR2 and pLR3 revealed two ORFs (ORF2 and ORF3, Fig. 2) encoding proteins of unknown function. A short ORF (ORF1) preceding ispA is cotranscribed with this gene (21) (Fig. 2). ORF2 starts at the ATG codon located 24 bp downstream of the stop codon of ispA and extends over 1863 bp. ORF3 starts 179 bp downstream of ORF2 and extends over 975 bp. The stop codon of ORF3 is located 80 bp upstream of the translation start codon of the previously characterized pgpA gene, which encodes a phospholipid phosphatase (32). ORF2 and ORF3 are preceded by consensus Shine-Dalgarno sequences.
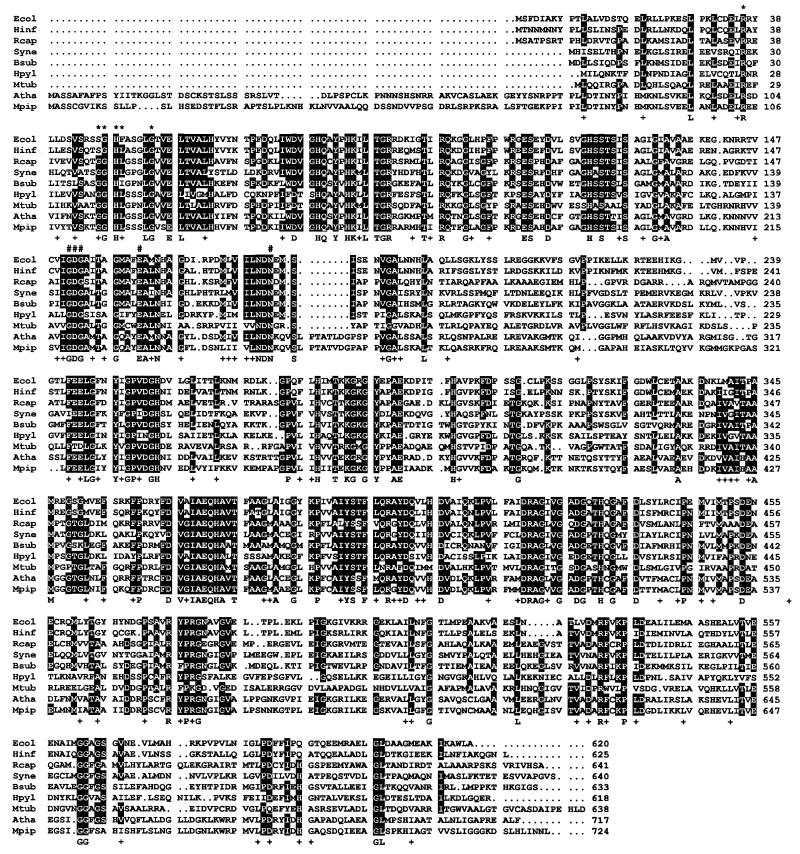
The protein predicted from ORF2 has 620 amino acid residues and a molecular mass of 67.6 kDa. Homology searches revealed that the ORF2-encoded protein shows high similarity to a putative transketolase encoded by ORF C2814 present in the 3′-region of a photosynthetic gene cluster of Rhodobacter capsulatus (33) (Fig. 3). The protein encoded by ORF2 is also highly similar to proteins of unknown function encoded in the genome of Haemophilus influenzae, Bacillus subtilis, Synechocystis sp. (PCC 6803), Helicobacter pylori, and Mycobacterium tuberculosis (Fig. 3). Interestingly, the protein encoded by ORF2 also shows high level of similarity with Arabidopsis thaliana CLA1 gene product (Fig. 3), which has recently been implicated in chloroplast development (34). A significant level of similarity was also found with transketolases and other thiamin-requiring enzymes. The protein predicted from ORF3 contains 324 amino acid residues and shows similarity to proteins of the aldo–keto reductase superfamily (35).
Figure 3.
Amino acid sequence alignment of d-1-deoxyxylulose-5-phosphate synthase from different organisms. Dots indicate the absence of particular amino acid residues. Numbers indicate the position of amino acid residues in the sequences. White-on-black letters denote amino acid residues common to at least six polypeptides. A consensus sequence denoting identical and conserved amino acid residues in all polypeptides is shown below the alignment. Conserved residues are indicated with the symbol +. Amino acid residues putatively involved in the binding of thiamin diphosphate are indicated with the symbol #. The conserved motif containing the histidine residue putatively involved in proton transfer during catalysis is indicated with ∗. Ecol, E. coli (GenBank AF035440); Hinf, H. influenzae (Swiss-Prot P45205); Rcap, R. capsulatus (Swiss-Prot P26242); Syne, Synechocystis sp. (PCC6803) (GenBank D90903); Bsub, B. subtilis (Swiss-Prot P54523); Hpyl, H. pylori (GenBank AE000552); Mtub, M. tuberculosis (GenBank Z96072); Atha, A. thaliana (GenBank U27099); Mpip, Mentha × piperita [GenBank AF019383; Lange et al. (40)].
ORF2 and ORF3 Are Cotranscribed with the ispA Gene.
To study whether ispA, ORF2, and ORF3 were included in the same transcription unit we performed reverse transcription–PCR analysis by using E. coli DNA-free RNA and gene-specific primers located within the coding regions (for details, see Materials and Methods and Fig. 2). Amplification products showing the predicted size were obtained in all cases. The amplification products were further characterized by restriction mapping, and in all cases we obtained the pattern of fragments expected from the restriction sites deduced from the nucleotide sequence (data not shown). No amplification products were obtained in the control reactions lacking reverse transcriptase, thus confirming the absence of contaminating genomic DNA in the RNA samples.
ORF2 Encodes d-1-Deoxyxylulose-5-Phosphate Synthase.
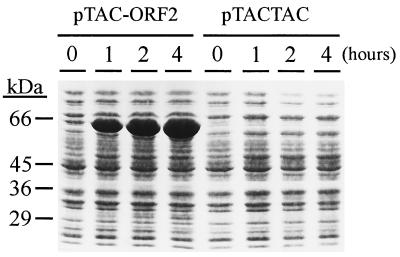
It has been reported that the first reaction of the novel mevalonate-independent isoprenoid pathway is the condensation of (hydroxyethyl)thiamin derived from pyruvate on the carbonyl group of d-glyceraldehyde 3-phosphate (5, 12). This reaction resembles that catalyzed by transketolases and yields d-1-deoxyxylulose 5-phosphate (5, 12). Because the protein encoded by ORF2 shows similarity with transketolases, we studied whether this putative transketolase was able to catalyze the synthesis of d-1-deoxyxylulose 5-phosphate from pyruvate and d-glyceraldehyde 3-phosphate. For this purpose, we constructed plasmid pTAC-ORF2, in which the ORF2 nucleotide sequence was cloned into the expression vector pTACTAC. After induction with isopropyl β-d-thiogalactoside, a protein showing an apparent molecular mass of 65 kDa was detected by SDS/PAGE and Coomassie blue staining (Fig. 4).
Figure 4.
Overexpression of the dxs gene product in E. coli. (A) E. coli XL1-Blue cells harboring plasmid pTAC-ORF2 or pTACTAC were induced with isopropyl β-d-thiogalactoside, and samples were withdrawn at the indicated times. Proteins were analyzed by SDS/PAGE and Coomassie blue staining. The position of molecular mass markers is indicated on the left.
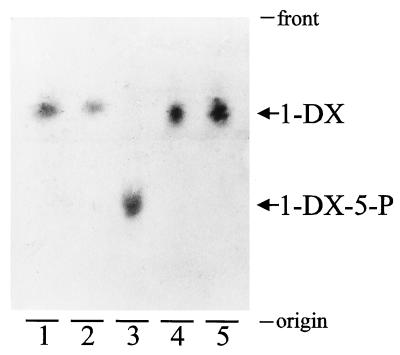
Cell-free extracts prepared from induced E. coli XL1-Blue cells harboring plasmid pTAC-ORF2 were incubated with pyruvate and d-glyceraldehyde or dl-glyceraldehyde 3-phosphate, and the reaction products were analyzed by TLC. A product showing an Rf identical to that of a chemically synthesized d-1-deoxyxylulose standard was detected in the reaction mixture containing pyruvate and d-glyceraldehyde (Fig. 5, lanes 1 and 2). When dl-glyceraldehyde 3-phosphate was included in the reaction mixture instead of d-glyceraldehyde, a more polar product, consistent with a phosphorylated compound, was detected (Fig. 5, lane 3). Further incubation of an aliquot of the reaction mixture corresponding to lane 3 with alkaline phosphatase resulted in the dephosphorylation of the reaction product to yield a compound showing the same migration as d-1-deoxyxylulose (Fig. 5, lane 4). Treatment with alkaline phosphatase had no effect on the d-1-deoxyxylulose obtained from pyruvate and d-glyceraldehyde (Fig. 5, lane 5). These results indicate that the product obtained in the reaction mixture containing pyruvate and dl-glyceraldehyde 3-phosphate was a phosphorylated derivative of d-1-deoxyxylulose that, according to the reaction mechanism proposed (5, 12), should correspond to d-1-deoxyxylulose 5-phosphate. In all cases, the reaction was dependent on the presence of pyruvate and d-glyceraldehyde or dl-glyceraldehyde 3-phosphate. However, the reaction was not dependent on the presence of thiamin diphosphate.
Figure 5.
Thin layer chromatograms of the reaction products obtained from pyruvate and d-glyceraldehyde or dl-glyceraldehyde 3-phosphate. Cell-free extracts from induced E. coli XL1-Blue cells harboring plasmid pTAC-ORF2 (25 μg of protein) were incubated for 2 h under the conditions described in Materials and Methods with the following substrates: pyruvate and d-glyceraldehyde (lanes 2 and 5) and pyruvate and dl-glyceraldehyde 3-phosphate (lanes 3 and 4). The reactions were stopped, and reaction mixture aliquots (10 μl) were treated with 1.25 units of calf intestine alkaline phosphatase for 1 h (lanes 4 and 5). Lane 1 corresponds to a standard of chemically synthesized d-1-deoxyxylulose. Reaction products were detected by staining with p-anisaldehyde/sulfuric acid. The positions of d-1-deoxyxylulose (1-DX) and d-1-deoxyxylulose 5-phosphate (1-DX-5-P) are indicated.
By using a quantitative assay in vitro, based on the inclusion of [2-14C]pyruvate in the reaction mixture and the autoradiographic analysis of the reaction product after separation by TLC, we found that the d-deoxyxylulose-5-phosphate synthase activity measured in the cell-free extracts of the induced strain was 150-fold higher than that detected in the control strain harboring plasmid pTACTAC (1.1 ± 0.2 μmol/min⋅mg and 0.0068 ± 0.0008 μmol/min⋅mg, respectively). When d-glyceraldehyde was used as substrate instead of dl-glyceraldehyde 3-phosphate, a lower enzyme activity was measured in the cell-free extract of the induced strain (0.58 ± 0.08 μmol/min⋅mg), suggesting that dl-glyceraldehyde 3-phosphate is a better substrate for the enzyme. The gene corresponding to ORF2 was designated dxs for d-1-deoxyxylulose 5-phosphate synthase.
NMR Structural Analysis of d-1-Deoxyxylulose Synthesized in Vitro.
Two different ways of isolating d-1-deoxyxylulose were used. The first involved acetylation of the crude reaction product and isolation of the triacetate of d-1-deoxyxylulose. According to TLC and 1H NMR data on the crude product after acetylation, d-3,4,5-triacetyl-1-deoxyxylulose was present as the major compound (approximately 75% as estimated from GLC and 1H NMR) together with two other compounds. These were not artifacts produced during the acetylation reaction, as they were not detected after acetylation of a d-1-deoxyxylulose sample obtained by chemical synthesis. When we isolated free d-1-deoxyxylulose, the same pattern of two accompanying products was observed. The identity of these possible metabolites is at present under study. Purification of either free d-1-deoxyxylulose or d-3,4,5-triacetyl-1-deoxyxylulose by chromatography was difficult. The two by-products migrated before or after acetylation with Rf values, respectively, slightly higher and lower than those of the free or acetylated d-1-deoxyxylulose. Cutting of zones was therefore necessary to obtain a pure compound, thus making it difficult to measure the accurate yield of the reaction. Identity and purity of d-1-deoxyxylulose were tested by 1H and 13C NMR spectroscopy on the free and the acetylated carbohydrate and by GC–MS of the triacetate. All data were identical to results reported in the literature (36) or to data directly measured on a synthetic reference sample obtained by the method of Broers (12).
DISCUSSION
d-1-Deoxyxylulose 5-phosphate has been proposed as the first intermediate of the mevalonate-independent pathway for isoprenoid biosynthesis recently reported in bacteria, green algae, and higher plants (5, 12). Although in E. coli d-1-deoxyxylulose is readily incorporated into isoprenoids (12, 13), thiamin (15, 16), and pyridoxol (18–20), it is likely that its phosphorylated form, d-1-deoxyxylulose 5-phosphate, may represent the metabolic intermediate utilized in vivo. The requirement of d-1-deoxyxylulose (or d-1-deoxyxylulose 5-phosphate) as the common precursor of three different essential biosynthetic pathways in E. coli could explain why mutations affecting the gene(s) involved in the synthesis of this metabolite have not been previously described.
Here we report the cloning and characterization of a novel gene from E. coli that encodes d-1-deoxyxylulose-5-phosphate synthase. This gene, designated dxs, has been identified as part of an operon that includes ispA, the gene that encodes farnesyl-diphosphate synthase (21) and a third gene encoding a protein of unknown function that shows similarity to proteins of the aldo–keto reductase superfamily. This enzyme family includes monomeric NADP(H) oxidoreductases that play a variety of roles, some acting on a broad range of substrates (35). Because the conversion of d-1-deoxyxylulose 5-phosphate to isopentenyl diphosphate requires the reduction of this molecule and/or some derived intermediate(s), it is plausible that this novel protein could be involved in this process. Work is currently in progress to assess the role of this putative oxidoreductase in the mevalonate-independent isoprenoid pathway.
An enzymatic acyloin-type condensation reaction between pyruvate and d-glyceraldehyde catalyzed by cell-free extracts of different microorganisms, including E. coli, has been previously reported (37). The reaction product was isolated and determined to be d-1-deoxyxylulose (37), and it was proposed that pyruvate dehydrogenase was involved in this reaction (38). However, the observation that E. coli mutants carrying deletions of the pyruvate dehydrogenase complex only show auxotrophy for acetate (39) suggests that pyruvate dehydrogenase activity might not be essential for the synthesis of d-1-deoxyxylulose 5-phosphate in vivo. However, further genetic and biochemical studies are needed before establishing the specific role of the dxs gene product in the biosynthesis of this essential metabolite.
Based on the data derived from the bacterial genomes currently sequenced, homologs of the E. coli dxs gene have been found in the eubacteria H. influenzae, H. pylori, and Synechocystis sp. (PCC 6803). Searches in nucleotide sequence data banks have detected homologs of the dxs gene in other eubacteria (R. capsulatus, B. subtilis, M. tuberculosis, and Mycobacterium leprae) and plants (the CLA1 gene of A. thaliana and partial expressed sequence tags from Oryza sativa, Ricinus communis and Pinus taeda). A cDNA homolog of the dxs gene has also been identified in peppermint (Mentha × piperita) (40). Nevertheless, no homologs of the dxs gene have been identified in other organisms whose complete genome has been sequenced (the archaebacteria Methanococcus jannaschii, the mycoplasmas Mycoplasma genitalium and Mycoplasma pneumoniae, and the yeast Saccharomyces cerevisiae). No homolog of the dxs gene has been found in animals. The amino acid sequence alignment of the bacterial enzymes shows levels of identity between 39.8 and 73.1% (similarity between 61.6 and 86.6%). The E. coli and the plant enzymes show levels of identity of about 49% (58% similarity). The amino acid sequence alignment shown in Fig. 3 reveals that the conserved residues are distributed along the entire protein. This high level of sequence conservation suggests a strong evolutionary pressure to maintain these amino acid residues at specific positions, thus indicating that they might play an important role in the structural conformation and/or in the catalytic properties of the enzyme. All enzymes show a conserved motif with the typical features of the binding site for thiamin diphosphate (41, 42) (Fig. 3). Another motif common to d-1-deoxyxylulose-5-phosphate synthase and transketolases includes the histidine residue that has been proposed to participate in proton transfer in the reaction catalyzed by transketolases (41, 42) (Fig. 3). However, the motif that appears to involved in substrate binding in transketolases (41, 42) is not conserved in d-1-deoxyxylulose-5-phosphate synthase. The enzyme d-1-deoxyxylulose-5-phosphate synthase reported in this paper defines a novel family of transketolase-like proteins that are highly conserved in evolution.
The A. thaliana homolog of the bacterial dxs gene corresponds to the recently cloned CLA1 gene (31). Interestingly, disruption of this gene results in an albino phenotype. The mutant plants show an arrest of chloroplast development at an early stage and an absence of accumulation of carotenoids and chlorophylls (34). The presence of an N-terminal extension having the typical features of plastid transit peptides (34), together with the high similarity between the A. thaliana and the bacterial enzyme, suggest that the plant enzyme could be involved in the synthesis of isoprenoid precursors in the chloroplast. Preliminary results indicate that the CLA1 gene product shows d-1-deoxyxylulose-5-phosphate synthase activity when expressed in E. coli (N.C., L.M.L., and A.B., unpublished results). The involvement of d-1-deoxyxylulose-5-phosphate synthase in the biosynthesis of plastid-derived isoprenoids is further supported by the work of Lange et al. (40).
Acknowledgments
We thank Dr. Y. Kohara for providing the clone λ19F6. Critical reading of the manuscript by Dr. A. Ferrer and C. Marín is also appreciated. This study was supported by Grants PB93-0753 from the Dirección General de Investigación Científica y Técnica and 1995SGR-00457 from the Comissió Interdepartamental de Recerca i Innovació Tecnològica de la Generalitat de Catalunya (to A.B.). L.M.L. is the recipient of a predoctoral fellowship from the Ministerio de Educación y Cultura, Spain. S.R.P. was on leave from the Institute of Technology “10th November” (Surabaya, Indonesia) and was supported by a grant from the Indonesian High Education Project and Asian Development Bank (Nb 1253-INO, 1994).
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF035440).
References
- 1.Wright D L. Annu Rev Biochem. 1961;20:525–548. [Google Scholar]
- 2.Spurgeon S L, Porter J W. In: Biosynthesis of Isoprenoid Compounds. Porter J W, Spurgeon S L, editors. Vol. 1. New York: Wiley; 1981. pp. 1–46. [Google Scholar]
- 3.Horbach S, Sahm H, Welle R. FEMS Microbiol Lett. 1993;111:135–140. doi: 10.1111/j.1574-6968.1993.tb06375.x. [DOI] [PubMed] [Google Scholar]
- 4.Rohmer M, Knani M, Simonin P, Sutter B, Sham H. Biochem J. 1993;295:517–524. doi: 10.1042/bj2950517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rohmer M, Seemann M, Horbach S, Bringer-Meyer S, Sahm H. J Am Chem Soc. 1996;118:2564–2566. [Google Scholar]
- 6.Schwender J, Seemann M, Lichtenthaler H K, Rohmer M. Biochem J. 1996;316:73–80. doi: 10.1042/bj3160073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schwarz M K. Ph.D. thesis. Zürich, Switzerland: Eidgenössische Technische Hochschule; 1994. [Google Scholar]
- 8.Eisenreich W, Menhard B, Hylands P J, Zenk M H, Bacher A. Proc Natl Acad Sci USA. 1996;93:6431–6436. doi: 10.1073/pnas.93.13.6431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lichtenthaler H K, Schwender J, Disch A, Rohmer M. FEBS Lett. 1997;400:271–274. doi: 10.1016/s0014-5793(96)01404-4. [DOI] [PubMed] [Google Scholar]
- 10.Eisenreich W, Sagner S, Zenk M H, Bacher A. Tetrahedron Lett. 1997;38:3889–3892. [Google Scholar]
- 11.Zeidler J G, Lichtenthaler H K, May H U, Lichtenthaler F W. Z Naturforsch. 1997;52c:15–23. [Google Scholar]
- 12.Broers S T J. Ph.D. thesis. Zürich, Switzerland: Eidgenössische Technische Hochschule; 1994. [Google Scholar]
- 13.Rosa Putra S, Lois L M, Campos N, Boronat A, Rohmer M. Tetrahedron Lett. 1998;39:23–26. [Google Scholar]
- 14.Arigoni D, Sagner S, Latzel C, Eisenreich W, Bacher A, Zenk M H. Proc Natl Acad Sci USA. 1997;94:10600–10605. doi: 10.1073/pnas.94.20.10600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thérisod M, Fischer J C, Estramareix B. Biochem Biophys Res Commun. 1981;98:374–379. doi: 10.1016/0006-291x(81)90850-0. [DOI] [PubMed] [Google Scholar]
- 16.David S, Estramareix B, Fischer J C, Thérisod M. J Am Chem Soc. 1981;103:7341–7342. [Google Scholar]
- 17.Julliard J H, Douce R. Proc Natl Acad Sci USA. 1991;88:2042–2045. doi: 10.1073/pnas.88.6.2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hill R E, Sayer B G, Spenser I D. J Am Chem Soc. 1989;111:1916–1917. [Google Scholar]
- 19.Kennedy I A, Hill R E, Pauloski R M, Sayer B G, Spenser I D. J Am Chem Soc. 1995;117:1661–1662. [Google Scholar]
- 20.Hill R E, Himmeldirk K, Kennedy I A, Pauloski R M, Sayer B G, Wolf E, Spenser I D. J Biol Chem. 1996;271:30426–30435. doi: 10.1074/jbc.271.48.30426. [DOI] [PubMed] [Google Scholar]
- 21.Fujisaki S, Hara H, Nishimura Y, Horiuchi K, Nishino T. J Biochem (Tokyo) 1990;108:995–1000. doi: 10.1093/oxfordjournals.jbchem.a123327. [DOI] [PubMed] [Google Scholar]
- 22.Asai K, Fujisaki S, Nishimura Y, Nishino T, Okada K, Nakagawa T, Kawamukai M, Matsuda H. Biochem Biophys Res Commun. 1994;202:340–345. doi: 10.1006/bbrc.1994.1933. [DOI] [PubMed] [Google Scholar]
- 23.Bachmann B J. Bacteriol Rev. 1972;36:525–557. doi: 10.1128/br.36.4.525-557.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kohara Y, Akiyama K, Isono K. Cell. 1987;50:495–508. doi: 10.1016/0092-8674(87)90503-4. [DOI] [PubMed] [Google Scholar]
- 25.Browner M F, Rasor P, Tugendreich S, Fletterick R J. Protein Eng. 1991;4:351–357. doi: 10.1093/protein/4.3.351. [DOI] [PubMed] [Google Scholar]
- 26.Kunkel T A, Roberts J D, Zakour R A. Methods Enzymol. 1987;154:367–389. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 27.Sanger F, Nicklen S, Coulson A R. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garrido T, Sánchez M, Palacios P, Aldea M, Vicente M. EMBO J. 1993;12:3957–3965. doi: 10.1002/j.1460-2075.1993.tb06073.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dumas Milne Edwards J B, Ravassard P, Icard-Liepkalsns C, Mallet J. In: PCR2. A Practical Approach. McPherson M J, Hames B D, Taylor G R, editors. New York: Oxford Univ. Press; 1995. pp. 89–118. [Google Scholar]
- 30.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current Protocols in Molecular Biology. New York: Wiley; 1989. [Google Scholar]
- 31.Touchstone J C, Dobbins M F. Practice of Thin Layer Chromatography. New York: Wiley; 1978. p. 182. [Google Scholar]
- 32.Icho T. J Bacteriol. 1988;170:5110–5116. doi: 10.1128/jb.170.11.5110-5116.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Youvan D C, Bylina E J, Alberti M, Begusch H, Hearst J E. Cell. 1984;37:949–957. doi: 10.1016/0092-8674(84)90429-x. [DOI] [PubMed] [Google Scholar]
- 34.Mandel M A, Feldmann K A, Herrera-Estrella L, Rocha-Sosa M, León P. Plant J. 1996;9:649–658. doi: 10.1046/j.1365-313x.1996.9050649.x. [DOI] [PubMed] [Google Scholar]
- 35.McCormack T, McCormack K. Cell. 1994;79:1133–1135. doi: 10.1016/0092-8674(94)90004-3. [DOI] [PubMed] [Google Scholar]
- 36.Kennedy I A, Hemscheidt T, Britten J F, Spencer I D. Can J Chem. 1995;73:1329–1337. [Google Scholar]
- 37.Yokota A, Sasajima K-I. Agric Biol Chem. 1984;48:149–158. [Google Scholar]
- 38.Yokota A, Sasajima K-I. Agric Biol Chem. 1986;50:2517–2524. [Google Scholar]
- 39.Langley D, Guest J R. J Gen Microbiol. 1977;99:263–276. doi: 10.1099/00221287-99-2-263. [DOI] [PubMed] [Google Scholar]
- 40.Lange B M, Wildung M R, McCaskill D, Croteau R. Proc Natl Acad Sci USA. 1998;95:2100–2104. doi: 10.1073/pnas.95.5.2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lindqvist Y, Schneider G, Ermler U, Sundström M. EMBO J. 1992;11:2373–2379. doi: 10.1002/j.1460-2075.1992.tb05301.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reizer J, Reizer A, Bairoch A, Saier M H., Jr Res Microbiol. 1993;144:341–347. doi: 10.1016/0923-2508(93)90191-4. [DOI] [PubMed] [Google Scholar]