Abstract
Papillomavirus late gene expression is tightly linked to the differentiation state of the host cell. Levels of late mRNAs are only in part controlled by regulation of the late promoter, other posttranscriptional mechanisms exist that reduce the amount of late mRNA in undifferentiated cells. Previously we described a negative regulatory element (NRE) located upstream of the human papillomavirus type 16 late poly(A) site. We have delineated the NRE to a 79-nt region in which a G+U-rich region was the major determinant of NRE activity. UV-crosslinking assays identified a prominent nuclear protein of 65 kDa as the only factor in close contact with the NRE, and a complex of at least five proteins, including the 65-kDa protein, was enriched on NRE–RNA. Binding of the 65-kDa protein was depleted by preincubation with poly(U) Sepharose in high salt, a property characteristic of the U2 small nuclear ribonucleoprotein auxiliary factor U2AF65 and bacterially expressed U2AF65 exhibited NRE binding. The 65-kDa protein bound to the G+U-rich NRE 3′ half which shows homology to the B2P2 sequence a known U2AF65 binding site in the α-tropomyosin gene, and the G+U-rich element can be replaced by B2P2 in the binding assay. Treatment of cells with phorbol 12-myristate 13-acetate reduced binding of the 65-kDa protein, induced NRE binding of a cytoplasmic protein, and relieved the NRE block on reporter gene expression.
Papillomaviruses infect basal cells of the skin or mucosal membranes. Viral DNA persists extrachromosomally and virus early gene functions lead to hyperproliferation of the infected cells however vegetative viral DNA replication, late mRNA formation, translation of late protein, and capsid formation are suppressed until cells in upper layers of the stratified epithelium have differentiated (1, 2). Differentiation of human papillomavirus containing keratinocytes in organotypic culture permits vegetative viral DNA replication, increases polyadenylylated late mRNA, and can lead to virus production (3, 4) which is stimulated by induction with phorbol 12-myristate 13-acetate [PMA (2)] or grafting onto nude mice (5) allows translation of these mRNAs into late proteins.
Primary transcripts of early and late virus RNA’s overlap and are processed by complex splicing patterns with polyadenylylation occurring either at the early (5′ proximal) or late poly(A) site. The switch from early to late virus gene expression could be regulated at the level of gene dosage, change from early to late promoter(s), alteration of RNA splicing patterns, up-regulation of 3′ processing at the late poly(A) site, cytoplasmic transport of late RNA, and changes in cytoplasmic RNA stability. Regulation by the latter posttranscriptional mechanisms has been suggested to be mediated by sequences within the 3′ untranslated region of papillomavirus late mRNA, and this is the aspect examined here.
Our analysis of human papillomavirus type 16 (HPV-16) late poly(A) sites led to the discovery of a negative regulatory element (NRE) in the late RNA 3′ untranslated region that exerted a strong negative effect on expression of a reporter gene (6), and which could act at the level of RNA stability (7). A similar NRE element was identified in bovine papillomavirus type 1 [BPV-1 (8)] where studies demonstrated that a sequence homologous to a 5′ splice site was necessary and sufficient for inhibitory activity (9); base pairing between the element and the 5′ end of U1 small nuclear RNA was required and the proposed mode of inhibitory action was reduction of polyadenylylation efficiency. Similar analysis of the HPV-16 NRE identified a 51-nt NRE fragment that exerted 6-fold inhibition in the context of the simian virus 40 poly(A) site and contained four sequence motifs with partial homology to 5′ splice sites; only 5′ splice site 2 made a significant contribution to inhibitory activity, and it was suggested that other motifs may contribute to full HPV-16 NRE activity. The 51-nt HPV-16 fragment analyzed (9) formed part of a 99-nt region characterized by us (7) which exerted a 100-fold inhibition of reporter gene expression in the context of the HPV-16 late poly(A) site located more than 90-nt downstream and 5′ splice site 2 accounted only for 13% of the inhibition.
We have delineated the minimal NRE to 79-nt region located between HPV-16 nucleotides 7128–7206. The major determinant of NRE activity was a G+U-rich region located downstream of splice site homology 2, and the 5′ splice site homologies made only a minor contribution to and were not sufficient for NRE activity. A nuclear 65-kDa protein, present in HeLa cells and the HPV-16 containing W12 keratinocyte cell line, sharing characteristic properties with the splicing auxiliary factor U2AF65 (10), was cross-linked efficiently to the NRE, and a 36-nt sequence located downstream of the 5′ splice site homologies was sufficient for binding of the 65-kDa protein. The G+U-rich element and the B2P2 sequence, a known U2AF65 binding site (10, 11), display homology and were interchangeable in the binding assay. A complex containing at least four additional proteins could assemble over the NRE by protein–protein interactions. Treatment of W12 cells with PMA gave reduced 65-kDa protein binding, induced NRE binding of a predominantly cytoplasmic 40-kDa protein and abrogated NRE inhibition of reporter gene activity.
MATERIALS AND METHODS
Plasmids.
Plasmids CAT SE227 [C-SE227, containing an HPV-16 SspI–EcoRI fragment nt 7226–7453 of the prototype DNA sequence (12)] and CAT PE445Δ121 (CΔ121, derived from an HPV-16 PstI–EcoRI fragment nt 7008–7453 by deletion of nt 7008–7128) were as described (7) and respectively lack or contain the NRE. HPV-16 sequences of these plasmids were cloned into pGEM-1 (Promega) for synthesis of riboprobes. Plasmid pG44 contained the herpes simplex virus 1 (HSV-1) UL44 gene poly(A) site (13). Plasmid BS-LPA was pBluescript SK+ (Stratagene) with HPV-16 nt 7008–7453 cloned after PCR amplification of viral DNA from W12 cells. For construction of 3′ deletions, plasmid CΔ121 was partially digested with DraI, a clone (Δ121X) with a XhoI linker in the NRE DraI site (nt 7282–7287) was digested with XhoI and treated with Bal31 nuclease. HindIII–XhoI fragments containing the shortened NRE were cloned into HindIII–XhoI digested Δ121X to reconstitute the HPV-16 late poly(A) signal. Point mutations substituting A for G in the putative first intron base of either 5′ splice site 1 or both 5′ splice sites 3 and 4 were generated from GΔ121 using the Doubletake kit (Stratagene). HPV-16 sequences with the different mutations were recloned into CΔ121 for analysis of CAT expression.
Cell Procedures.
HeLa cells were grown in DMEM supplemented with 5% newborn calf serum. W12 cells (14) were grown in fibroblast feeder supported cultures as described (15); as appropriate, PMA was added to W12 cells at a concentration of 16 nM (2). Cells were grown in 9-cm-diameter dishes and DNA transfections/chloramphenicol acetyltransferase (CAT) assays were carried out (7) at 70% confluence. Cells were incubated with DNA for 16 hr (HeLa) or 6 hr (W12), fresh medium was added, and after incubation for further 48 hr the cells were washed and harvested.
Nuclear extracts were prepared (16); for W12 cells, feeder cells were removed first by rinsing with 0.01% EDTA (15). Cells were washed in 30 packed cell volumes (pcv) of PBS at 4°C, resuspended in 1 pcv of 10 mM Hepes (pH 8.0), 1.5 mM MgCl2, 10 mM KCl, and 1 mM DTT. After 15 min on ice cells were lysed by drawing five times through a 26-gauge needle. Nuclei were pelleted and the supernatant “cytoplasmic extract” was dialyzed against binding buffer [60 mM KCl/20 mM Hepes, pH 7.6/1 mM MgCl2/10% (vol/vol) glycerol]. Nuclear pellets were resuspended in 0.7 pcv of buffer C [20 mM Hepes, pH 8.0/1.5 mM MgCl2/25% (vol/vol) glycerol/420 mM NaCl/0.2 mM EDTA/1 mM DTT/0.5 mM phenylmethylsulfonyl fluoride] and incubated on ice with stirring for 30 min. Nuclear debris was removed and the supernatant “nuclear extract” was dialyzed against binding buffer and used directly or stored at −70°C.
Recombinant U2AF65 expressed in Escherichia coli was provided by C. Oubridge (Medical Research Council Laboratory of Molecular Biology, Cambridge, U.K.). U2AF65 partially purified by chromatography on heparin Sepharose and eluted with 1 M urea, 50 mM Tris·acetate (pH 6.0), and 1 mM EDTA was at least 50% U2AF65 as visualized on a Coomassie stained polyacrylamide gel.
PCR and Riboprobes.
Templates for wild-type and mutated NRE riboprobes were generated by PCR. Forward primers are as follows: SP6-1, 5′-ATCTGATCACGATTTAGGTGACACTATAGATCTGCTAACGC-3′; SP6-2, 5′-GATCGATTTAGGTGACACTATAGTATTGTATGTATGTTGAATTAGTGTTG-3′; SP6-3, 5′-GATCGATTTAGGTGACACTATAGAATTAGTGTTGTTTGTTGTG-3′ (nucleotides corresponding to position 1 of the RNA transcripts are underlined); and wt1234, 5′-GCTAAACGCAAAAAACGTAAGCTGTAAGTATTGTATG-3′. Reverse primersare as follows: NE-5, 5′-GTGAACATACATACAATACTT-3′; NE-6, 5′-GTGTACACAACAAACAACACTAA-3′; NE-R, 5′- GTGACCGGTACATACAAACATATACACAACAAACAACACTAATTC-3′; SD1234, 5′-CACAACAAACAACACTAATTCAACATATATATAATACTTATAGCTTATGTTTTTTGCGTTTAGCATAGTG-3′ (nucleotides that introduce G to A mutations in the first base of putative intron sequences in all four of the 5′ splice site homologies are underlined); B2P2-1, 5′-AACGGGTGCCAGGTTGGCATACATACAATACTTACAGC-3′; B2P2-2, 5′-GTGACCGGTGTGAGACACACACAACAAACGGGTGCCAGGT-3′; GUA, 5′- GCACATACATACATATACTCATCATACTACTCTAATTCAACA-3′ (nucleotides that introduce U to A mutations in the G+U-rich element are underlined). Overlapping forward and reverse primers were filled in during 30 cycles of 20-sec denaturing at 94°C, 20-sec annealing at 45°C, and 10-sec extension at 72°C in 100 μl DeepVent buffer (Stratagene), in the presence of 0.1 mM dNTPs, 20 pmols each primer, and 2 units DeepVent DNA polymerase. Longer templates, where primers did not overlap, were synthesized during 40 cycles in the presence of 1 ng BS-LPA plasmid or 0.2 pmol bridging oligonucleotide. Generation of probes was as follows: probe 5′ with primers SP6-1 and NE5, template wt1234; probe m with primers SP6-2 and NE-6 (overlap); probe 3′ with primers SP6-3 and NE-R (overlap); probe WT with primers SP6-1 and NE-R, template BS-LPA; probe B2 with primers SP6-1 and B2P2-2, templates wt1324 and B2P2-1; probe GA with primers SP6-1 and NE-R, template SD1234; and probe UA with primers SP6-1 and GUA, template BS-LPA.
Synthesis of probe and competitor RNA (17) for UV-crosslinking assays utilized the Promega riboprobe system. Unlabeled competitors or 32P-labeled probes for the UV-crosslinking assay were transcribed with SP6 RNA polymerase from 1 μg linearized plasmid or from templates generated by PCR (70–150 ng). Biotin-labeled transcripts were synthesized as 32P-labeled probes, but in the presence of 15 μM UTP and 1 μM biotin-16-uridine-5′-triphosphate (bio-UTP; Boehringer Mannheim).
UV-Crosslinking.
Varying amounts of unlabeled competitor RNA, 1 μl (100 counts/sec) of 32P-labeled riboprobe and nuclear extract corresponding to 10 μg of protein were added in a final volume of 10 μl binding buffer and incubated at 20°C for 15 min. RNA and protein were UV-crosslinked on ice in a Stratalinker (Stratagene) at a setting of 250 mJ. Unbound probe was digested with 1 μl of RNase (10 μg/ml) at 37°C for 15 min. After addition of 10 μl boiling mix and incubation at 100°C for 5 min, samples were electrophoresed on SDS/polyacrylamide gels. Gels were vacuum dried and autoradiographed on x-ray film or PhosphorImager (Molecular Dynamics) screens.
Binding to Poly(U) Sepharose or Streptavidin Agarose.
For depletion experiments, nuclear extracts were preincubated with poly(U) Sepharose 4B (Pharmacia). Typically, 200 μl poly(U) Sepharose (0.15 mg/ml, equilibrated in binding buffer with 2 M KCl) were pelleted and incubated for 30 min at 20°C with 80 μg nuclear proteins in binding buffer with 2 M KCl. The supernatant was removed and dialyzed against binding buffer. For depletion or purification of the NRE binding activity, nuclear extracts (100 μg protein) were incubated with 1 μg biotinylated probe N in 150 μl binding buffer for 30 min at 4°C. The RNA and associated proteins were then bound for 20 min at 20°C to 200 μl 4% streptavidin-beaded agarose (Sigma) equilibrated in binding buffer. The unbound fraction was removed, the beads were washed twice in 500 μl binding buffer, and once in binding buffer with 300 mM KCl. For elution of bound proteins, beads were incubated in 100 μl binding buffer with 1 M KCl, or with RNase A (0.2 μg/ml, 15 min at 37°C), or with 200 mM poly(U) (Boehringer Mannheim). Undepleted or depleted extracts corresponding to 10 μg extract input and wash or eluate fractions corresponding to 50 μg extract input were separated by electrophoresis on SDS/polyacrylamide gels and silver stained.
RESULTS
The NRE Extends Downstream of Four Weak 5′ Splice Site Homologies.
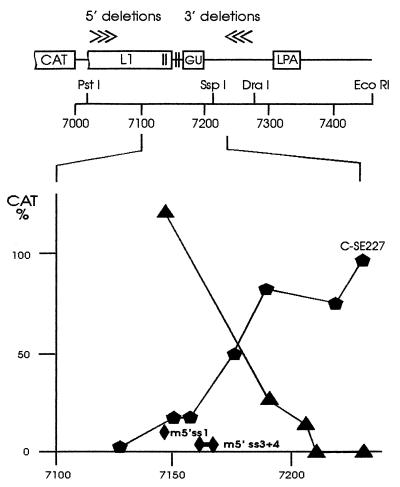
A series of 3′ deletions from a DraI site in HPV-16 sequences of plasmid CΔ121 was constructed and CAT activity measured after transfection of HeLa cells. CAT activity remained low for deletions up to 67 nt upstream of the DraI site, and further deletions progressively relieved the negative effect (Fig. 1). This mapped the NRE 3′ boundary to 79 nt downstream from the endpoint identified with the 5′ deletion series (between nt 7128–7206). Thus, the NRE extended 37 nt downstream of four weak 5′ splice site homologies present in its 5′ portion. In the context of the HPV-16 poly(A) site, located some 110 nt downstream, the 79-nt NRE reduced CAT expression by 99%. Deletion of splice site 1 (AACguaagc, lowercase letters denote hypothetical intron sequences) together with the crucial splice site 2 (GCUguaagu), the only element that significantly suppressed CAT activity in a previous analysis (9), still resulted in an 86% reduction of CAT expression. Continued deletion of the weak splice site homologies 3 and 4 (including 2 nt of a downstream G+U-rich region) reduced CAT activity by 50%. A retained splice site 1 showed no inhibition, and the presence of point mutations in splice site 1 or in both splice sites 3 and 4 had little effect on CAT expression (Fig. 1). The fragment containing splice sites 1 and 2 is 33% in G+U and 30% in pyrimidine content, whereas the region downstream of splice site 2 is 83.7% G+U and 57% pyrimidine. Deletions from either side into the G+U-rich region progressively alleviated the block on CAT expression.
Figure 1.
Boundaries of the HPV-16 NRE. The 5′ deletion series described earlier (6) was complemented by a 3′ deletion series. Effects on NRE activity were assayed using CAT reporter constructs with different NRE portions present upstream of the HPV-16 late poly(A) site. CAT activities relative to plasmid C-SE227 (100%) are indicated. ▴, 3′ deletion series; , 5′ deletion series; ♦, point mutations in 5′ splice site homologies 1 (m5′ss1) or 3 and 4 (m5′ss3+4).
A 65-kDa Cellular Protein Binds to an NRE Riboprobe.
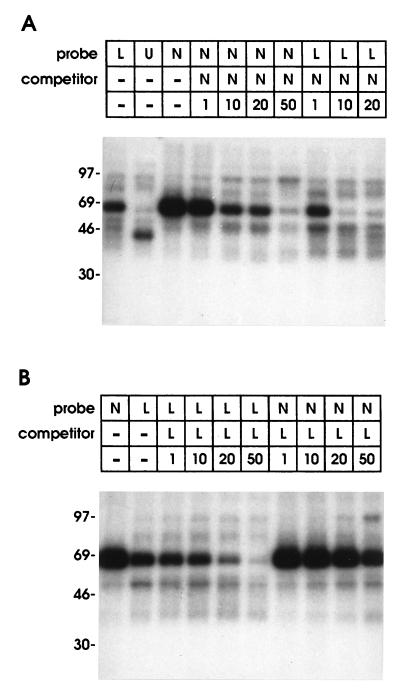
The mechanisms proposed to explain NRE function all imply that nuclear or cytoplasmic proteins bind to RNA containing this sequence motif. Thus, we examined binding of 32P-labeled NRE riboprobes to proteins from HeLa cells using a UV-crosslinking assay. Initially we focused on nuclear proteins as previous studies had implicated interference of splicing factors with polyadenylylation and/or mRNA transport. The only nuclear protein consistently and efficiently identified by the NRE riboprobe was represented by a band of 65–70 kDa apparent molecular weight on denaturing polyacrylamide gels; a protein of the same size bound to a region downstream of the NRE containing the HPV-16 late poly(A) site. To examine strength of binding, we performed cross-competition assays. The 99-nt NRE riboprobe (N in Fig. 2) was strongly bound by a 65- to 70-kDa protein (Fig. 3A) and the same unlabeled RNA competed with the labeled probe. Probe N competed even more efficiently with a labeled RNA from the adjacent region (L in Fig. 2), which contained the late poly(A) site and exhibited weaker 65- to 70-kDa binding. The converse was found with unlabeled probe L which competed efficiently against its radiolabeled counterpart but was not a good competitor for labelled probe N (Fig. 3B), suggesting a weak binding site for the 65-to 70-kDa protein downstream of the functional NRE. A riboprobe containing the HSV-1 UL44 gene poly(A) site showed an unrelated binding pattern (Fig. 3A, lane U); this and other HSV-1 poly(A) site riboprobes did not compete for binding of the 65- to 70-kDa protein (data not shown), indicating that the binding factor was specific for HPV-16 sequences and was not a poly(A) site binding protein.
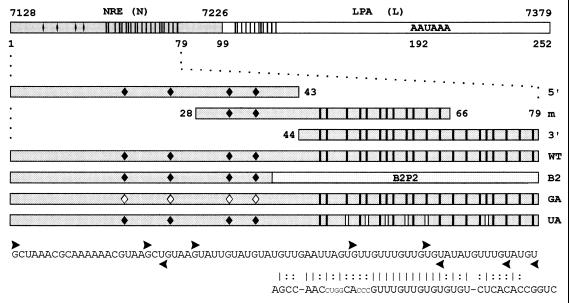
Figure 2.
Templates used to generate radiolabeled riboprobes and unlabeled competitor RNAs. Probe N: 79-nt minimal NRE and 20 nt downstream (HPV-16 nt 7128–7226). Probe L: 153-nt region (nt 7227–7379) containing the HPV-16 late poly(A) site (LPA). Probe 5′: 5′ portion of the 79-nt NRE containing four 5′ splice site homologies (♦). | = uridines present in the 3′ portion of probe N and within the putative weak binding site in L. Probe m: middle NRE section containing two 5′ splice site homologies and half of the G+U-rich element. Probe 3′: 3′ NRE portion containing a G+U-rich region with homology to a B2P2 binding site. Probe WT: 79-nt wild-type NRE. Probe B2: G+U-rich region substituted by the U2AF65 binding site B2P2 from the α-tropomyosin gene. Probe GA: HPV-16 NRE with G to A mutations in the putative first intron base of all four 5′ splice site homologies ♦. Probe UA: six U to A mutations in the G+U-rich region (| |). Vertical bars in the L fragment indicate partial homology to the G+U-rich element. ▸, first nucleotides retained in the 5′ deletion clones; ◂, last nucleotides retained in the 3′ deletion clones. The B2P2 sequence is shown; vertical bars indicate nucleotide homologies, and colons refer to conserved purine/pyrimidine patterns.
Figure 3.
Binding of HeLa cell nuclear proteins to the HPV-16 NRE. Nuclear proteins were UV-crosslinked to radiolabeled RNA probes spanning the NRE and 20 nt of downstream region (probe N), the adjacent 153-nt region (probe L), or the HSV-1 UL44 gene poly(A) site (probe U). A 1–50 molar excess of unlabeled probe was used for competition as indicated. (A) Competition of unlabeled riboprobe N against labeled probe N or labeled probe L. (B) Competition of unlabeled riboprobe L against labeled probe L or labeled probe N.
Binding Properties of the 65-kDa Protein and Comparison with U2AF65.
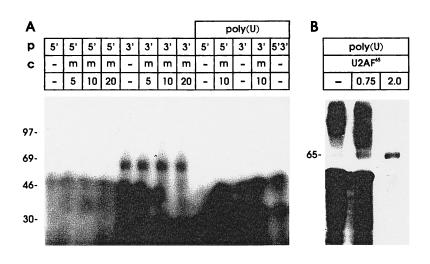
Riboprobes were synthesized from PCR templates covering either the 5′ portion of the NRE with the four 5′ splice site homologies (nt 7128–7170) or the G+U-rich 3′ portion that conferred most NRE activity (nt 7171–7206). No NRE-specific proteins were detected with the 5′ probe. In contrast, binding of 65- to 70-kDa protein was obtained with the G+U-rich 3′ probe (Fig. 4A) and this binding was not efficiently competed by cold probe m (nt 7155–7193) which contains 5′ splice sites 3 and 4 and extends midway into the G+U-rich region (Fig. 2). We noticed that the G+U-rich region showed a high degree of sequence homology to a U2AF65 binding site, B2P2, present in the default intron of α-tropomyosin pre-mRNA (10, 11).
Figure 4.
Binding properties of the 65-kDa protein and its target site within the NRE. (A) HeLa cell nuclear proteins were UV-crosslinked to probes corresponding to the 5′ half of the NRE (probe 5′) with four 5′ splice site homologies, or the 3′ half of the NRE (probe 3′) with a G+U-rich region with homology to a U2AF65 binding site. Poly(U) denotes nuclear extracts depleted by preincubation with poly(U) Sepharose in the presence of 2 M KCl. Unlabeled RNA corresponding to the middle section of the NRE (m, see Fig. 2) was used as competitor. (B) Reconstitution experiment with U2AF65. The indicated amounts (in micrograms) of partially purified, bacterial protein extract containing U2AF65 were added to HeLa cell nuclear extracts that were depleted of 65-kDa protein binding activity by preincubation with poly(U) Sepharose in 2 M KCl, probe N was used (Fig. 2).
The U1 small nuclear ribonucleoprotein (snRNP) 70-kDa protein (18) was a candidate for the 65- to 70-kDa protein but preincubation of nuclear extracts with anti-U1 70-kDa antibodies/protein A-Sepharose did not affect its binding (data not shown). Other splicing associated proteins or RNA binding proteins were considered, two proteins of the appropriate size, PTB [≈62 kDa (19)] and U2AF65 (10) bind to poly(U) on Sepharose beads. PTB is eluted from poly(U) Sepharose with 0.8M KCl (20) while U2AF65 remains bound even in 2 M KCl (10); apart from U2AF65, only the smaller C proteins remain bound efficiently to poly(U) in high salt (21). Nuclear extracts were depleted of NRE binding activity by preincubation in as high as 2 M KCl (Fig. 4A) which eliminated PTB as a candidate NRE binding factor (before UV-crosslinking, depleted extracts were redialyzed against binding buffer).
Of the factors essential for splicing, only U2AF65 is depleted from nuclear extracts by incubation with poly(U) in high KCl concentrations, and adding back bacterially expressed U2AF65 restores splicing (10). We tested the ability of bacterially expressed, partially purified U2AF65 protein to restore NRE binding (Fig. 4B). Addition of bacterially expressed U2AF65 to depleted nuclear extracts gave binding of a protein of identical size to the cellular NRE binding factor and allowed a better size estimate of 65 kDa. The bacterial extracts were contaminated with RNase; adding less RNase after crosslinking led to insufficient degradation of probe and unacceptable background while higher RNase concentrations reduced the signal. Thus, the weak signal obtained with bacterially expressed U2AF65 is explained by degradation of the probe during incubation before UV-crosslinking. Nevertheless, it can be clearly seen that the HeLa extract was mostly depleted of NRE binding activity (Fig. 4B, lane 1), and that increasing amounts of U2AF65 gave increased binding.
Mutations Expected to Affect U2AF65 Binding Also Weakened Binding of the 65-kDa Protein.
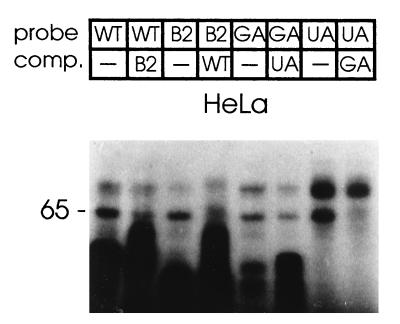
To elucidate the role of individual NRE components in 65-kDa protein binding, a series of riboprobe templates with the following features was generated by PCR. These were (Fig. 2) WT (a wild-type NRE), B2 (wild-type 5′ splice site homologies with the G+U-rich element replaced by the B2P2 sequence), GA (all four 5′ splice sites mutated G to A in the putative first intron base), and UA (wild-type 5′ splice sites with six U to A substitutions in the G+U-rich element). Polypyrimidine tract binding proteins are involved in a number of posttranscriptional regulatory processes, and though little is known about what determines their specificity (22), substitutions of U with A have been shown to weaken U2AF65 binding (10). We tested riboprobes synthesized from these templates using HeLa cell nuclear extracts for binding and cross competition (Fig. 5). The 65-kDa protein bound to the NRE with the G+U-rich element or the B2P2 binding site and B2 competed with WT for 65-kDa binding and vice versa. The GA probe with all four 5′ splice sites mutated and the UA probe with mutations in the G+U-rich region still bound to the 65-kDa protein; however, differences were seen in competition binding experiments. The probe UA competed less efficiently against the probe GA, whereas GA competed more efficiently against UA; absolute differences in signal strength between GA and UA probes are due to differential labeling efficiencies. These results demonstrated that the putative 5′ splice sites were not required for 65-kDa binding and mutations expected to reduce binding of U2AF65 weakened binding of the HeLa cell nuclear 65-kDa protein.
Figure 5.
Effects of mutations and substitutions in the NRE on 65-kDa protein binding. Wild-type and B2P2 substitutions (probes as in Fig. 2) were assayed for cross-competition with 10-fold molar excess of the respective unlabeled RNA, similarly the 5′ splice sites and G+U mutants were tested for cross-competition with their unlabeled counterparts with HeLa nuclear extract.
The 65-kDa Protein Forms Part of a Complex That Can Be Enriched on Biotinylated NRE–RNA.
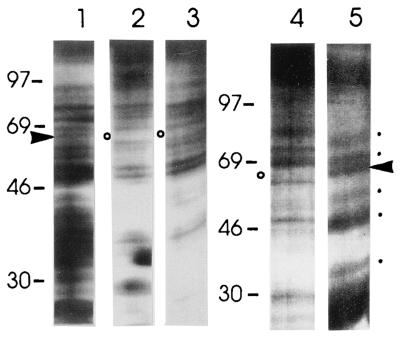
Comparison of HeLa cell undepleted nuclear extracts with the unbound fractions of nuclear extracts preincubated with either the biotinylated NRE riboprobe-streptavidin beads or with poly(U) Sepharose beads in the presence of 2 M KCl, revealed that a 65-kDa protein was depleted by both treatments (Fig. 6; compare lane 1 with lanes 2 and 3). Several proteins were eluted by a step gradient from 0.3 to 2 M KCl; however, the 65-kDa protein was not eluted (data not shown). Release of the bound proteins was achieved either by treatment with RNase A (Fig. 6, lane 5) or, as U2AF65 has a higher affinity for poly(U) than for its natural binding sites, using soluble poly(U). In addition to the 65-kDa protein, four eluted bands of ≈90, 70, ≈50, and ≈30 kDa were detected on silver stained polyacrylamide gels: other proteins depleted from the extracts and not enriched in the eluate fraction represent either unspecific or low affinity factors lost during the washes. Apart from 65-kDa band, none of the other protein bands eluted was consistently detected in the UV-crosslinking assay; hence, none of these is in close contact with NRE–RNA. These data indicated that a larger complex, which is stable to at least 0.3 M KCl, could assemble over the NRE presumably through protein–protein interactions.
Figure 6.
Purification of a larger NRE-associated protein complex. (A) Comparison of complete HeLa cell nuclear extract (lane 1) with extracts depleted by preincubation with biotinylated NRE–RNA and adsorption on streptavidin beads (lanes 2 and 4) or depleted by preincubation with poly(U) Sepharose in the presence of 2 M KCl (lane 3). Release of NRE-bound proteins from streptavidin beads by RNase treatment (lane 5). ▸, 65-kDa protein; ○, depleted 65-kDa protein; [cirf], enriched proteins; R, RNase A.
The NRE Confers PMA Responsiveness to CAT Expression and PMA Down-Regulates the 65-kDa Protein.
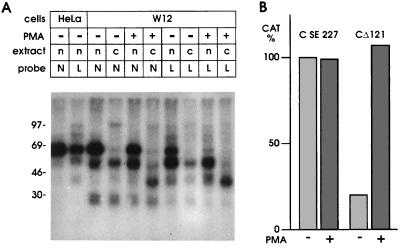
In a cell line containing HPV-31 DNA, PMA induces cellular differentiation which in turn allows viral late mRNA synthesis, translation of late proteins, and viral capsid formation (2). We asked if the NRE could play a regulatory role in the HPV-16 containing W12 cell line, and what would be the effect of treating these cells with PMA. CAT activity of the C-SE227 reporter plasmid that contains the HPV-16 late poly(A) site but lacks the NRE showed a high basal expression level which was unaffected by PMA treatment (Fig. 7B). By contrast, expression from the CΔ121 construct which contains the NRE was suppressed in untreated cells and, typically, CAT activity was stimulated by PMA to virtually the same level as the NRE lacking plasmid (Fig. 7B). Higher basal CAT levels from the NRE containing plasmid in the absence of PMA are explained by leakiness of the W12 cells for late gene expression (23); only 5- to 6-fold stimulation of CAT activity by PMA was observed (Fig. 7B) as compared with the 100-fold difference in HeLa cells in the absence or presence of the NRE (Fig. 1).
Figure 7.
Effect of PMA treatment on NRE binding proteins and reporter gene expression. (A) Changes in protein binding of nuclear (n) or cytoplasmic (c) extracts from HeLa cells or W12 cells following treatment with PMA. (B) Changes of CAT activity in W12 cells transfected with CD121 or C-SE227 plasmids following treatment with PMA.
In nuclear extracts, PMA treatment of W12 cells led to a consistent 25% reduction in binding of 65-kDa protein to an NRE riboprobe (Fig. 7A). The estimated ratio of 65-kDa to a ≈50-kDa band was 1.8-fold higher in untreated as compared with treated W12 cells. Reduction of 65-kDa protein binding on PMA treatment was readily apparent at the weaker binding site in probe L (Fig. 7A). Thus, binding of the 65-kDa protein does respond to PMA treatment in a manner consistent with a possible involvement in NRE regulation in W12 cells. Interestingly, PMA treatment resulted in a readily detectable ≈40-kDa binding protein present predominantly in the cytoplasm and which bound to both NRE and L probes (Fig. 7A).
DISCUSSION
The HPV-16 NRE maps to a 79-nt segment [nt 7128–7206 of the prototype sequence (12)] which contains four 5′ splice site homologies in its 5′ portion which deviate significantly from the consensus sequence together with a G+U-rich region in its 3′ portion. Other studies (9) had indicated that only the second of the four 5′ splice site homologies made a major contribution to the 6-fold inhibitory effect of a 51-nt NRE segment (nt 7128–7178) in the context of the simian virus 40 early poly(A) site; when sites 1 and 2 were mutated, reduction of CAT expression went from 83% to 10%. By contrast, we have shown that the 79-nt-long NRE in the context of the HPV-16 poly(A) site reduced CAT expression by 99%, and after deletion of sites 1 and 2 CAT expression was still reduced by 86%. The major determinant of NRE function was located downstream of site 2 in a region of 84% G+U and deletions from either side into this G+U-rich region progressively relieved the block on CAT expression.
Similar to these data, deletion of the single 5′ splice site homology in the HPV-1a NRE did not affect its inhibitory properties (24). This situation contrasts with BPV-1 where virtually all the NRE activity was attributable to a single consensus 5′ splice site sequence present 10 nt upstream of the late poly(A) site and NRE mutations expected to interfere with binding of U1 snRNP alleviated the block which was restored by compensatory mutations in U1 RNA. G+U-rich sequence elements similar to the NRE were found in the late 3′ untranslated regions of HPV-1a (25), HPV-11 (26), HPV-18 (27), HPV-31 (28), HPV-33 (29), HPV-35 (30), HPV-57 (31), and HPV-58 (32) located upstream of their late poly(A) sites. BPV-1 lacks G+U-rich elements upstream from the late poly(A) site and the HPV-16.
Three mechanisms have been proposed to explain the functioning of papillomavirus NRE elements; namely, interference of nuclear factors with efficient 3′ RNA processing, inhibition of late mRNA nuclear export, and decreased cytoplasmic stability of mRNA. All require NRE recognition by nuclear or cytoplasmic factors, most likely proteins or ribonucleoprotein complexes. UV-crosslinking assays identified a nuclear 65-kDa protein that bound to the G+U-rich NRE portion; a protein of identical size also bound to the region downstream that contains the HPV-16 late poly(A) site; however, binding was much weaker and deletion of this binding site had little effect on inhibition of CAT expression. A major difference between the strong functional and the weaker nonfunctional binding sites is the presence of a tandem UGUUUGU repeat in the stronger binding site that is present only once in the weaker binding region: the B2P2 binding site contains a single CGUUUGU motif but it also contains the related sequence UGUGUGU and other clusters of pyrimidines which could be relevant for U2AF65 binding. No proteins were efficiently crosslinked to the NRE 5′ portion containing the 5′ splice site homologies.
A strong 5′ splice site present upstream of a poly(A) site would be expected to interfere with exon definition (33, 34) and efficient processing at the poly(A) site (35, 36). In contrast, weak 5′ splice sites are associated with stimulation of an adjacent poly(A) site (35, 36), perhaps through direct interaction of U1A protein with poly(A) polymerase (37). Although no such inhibitory role has been described so far for U2AF65, interactions between the U1 snRNP and U2AF65 have been described (38) and U1 snRNP acts to target U2AF65 across an exon to an upstream 3′ splice site (39). Alternatively, NRE binding of a U2AF65 like factor could affect late RNA processing or transport by changing RNA secondary structure through annealing of complementary sequences, a property reported for U2AF65 (40). Recently, a 65-kDa protein, speculated to be U2AF, has been shown to bind near the termini of hepatitis B virus pregenomic RNA at sites which are important for polyadenylylation; functional analysis indicated little involvement with polyadenylylation, and effects on RNA transport and stability were proposed (41).
For several reasons the splicing auxiliary factor U2AF65 is a likely candidate for the 65-kDa NRE binding factor although identity with U2AF65 was not definitively proven, an available antibody did not work in immunodepletion experiments and showed too high crossreactivity on Western blots. Like U2AF65 (10), the 65-kDa NRE binding protein was removed from nuclear extracts by poly(U) in the presence of high salt concentrations. NRE binding was obtained with recombinant U2AF65 and the 65-kDa protein bound to a known U2AF65 binding site; strong NRE binding did not require the 5′ splice site homologies. Interaction between U2AF65 and U1 snRNP occurs in the earliest prespliceosome complex (E) in mammals (38) and, similar to the 65-kDa protein in our study, only U2AF65 was crosslinked to RNA despite the presence of spliceosome associated proteins and U1 RNA-associated proteins. The sizes of the other bands eluted from the biotinylated NRE–RNA column are compatible with proteins present in the U1 snRNP (42) or the E complex (43).
Direct evidence for involvement of U2AF65 in NRE regulation is under study; however, deletions from either side into the binding region greatly reduced NRE activity. If the 65-kDa protein was involved in HPV-16 late gene regulation in vivo, changes might be expected upon induction of keratinocyte differentiation as increased late mRNA levels could occur by a reduction in 65-kDa binding. This effect was observed following stimulation of W12 cells with PMA, which also relieved the NRE block on reporter gene expression. While the changes in 65-kDa protein binding observed were moderate, in the natural system a combination of increased late transcription and a reduction in NRE binding protein could prevail. Similar down-regulation of U2AF65 levels takes place during neuronal and glial cell differentiation (44). In HPV-1, supply of HIV-1 Rev protein and presence of the Rev response element counteracted the inhibitory effect of the late 3′ untranslated region (24). We see an induction by PMA of a 40-kDa cytoplasmic NRE binding protein, and this is a candidate to facilitate export/translation of HPV-16 late RNAs.
In vitro assays showed that sequences at the 3′ end of HPV-16 late mRNA could mediate cytoplasmic instability (7); however, no such effect was found with BPV-1 late mRNA (9). Cytoplasmic instability is associated with AUUUA motifs at the 3′ end of mRNAs (45) but may require the longer UUAUUU(U/A)(U/A) motif (46) and in some motifs G can substitute for A (47). Thus, the (U/A)UGUUUGU(U/A) motifs present at the core of the 65-kDa protein binding site could also act as cytoplasmic RNA instability elements. Additional regulation at this level remains an intriguing possibility; it is conceivable that the HPV-16 NRE acts at multiple levels.
Acknowledgments
We would like to thank Drs. Michael Green and Chris Oubridge for providing U2AF65 protein and Dr. Walter Keller for anti-U1 70-kDa protein. We also wish to thank Jeanette Haddow for expert technical assistance. This work was supported by a grant from the Wellcome Trust (No. 032508/2/90/B).
Footnotes
Abbreviations: PMA, phorbol 12-myristate 13-acetate; HPV-16, human papillomavirus type 16; NRE, negative regulatory element; BPV-1, bovine papillomavirus type 1; CAT, chloramphenicol acetyltransferase; HSV-1, herpes simplex virus 1; snRNP, small nuclear ribonucleoprotein.
References
- 1.Bedell M A, Hudson J B, Golub T R, Turyk M E, Hosken M, Wilbanks G D, Laimins L A. J Virol. 1991;65:2245–2260. doi: 10.1128/jvi.65.5.2254-2260.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meyers C, Frattini M G, Hudson J B, Laimins L A. Science. 1992;257:971–973. doi: 10.1126/science.1323879. [DOI] [PubMed] [Google Scholar]
- 3.Dollard S C, Wilson J L, Demeter L M, Bonney W, Reichman R C, Becker T R, Chow L T. Genes Dev. 1992;6:1131–1142. doi: 10.1101/gad.6.7.1131. [DOI] [PubMed] [Google Scholar]
- 4.Frattini M G, Lim H B, Laimins L A. Proc Natl Acad Sci USA. 1996;93:3062–3067. doi: 10.1073/pnas.93.7.3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sterling J, Stanley M, Gatward G, Minson T. J Virol. 1990;64:6305–6307. doi: 10.1128/jvi.64.12.6305-6307.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kennedy I M, Haddow J K, Clements J B. J Virol. 1990;64:1825–1829. doi: 10.1128/jvi.64.4.1825-1829.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kennedy I M, Haddow J K, Clements J B. J Virol. 1991;65:2093–2097. doi: 10.1128/jvi.65.4.2093-2097.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Furth P A, Baker C C. J Virol. 1991;65:5806–5812. doi: 10.1128/jvi.65.11.5806-5812.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Furth P A, Choe W-T, Rex J H, Byrne J C, Baker C C. Mol Cell Biol. 1994;14:5278–5289. doi: 10.1128/mcb.14.8.5278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zamore P D, Patton J G, Green M R. Nature (London) 1992;355:609–614. doi: 10.1038/355609a0. [DOI] [PubMed] [Google Scholar]
- 11.Mullen M P, Smith C W J, Patton J G, Nadal-Ginard B. Genes Dev. 1991;5:642–655. doi: 10.1101/gad.5.4.642. [DOI] [PubMed] [Google Scholar]
- 12.Seedorf K, Krammer G, Durst M, Suhai S, Rowekamp W G. Virology. 1985;145:181–185. doi: 10.1016/0042-6822(85)90214-4. [DOI] [PubMed] [Google Scholar]
- 13.McGregor F, Phelan A, Dunlop J, Clements J B. J Virol. 1996;70:1931–1940. doi: 10.1128/jvi.70.3.1931-1940.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stanley M A, Browne H M, Appleby M, Minson A C. Int J Cancer. 1989;43:672–676. doi: 10.1002/ijc.2910430422. [DOI] [PubMed] [Google Scholar]
- 15.Stanley M A, Greenfield I M. In: Culture of Epithelial Cells. Freshney R I, editor. New York: Wiley; 1992. pp. 135–158. [Google Scholar]
- 16.Lee K A W, Green M R. Methods Enzymol. 1990;181:20–30. doi: 10.1016/0076-6879(90)81108-7. [DOI] [PubMed] [Google Scholar]
- 17.Melton D A, Krieg P A, Rebagliati M R, Maniatis T, Zinn K, Green M R. Nucleic Acids Res. 1984;12:7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Patton J R, Pederson T. Proc Natl Acad Sci USA. 1988;85:747–751. doi: 10.1073/pnas.85.3.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gil A, Sharp P A, Jamison S F, Garciablanco M A. Genes Dev. 1991;5:1224–1236. doi: 10.1101/gad.5.7.1224. [DOI] [PubMed] [Google Scholar]
- 20.Mulligan G J, Guo W, Wormsley S, Helfman D M. J Biol Chem. 1992;267:25480–25487. [PubMed] [Google Scholar]
- 21.Swanson M S, Dreyfuss G. Mol Cell Biol. 1988;8:2237–2241. doi: 10.1128/mcb.8.5.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morris D R, Kakegawa T, Kaspar R L, White M W. Biochemistry. 1993;32:2931–2937. doi: 10.1021/bi00063a001. [DOI] [PubMed] [Google Scholar]
- 23.Doorbar J, Parton A, Hartley K, Banks L, Crook T, Stanley M, Crawford L. Virology. 1990;178:254–262. doi: 10.1016/0042-6822(90)90401-c. [DOI] [PubMed] [Google Scholar]
- 24.Tan W, Schwartz S. J Virol. 1995;69:2932–2945. doi: 10.1128/jvi.69.5.2932-2945.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Danos O, Katinka M, Yaniv M. EMBO J. 1982;1:231–236. doi: 10.1002/j.1460-2075.1982.tb01152.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dartmann K, Schwarz E, Gissmann L, Zur Hausen H. Virology. 1986;151:124–130. doi: 10.1016/0042-6822(86)90110-8. [DOI] [PubMed] [Google Scholar]
- 27.Cole S T, Danos O. J Mol Biol. 1987;193:599–608. doi: 10.1016/0022-2836(87)90343-3. [DOI] [PubMed] [Google Scholar]
- 28.Goldsborough M D, DiSilvestre D, Temple G F, Lorincz A T. Virology. 1989;171:306–311. doi: 10.1016/0042-6822(89)90545-x. [DOI] [PubMed] [Google Scholar]
- 29.Cole S T, Streeck R E. J Virol. 1986;58:991–995. doi: 10.1128/jvi.58.3.991-995.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marich J E, Pontsler A V, Rice S M, McGraw K A, Dubensky T W. Virology. 1992;186:770–776. doi: 10.1016/0042-6822(92)90045-q. [DOI] [PubMed] [Google Scholar]
- 31.Hirsch-Behnam A, Delius H, de Villiers E M. Virus Res. 1990;18:81–97. doi: 10.1016/0168-1702(90)90091-o. [DOI] [PubMed] [Google Scholar]
- 32.Kirii Y, Iwamoto S, Matsukura T. Virology. 1991;185:424–427. doi: 10.1016/0042-6822(91)90791-9. [DOI] [PubMed] [Google Scholar]
- 33.Niwa M, MacDonald C C, Berget S M. Nature (London) 1992;360:277–280. doi: 10.1038/360277a0. [DOI] [PubMed] [Google Scholar]
- 34.Robberson B L, Cote G J, Berget S M. Mol Cell Biol. 1990;10:84–94. doi: 10.1128/mcb.10.1.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peterson M L, Perry R P. Proc Natl Acad Sci USA. 1986;83:8883–8887. doi: 10.1073/pnas.83.23.8883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wassarman K M, Steitz J A. Genes Dev. 1993;7:647–659. doi: 10.1101/gad.7.4.647. [DOI] [PubMed] [Google Scholar]
- 37.Gunderson S I, Beyer K, Martin G, Keller W, Boelens W C, Mattaj I W. Cell. 1994;76:531–541. doi: 10.1016/0092-8674(94)90116-3. [DOI] [PubMed] [Google Scholar]
- 38.Staknis D, Reed R. Mol Cell Biol. 1994;14:7670–7682. doi: 10.1128/mcb.14.11.7670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hoffman B E, Grabowski P J. Genes Dev. 1992;6:2554–2568. doi: 10.1101/gad.6.12b.2554. [DOI] [PubMed] [Google Scholar]
- 40.Lee C-G, Zamore P D, Green M R, Hurwitz J. J Biol Chem. 1993;268:13472–13478. [PubMed] [Google Scholar]
- 41.Peiri S, Ganem D. J Virol. 1996;70:6803–6809. doi: 10.1128/jvi.70.10.6803-6809.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Petterson I, Hinterberger M, Mimori T, Gottlieb E, Steitz J A. J Biol Chem. 1984;259:5907–5914. [PubMed] [Google Scholar]
- 43.Michaud S, Reed R. Genes Dev. 1993;7:1008–1020. doi: 10.1101/gad.7.6.1008. [DOI] [PubMed] [Google Scholar]
- 44.Tsukahara T, Kunika N, Momoi T, Arahata K. Brain Res. 1995;679:178–183. doi: 10.1016/0006-8993(95)00216-d. [DOI] [PubMed] [Google Scholar]
- 45.Lairdoffringa I A. Bioessays. 1992;14:119–124. [Google Scholar]
- 46.Lagnado C A, Brown C Y, Goodall G J. Mol Cell Biol. 1994;14:7984–7995. doi: 10.1128/mcb.14.12.7984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Levine T D, Gao F B, King P H, Andrews L G, Keene J D. Mol Cell Biol. 1993;13:3494–3504. doi: 10.1128/mcb.13.6.3494. [DOI] [PMC free article] [PubMed] [Google Scholar]