Abstract
CREB-binding protein (CBP) is a transcriptional coregulator that interacts with different DNA binding proteins and components of the general transcription machinery. CBP enhanced androgen receptor (AR)-dependent transcription under transient transfection conditions in CV-1 cells. The ligand binding domain (LBD) and residues 38–296 of the N-terminal region of AR are not required because the activity of a receptor mutant devoid of these domains was augmented by coexpressed CBP. There is physical interaction between AR and CBP in vivo, as judged by coimmunoprecipitation experiments from cell extracts. Consistent with the role of CBP as a coactivator for AR, the 12S E1A adenoviral protein that inactivates CBP function strongly inhibited AR-dependent transactivation. Exogenous CBP was also capable of overcoming the inhibitory effect of AR on AP-1 activity and diminished the mutual transcriptional repression between AR and NF-κB (RelA). Collectively, these data imply that transcriptional interference between AR and AP-1 or NF-κB is mediated, at least in part, through competition for intracellular CBP and that this coactivator serves as an integrator between androgen-mediated and other signaling pathways.
Androgens, like other steroid hormones, act through intracellular receptors that belong to the superfamily of ligand-activated transcription factors, the nuclear receptors (1, 2). Subsequent to hormone binding, androgen receptor (AR) acquires a new conformation that is capable of interacting not only with specific androgen response elements (AREs) located near or within the promoter regions of regulated genes but also with other transcription-regulating proteins and cofactors. Depending on the physiological context, these interactions promote either activation or repression of specific genes or gene networks that govern the regulation of development, differentiation, and maintenance of male reproductive functions.
Transcriptional regulation requires the participation of at least three classes of proteins: (i) proteins that recognize specific DNA motifs, (ii) proteins that are recruited to promoters by protein–protein interactions and act as transcriptional coactivators or corepressors, and (iii) proteins that alter the architecture of chromatin (3). To initiate transcription, nucleotide-sequence-specific factors such as steroid receptors have to communicate with components of the basal transcription apparatus. The mechanisms by which these proteins convey their activating and/or repressing functions to the basal transcription machinery are not fully understood. Several nuclear receptors have been reported to interact directly with components of the basal transcription apparatus, such as TFIIB, TFIIF, and the TATA box binding protein (4–7). Alternatively, coactivators or corepressors can act as bridging molecules between steroid receptors and general transcription factors (for review, see ref. 8).
The CREB-binding protein (CBP) was originally identified as a coactivator for CREB, the cAMP response element binding protein. CREB binds to CBP only when it is phosphorylated by protein kinase A (9). In addition to CREB, CBP interacts directly with TFIIB and may thereby act as a bridging factor between CREB and the basal transcription apparatus (10). CBP has also been reported to associate with RNA polymerase II and to recruit the polymerase to phosphorylated CREB (11). In addition to being a transcriptional adapter, CBP/p300 was recently shown to have histone acetyltransferase activity (12) and to associate with another cellular factor (P/CAF) with histone acetylase activity (13).
In addition to CREB, CBP/p300 associates with several other sequence-specific factors, including c-Jun (14, 15), c-Fos (16), c-Myb (17, 18), v-Myb (18), Sap-1a (19), Elk-1 (20), SREBP (21), NF-κB (22, 23), and members of the nuclear receptor superfamily (24–28). Transcriptional activity of retinoic acid receptor (RAR), thyroid hormone receptor, retinoid X receptor, glucocorticoid receptor (GR), and estrogen receptor is enhanced by CBP (24–28). CBP/p300 forms a complex with the coactivator SRC-1/ERAP160 and this complex, in turn, interacts with the ligand binding domain (LBD) of the RAR, thyroid hormone receptor, and estrogen receptor (24, 26–28). Moreover, competition for limiting amounts of intracellular CBP has been observed to result in inhibition of AP-1 activity by RAR and GR (24). Thus, CBP and p300 are coactivators that are used by many transcriptional regulators and that appear to be involved in cross-talk between different signaling pathways.
In the present work, we demonstrate that CBP is a coactivator for AR-dependent transactivation. Moreover, competition for intracellular CBP is responsible, at least in part, for repression of AP-1 activity by AR and for mutual transcriptional interference between AR and NF-κB (RelA).
MATERIALS AND METHODS
Materials.
Rat (r) and human (h) AR expression vectors have been described (29, 30). Deletion mutants rARΔ641–902 and rARΔ38–296/Δ641–902 were constructed from pSG5-rAR by PCR (31). pSG5-hGR was created by inserting human GR coding sequence from pGH0 (a gift from Pierre Chambon, University of Strasbourg 1, Illkirch, France) as a BamHI fragment into the BamHI site of pSG5 (Stratagene). CBP expression vector pRSV-CBP (9) was a gift form Richard H. Goodman (Vollum Institute, Oregon Health Sciences University, Portland, OR). pSG5-CBP was constructed by inserting a BamHI fragment of pRSV-CBP into the polylinker region of pSG5. pGRE2-E1b-CAT (termed pARE2-E1b-CAT in this report) was obtained from John A. Cidlowski (National Institute of Environmental Health Sciences, Research Triangle Park, NC). pARE4tk-LUC was made by inserting a duplicated C3(1) ARE into the SmaI site of pARE2-tk-LUC vector (30, 31). FLAG-tagged AR expression vector (pcDNA3.1-FLAG-rAR) was constructed as described (A-M. Moilanen, Institute of Biomedicine, University of Helsinki, Helsinki, Finland, personal communication). Expression vector encoding hRelA (pCMV-p65) and pκB6tk-LUC reporter were gifts from Patrick Baeuerle (Alberts-Ludwig Universität, Freiburg, Germany) (32). p75–1050CAT was provided by Mart Saarma (Institute of Biotechnology, University of Helsinki, Helsinki, Finland). [3H]Acetyl-CoA was obtained from New England Nuclear and the luciferase (LUC) assay reagent was from Promega. Affinity-purified rabbit polyclonal antibody against human CBP (A-22) was purchased from Santa Cruz Biotechnology. Mouse monoclonal antibody (M2) against the FLAG epitope was purchased from Kodak.
Cell Culture and Transfections.
CV-1 cells (American Type Culture Collection) were maintained in DMEM containing penicillin (25 units/ml), streptomycin (25 units/ml), and 10% fetal bovine serum (FBS). Transfections were performed by using the calcium phosphate precipitation method (29, 33). In short, 0.5 × 106 or 1.5 × 106 cells were plated on a 60-mm or a 10-cm dish, respectively, 24 h before adding the precipitate containing the amounts of expression and reporter vectors indicated. Eighteen hours after transfection, the cells received fresh medium containing 2% charcoal-stripped FBS with or without 25 nM testosterone or dexamethasone. Chloramphenicol acetyltransferase (CAT) and LUC activities were determined as described (34). For whole cell extracts, 5 × 106 COS-1 cells were transfected by electroporation with 10 μg of pcDNA 3.1-FLAG-rAR and 10 μg of pSG5-CBP as described (35). Statistical analyses were carried out with two-tailed Student’s t test.
Immunoprecipitation and Immunoblotting.
Whole cell extracts were prepared by using modified RIPA buffer containing 20 mM Tris⋅HCl (pH 7.8), 140 mM NaCl, 1 mM EDTA, 0.5% sodium deoxycholate, 0.5% Nonidet P-40, 1 mM phenylmethylsulfonyl fluoride, 5 mM dithiobis(succinimidylpropionate), and aprotinin (10 μg/ml). The extracts were first cleared with nonimmune rabbit serum and protein A-Sepharose and then incubated with mouse monoclonal anti-FLAG antibody for 1 h at 4°C, followed by an 1-h incubation with 15 μl of protein A-Sepharose. After four washes with 0.5 ml of modified RIPA buffer, the pellets were resuspended in electrophoresis sample buffer, boiled for 5 min, and analyzed on a SDS/7.5% polyacrylamide gel. Proteins were transferred to an Immobilon-P membrane and blotted with rabbit anti-CBP antibody (35). Immunoblotting of AR was performed with ARp3 antibody raised against a synthetic peptide corresponding to residues 14–32 of rAR (30).
RESULTS
CBP Stimulates AR-Mediated Transcription.
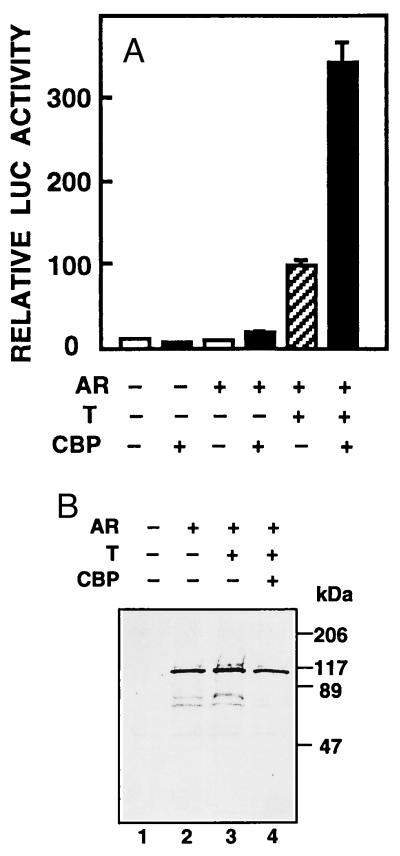
The ability of CBP to modulate AR-mediated transcription was studied in CV-1 cells, which do not express endogenous receptor protein. Coexpression of CBP with rAR increased the activity of a reporter gene driven by four AREs in front of the minimal thymidine kinase promoter (pARE4tk-LUC) by 3.5-fold over that with rAR alone in the presence of testosterone (P < 0.005; Fig. 1A). Overexpression of CBP also enhanced slightly transcriptional activity of AR in the absence of hormone. Basal promoter activity was not affected by CBP (Fig. 1A). Comparable results were obtained with other reporters regulated by AR, such as pARE2-E1b-CAT and the rat probasin gene proximal promoter (ref. 31 and unpublished results), indicating that the effect of CBP on AR function was not a feature specific for a given promoter/reporter construct. One possible mechanism for CBP-mediated enhancement of AR function is an increase in the amount of receptor protein. This was, however, not the case, as illustrated by immunoblotting experiments on extracts isolated from cells transfected with pSG5-rAR in the presence or absence of pSG5-CBP (Fig. 1B).
Figure 1.
CBP stimulates AR-mediated transactivation without altering the amount of receptor protein. (A) CV-1 cells (0.5 × 106 cells per 60-mm dish) were transfected with pARE4tk-LUC (1.5 μg), AR expression vector (pSG5-rAR, 0.3 μg), and CBP expression plasmid (pSG5-CBP, 3 μg). The total amount of DNA was kept constant by adding empty pSG5 DNA when appropriate. Eighteen hours after transfection, the cells received fresh medium with vehicle or 25 nM testosterone (T) as indicated. After a 30-h culture, the cells were harvested and LUC activity was determined. Reporter gene activities are expressed relative to that achieved with pSG5-rAR in the presence of testosterone, and data are the mean ± SEM of three experiments given as percentages. (B) COS-1 cells were transfected by electroporation with pSG5-rAR with or without pSG5-CBP. Control cells received empty pSG5 DNA. After transfection, the cells were cultured for 30 h in the presence or absence of 25 nM testosterone, after which soluble cell extracts were prepared and subjected for immunoblotting with ARp3 antibody (30).
CBP Interacts Physically with AR in Vivo.
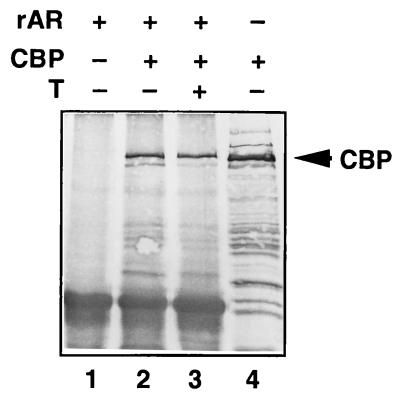
To study whether CBP interacts with AR in intact cells, COS-1 cells were transfected with CBP and FLAG-tagged AR expression vectors. Protein complexes associated with AR were first immunoprecipitated with anti-FLAG monoclonal antibody, and the precipitated proteins were then subjected to immunoblotting with anti-CBP antibody. A protein of >200 kDa was recognized in immunoprecipitates from cells transfected with both CBP and AR in the presence of testosterone but not from cells expressing CBP in the absence of AR (Fig. 2). The interaction between CBP and AR was not significantly altered by the presence of androgen, as judged by immunoprecipitation experiments (Fig. 2, lanes 2 and 3).
Figure 2.
AR interacts with CBP in transfected COS-1 cells. COS-1 cells were transfected by electroporation with pcDNA 3.1-FLAG-rAR and pSG5-CBP as indicated. After a 30-h culture in the presence or absence of 25 nM testosterone, whole cell extracts were prepared and subjected to immunoprecipitation with mouse monoclonal anti-FLAG antibody. Immunoprecipitated proteins were resolved by SDS/PAGE and subjected to immunoblotting with rabbit anti-CBP antibody. A portion of the cell extract (5%) was subjected to immunoblotting without prior immunoprecipitation with anti-FLAG antibody (lane 4).
Regions of AR Needed for Stimulation by CBP.
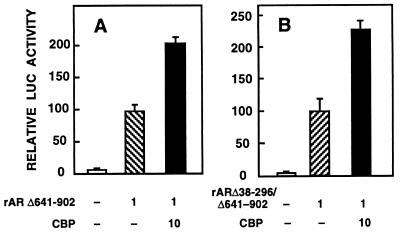
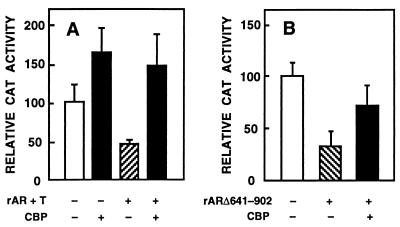
CBP has been shown to interact with LBDs of some nuclear receptors (24–28). To examine whether the same was true for AR, CV-1 cells were cotransfected with CBP and rAR mutants devoid of LBD (rARΔ641–902) or LBD and half of the N-terminal region (rARΔ38–296/Δ641–902). rARΔ641–902 and rARΔ38–296/Δ641–902 receptor forms exhibit a constitutive transactivation function that is comparable to that of the liganded wild-type rAR, when minimal promoters driven by ARE sequences are used (31). Coexpressed CBP enhanced the transactivation ability of the two mutant ARs by 2-fold with pARE4tk-LUC as the reporter (Fig. 3), as opposed to the 3.5-fold increase seen in the activity of wild-type rAR with the same reporter (Fig. 1A). CBP also stimulated rARΔ641–902-dependent transcriptional activation when pARE2-E1b-CAT was used as the reporter (31). Thus, the presence of LBD and residues 38–296 of the N-terminal region are not critical for AR–CBP interaction.
Figure 3.
Regions of AR involved in the interaction with CBP. CV-1 cells were transfected with 5 μg of pARE4tk-LUC, 1 μg of pSG5-rARΔ641–902 (A) or 1 μg of pSG5-rARΔ38–296/Δ641–902 (B), and 10 μg pSG5-CBP vectors as indicated. Reporter gene activities are expressed relative to that achieved with pSG5-rAR mutant alone, and data are the mean ± SEM of three experiments given as percentages.
AR-Dependent Transactivation Is Inhibited by 12S E1A.
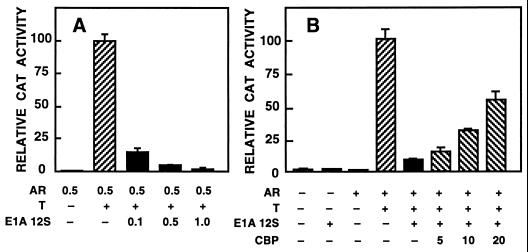
Adenoviral protein 12S E1A binds to and inactivates CBP (12, 13). Expression of exogenous 12S E1A should thus inhibit the activity of androgen-regulated reporters, if CBP were necessary for AR-dependent transactivation. To examine this, CV-1 cells were transfected with an AR expression vector, pARE2-E1b-CAT reporter, and increasing amounts of 12S E1A expression plasmid (Fig. 4A). Exogenous 12S E1A inhibited AR-dependent transactivation in a dose-dependent manner (P < 0.001). The 12S E1A did not influence basal reporter gene activity (Fig. 4B). The effect of 12S E1A on androgen-mediated transactivation was the same when another reporter (pARE4tk-LUC) was used (data not shown).
Figure 4.
AR-dependent transactivation is repressed by 12S E1A and the repression relieved by coexpressed CBP. (A) CV-1 cells (1.5 × 106 cells per 10-cm dish) were transfected with pARE2-E1b-CAT (5 μg), AR expression vector (pSG5-rAR, 1 μg), and the indicated amounts of 12S E1A expression vector. Eighteen hours after transfection, the cells received fresh medium containing 25 nM testosterone (T) and were harvested 30 h later to assay CAT activity. Reporter gene activities are expressed relative to that of pSG5-rAR plus testosterone. Data are the mean ± SEM of two experiments. (B) CV-1 cells were transfected with 5 μg of pARE2-E1b-CAT, 1 μg of pSG5-rAR, 0.1 μg of pRSV-E1A 12S, and the indicated amount of pSG5-CBP (in μg). For the relative CAT activity, the value from the pSG5-rAR plus testosterone result was set as 100. Data are the mean ± SEM of two experiments.
In addition to CBP and p300, 12S E1A associates with other proteins (36). To determine whether the inhibitory effect of 12S E1A on pARE2-E1b-CAT expression was indeed due to its interaction with CBP, CV-1 cells were transfected with an amount of 12S E1A that reduced AR-dependent transactivation to 15% of maximal and with increasing amounts of pSG5-CBP. Coexpression of CBP diminished the 12S E1A-mediated inhibition of AR function in a dose-dependent fashion, even though only a partial reversal was achieved with the amounts of CBP expression vector used (Fig. 4B).
AR and AP-1 Compete for CBP in Cultured Cells.
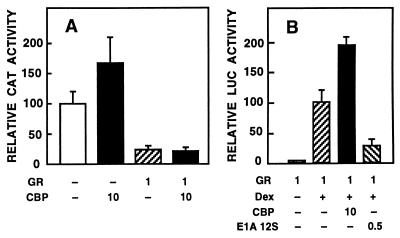
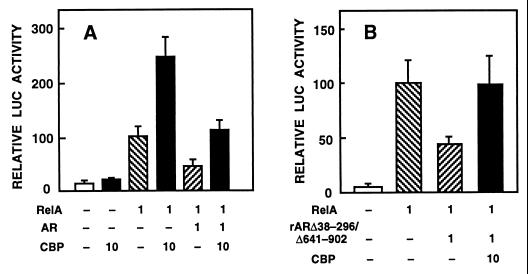
We have shown (37) by using extensively purified proteins that AR inhibits in vitro the binding of c-Jun to an AP-1 element without interacting itself with this motif. Because c-Jun interacts with CBP (14, 15) and competition for limiting amounts of intracellular CBP seems to be responsible for the repression of AP-1 activity by RAR and GR (24), it was of interest to examine whether CBP overcomes AR-mediated down-regulation of AP-1 activity. CV-1 cells were cotransfected with AR and CBP expression vectors and p75–1050CAT reporter. This reporter construct has in its vector backbone an AP-1 site whose activity is down-regulated by AR (37). The activity of p75–1050CAT was increased 1.5-fold by coexpression of CBP (Fig. 5A). Liganded AR repressed the reporter gene activity to one-half, and this repression was completely abrogated by a concomitant CBP expression. Under identical conditions, the AR mutant devoid of LBD (rARΔ641–902) down-regulated p75–1050CAT activity to one-third, and this repression was partially overcome by the presence of CBP (Fig. 5B). Thus, competition for available intracellular CBP is one of the mechanisms by which AR inhibits AP-1 function. Similar experiments performed with GR revealed that CBP could not rescue down-regulation of AP-1 activity by GR under the same conditions (Fig. 6A). GR-dependent transcriptional activation of a reporter gene (pARE4tk-LUC) was, however, stimulated by CBP and inhibited by 12S E1A (Fig. 6B) in a fashion similar to that determined for AR (compare Figs. 1 and 4A).
Figure 5.
Coexpression of CBP abrogates AR-mediated repression of AP-1 activity. (A) CV-1 cells were transfected with 5 μg of p75–1050CAT reporter, 1 μg of pSG5-rAR, and 10 μg of pSG5-CBP. Total amount of DNA was kept constant by adding empty pSG5 DNA when necessary. The cells were cultured in the presence of 25 nM testosterone (T) for 30 h. For the relative CAT activity, the value of p75–1050CAT alone was set as 100. Data are the mean ± SEM of four experiments. (B) CV-1 cells were transfected with 1 μg of pSG5-rARΔ641–902 instead of pSG5-rAR in experiments otherwise identical with those in A. Data are the mean ± SEM for two experiments.
Figure 6.
Coexpression of CBP does not relieve AP-1 repression brought about by GR but increases GR-mediated transcriptional activation. (A) CV-1 cells were transfected with 5 μg of p75–1050CAT reporter, 1 μg of pSG5-GR, and 10 μg of pSG5-CBP. The total amount of DNA was kept constant by adding pSG5 DNA when appropriate. The cells were cultured in the presence of 25 nM dexamethasone (DEX) for 30 h. For the relative CAT activity, the value of p75–1050CAT alone was set as 100. Data are the mean ± SEM values of three experiments. (B) CV-1 cells were transfected with 5 μg of pARE4-tk-LUC, 1 μg of pSG5-GR, 10 μg of pSG5-CBP, and 0.5 μg of pRSV-E1A 12S as indicated. The cells were treated and the reporter gene activity determined. For the relative LUC value, results of the pSG5-GR plus 25 nM dexamethasone experiment were set at 100. Data are the mean ± SEM of three experiments.
CBP Contributes to the Mutual Transcriptional Repression Between AR and NF-κB (RelA).
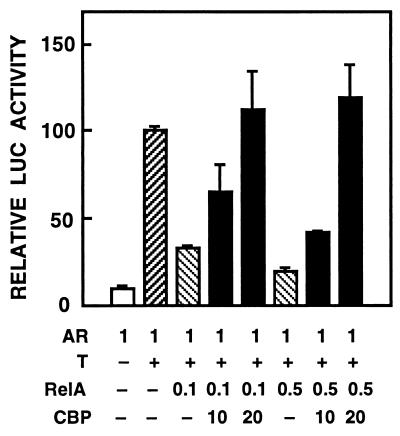
AR and RelA can inhibit each other’s transcriptional activity, and the mutual inhibition is not due to a strong physical interaction between the two proteins or their altered binding to cognate DNA elements (30). The mutual repression between AR and RelA could thus originate from competition for coregulators, such as CBP/p300. To test this possibility, CV-1 cells were cotransfected with AR, RelA, CBP expression vectors, and the pARE4tk-LUC reporter. Under these conditions, already low amounts of RelA inhibited transactivation by AR to one-third. The repression was abolished by cotransfected CBP (Fig. 7). The ability of CBP to abrogate inhibition of RelA activity by AR was next investigated. In CV-1 cells, expression of RelA elicited a 10-fold increase in pκB6tk-LUC activity, and this was enhanced 2.5-fold by coexpression of CBP (Fig. 8A). Liganded AR down-regulated RelA-induced reporter gene activity by more than 50%, and CBP coexpression rescued this repression (P < 0.05). CBP did not increase significantly basal reporter gene activity (Fig. 8A).
Figure 7.
CBP rescues RelA-mediated repression of AR function. CV-1 cells were transfected with pARE4tk-LUC reporter (5 μg) and indicated amounts (in μg) of pCMV-hAR, pCMV-RelA, and pSG5-CBP vectors in the absence or presence of 25 nM testosterone (T) as indicated. The cells were treated and harvested as described in Fig. 1. LUC activities are expressed relative to that of human AR plus testosterone, and data are the mean ± SEM of two experiments.
Figure 8.
CBP stimulates RelA-dependent transactivation and counteracts AR-mediated repression of RelA function. (A) CV-1 cells were transfected with pκB6tk-LUC reporter (5 μg), pCMV-hAR (1 μg), pCMV-RelA (1 μg), and pSG5-CBP (10 μg) expression vectors. For the relative LUC activity, RelA alone was set at 100. Testosterone (25 nM) was present in all cultures. Data are the mean ± SEM of four experiments are shown. (B) CV-1 cells were transfected with expression vectors for pCMV-RelA (1 μg), pSG5-rARΔ38–296/Δ641–902 (1 μg), and pSG5-CBP (10 μg) and with pκB6tk-LUC reporter. Data are the mean ± SEM of two experiments.
Our previous study (30) indicated that transrepression between rAR and RelA does not involve LBD or most of the N-terminal region of rAR but that the region between residue 297 and the DNA-binding domain of AR is mandatory (30). In view of this, we also tested an rAR mutant devoid of LBD and N-terminal residues 38–296 (rARΔ38–296/Δ641–902). The mutant AR form repressed RelA-induced pκB6tk-LUC expression to an extent similar to that of wild-type AR; this repression was relieved by coexpression of CBP (Fig. 8B). Thus, CBP is involved in the mutual transcriptional repression between AR and RelA. The results with rARΔ38–296/Δ641–902 also provided additional evidence for the notion that LBD of AR is not mandatory for the receptor’s interaction with CBP.
DISCUSSION
To regulate transcription, steroid receptors have to convey their activating or repressing signals to the basal transcription machinery. In addition to interacting directly with components of this apparatus, steroid receptors associate with various coactivators or corepressors that, in turn, may function as bridging factors to the basal transcription machinery. Several coregulators that activate steroid receptor function have been recently identified (38–41). With regard to negative transcriptional regulation by steroid receptors, the presence of so-called negative response elements has been postulated. However, it is not entirely clear whether these DNA motifs are true negative elements or just sequences overlapping with recognition sites for other sequence-specific proteins (42–46). During recent years, increasing body of evidence has emerged to indicate that steroid receptors can down-regulate expression of certain genes by interfering with the function of other transcription factors. Nuclear receptor interference with members of the AP-1 and NF-κB transcription factor families is well documented (47–49). This interference with heterologous transcription factors occurs either by direct protein–protein contacts or through competition for common coregulators.
In this study, we demonstrate that CBP is a coactivator for AR. In transfected cells, CBP activated expression of AR-dependent reporter genes without affecting basal promoter activity. In concert with this, the adenoviral protein 12S E1A that inactivates CBP function by preventing its association with RNA polymerase II complex and P/CAF histone acetyltransferase (12, 13, 50) was able to inhibit androgen-dependent transactivation. The fact that CBP and AR form a protein complex in vivo was demonstrated by their coimmunoprecipitation. Our results do not, however, distinguish whether AR–CBP interaction is direct or mediated by another yet unknown factor assembling a multiprotein complex with CBP and AR.
Previous studies have shown that CBP enhances transcriptional activity of several members of the nuclear receptor superfamily (24–28). In those experiments, CBP was found to interact with LBD of the receptors. This work shows, however, that CBP augments AR-mediated transactivation independent of the presence of LBD (mutant rARΔ641–902) or LBD and N-terminal residues 38–296 (mutant rARΔ38–296/Δ641–902). The two AR mutants devoid of LBD have an intact DNA binding domain and approximately one-half of the hinge region residues 607–659 (51). The LBD of AR was not mandatory for the receptor’s ability to compete for available intracellular CBP with members of the AP-1 and NF-κB transcription factor families. Even though these results do not necessarily rule out the possibility that LBD contacts CBP, they demonstrate nevertheless that LBD is not the only region of AR that interacts with CBP. Residues 38–296 that were also dispensable for AR–CBP interaction contain the AF-1 region of AR, which is mandatory for the transactivation ability of the native receptor (29) and possibly interacts with the general transcription factor TFIIF (7). We have previously shown that CBP is capable of facilitating the interaction between N and C termini of rAR (31), which may explain the result that CBP activated wild-type AR twice as much as LBD-deficient AR forms (compare Figs. 1 and 3). Thus, these findings indicate that the AR–CBP interaction is a complex event that may involve multiple interaction interfaces on both proteins.
Several nuclear receptors can repress the expression of genes regulated by AP-1 without binding to DNA (see ref. 47). For GR, RAR, and thyroid hormone receptor, the antagonism between the receptors and AP-1 is mutual (47). The situation with AR is less clear. With extensively purified proteins, we have shown (37) that full-length AR and AR DNA binding domain inhibit c-Jun–AP-1 element interaction under cell-free conditions, but c-Jun does not influence AR–ARE interaction (37). Although confirming our findings on the inhibition of c-Jun–AP-1 interaction by AR, Sato et al. (52) reported recently by using glutathione S-transferase fusion proteins that a large excess of c-Jun may also inhibit AR–ARE interaction in vitro. There seems to be an agreement that AP-1-dependent transactivation is down-regulated by hormone-occupied AR (37, 53); however, both potentiation and inhibition of AR-dependent transactivation by AP-1 family members have been reported (37, 52–54).
Coexpressed CBP was capable of rescuing down-regulation of p75–1050CAT activity brought about by ligand-occupied AR. Conversely, basal p75–1050CAT expression that is dependent on endogenous AP-1 activity was augmented by coexpressed CBP. Thus, in addition to inhibition of c-Jun–AP-1 element interaction (37), repression of AP-1 activity by AR involves competition for intracellular CBP. The two mechanisms need not to be mutually exclusive. Kamei et al. (24) have reported that the inhibitory action of both RAR and GR on AP-1 function could be abrogated by CBP. However, coexpression of CBP did not rescue AP-1 activity that was down-regulated by GR in our experiments, even though GR-mediated transactivation was stimulated by CBP. The reason for this discrepancy between our results and those of Kamei et al. (24) is not clear, but it may be related to the reporter constructs used and the distance of the AP-1 site from the proximal promoter elements which, for p75–1050CAT, is more than 1,000 nucleotides. Nevertheless, the molecular mechanisms governing the cross-talk of AR and GR with AP-1 were not identical under the experimental conditions used in this work.
We have demonstrated previously that AR and the RelA subunit of NF-κB can interfere with each other’s transcriptional activity (30). Similar observations have been reported for GR and estrogen receptor (55, 56). Already very low amounts of RelA inhibited AR-mediated transactivation and excess RelA was capable of rescuing the repression elicited by AR, whereas excess AR could not relieve the repression brought about by RelA (30). These results suggested that AR and RelA use a common coregulator, the affinity of which for RelA exceeds that for AR, and competition for this coregulator results in the mutual transcriptional repression. We show herein that CBP stimulates transcriptional activity of both AR and RelA and that CBP rescues the mutual transrepression. This implies that CBP is a coactivator present in limiting amounts in cells and subject to competition by AR and RelA. Indeed, while this work was in progress, it was reported that the transcriptional activation domain of RelA interacts with CBP/p300 (22, 23). In summary, CBP acts as an integrator between androgen-dependent and other nuclear signaling pathways and, therefore, regulation of CBP expression should be of great importance in determining transcriptional responses to androgens and other regulators of nuclear signaling.
Acknowledgments
We thank Drs. Patrik Baeuerle, Pierre Chambon, John A. Cidlowski, Richard H. Goodman, and Mart Saarma for plasmids. This work was supported by grants from the Medical Research Council (Academy of Finland), the Finnish Foundation for Cancer Research, the Jalmari and Rauha Ahokas Foundation, the Finnish Medical Society Duodecim, and the University of Helsinki.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: AR, androgen receptor; ARE, androgen responsive element; CAT, chloramphenicol acetyltransferase; CBP, CREB-binding protein; GR, glucocorticoid receptor; LBD, ligand binding domain; LUC, luciferase; RAR, retinoic acid receptor; r, rat.
References
- 1.Evans R M. Science. 1988;240:889–895. doi: 10.1126/science.3283939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Truss M, Beato M. Endocr Rev. 1993;14:459–479. doi: 10.1210/edrv-14-4-459. [DOI] [PubMed] [Google Scholar]
- 3.Tjian R, Maniatis T. Cell. 1994;77:5–8. doi: 10.1016/0092-8674(94)90227-5. [DOI] [PubMed] [Google Scholar]
- 4.Ing N H, Beekman J M, Tsai S Y, Tsai M-J, O’Malley B W. J Biol Chem. 1992;267:17617–17623. [PubMed] [Google Scholar]
- 5.Baniahmad A, Ha I, Reinberg D, Tsai S, Tsai M-J, O’Malley B W. Proc Natl Acad Sci USA. 1993;90:8832–8836. doi: 10.1073/pnas.90.19.8832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sadovsky Y, Webb P, Lopez G, Baxter J D, Fitzpatrick P M, Gizang-Ginsberg E, Cavailles V, Parker M G, Kushner P J. Mol Cell Biol. 1995;15:1554–1563. doi: 10.1128/mcb.15.3.1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McEwan I J, Gustafsson J-Å. Proc Natl Acad Sci USA. 1997;94:8485–8490. doi: 10.1073/pnas.94.16.8485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Horwitz K B, Jackson T A, Bain D L, Richer J K, Takimoto G S, Tung L. Mol Endocrinol. 1996;10:1167–1177. doi: 10.1210/mend.10.10.9121485. [DOI] [PubMed] [Google Scholar]
- 9.Chivira J C, Kwok R P S, Lamb N, Hagiwara M, Montminy M R, Goodman R H. Nature (London) 1993;365:855–859. doi: 10.1038/365855a0. [DOI] [PubMed] [Google Scholar]
- 10.Kwok R P, Lundblad J R, Chrivia J C, Richards J P, Bachinger H P, Brennan R G, Roberts S G, Green M R, Goodman R H. Nature (London) 1994;370:223–226. doi: 10.1038/370223a0. [DOI] [PubMed] [Google Scholar]
- 11.Kee B L, Arias J, Montminy M R. J Biol Chem. 1996;271:2373–2375. doi: 10.1074/jbc.271.5.2373. [DOI] [PubMed] [Google Scholar]
- 12.Ogryzko V V, Schiltz R L, Russanova V, Howard B H, Nakatani Y. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 13.Yang X-J, Ogryzko V V, Nishikawa J, Howard B H, Nakatani Y. Nature (London) 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 14.Bannister A J, Oehler T, Wilhelm D, Angel P, Kouzarides T. Oncogene. 1995;11:2509–2514. [PubMed] [Google Scholar]
- 15.Arias J, Alberts A S, Brindle P, Claret F X, Smeal T, Karin M, Feramisco J, Montminy M. Nature (London) 1994;370:226–229. doi: 10.1038/370226a0. [DOI] [PubMed] [Google Scholar]
- 16.Bannister A J, Kouzarides T. EMBO J. 1995;14:4758–4762. doi: 10.1002/j.1460-2075.1995.tb00157.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dai P, Akimaru H, Tanaka Y, Hou D-X, Yasukawa T, Kanei-Ishii C, Takahashi T, Ishii S. Genes Dev. 1996;10:528–540. doi: 10.1101/gad.10.5.528. [DOI] [PubMed] [Google Scholar]
- 18.Oelgeschläger M, Janknecht R, Krieg J, Schreek S, Lüscher B. EMBO J. 1996;15:2771–2780. [PMC free article] [PubMed] [Google Scholar]
- 19.Janknecht R, Nordheim A. Oncogene. 1996;12:1961–1969. [PubMed] [Google Scholar]
- 20.Janknecht R, Nordheim A. Biochem Biophys Res Commun. 1996;228:831–837. doi: 10.1006/bbrc.1996.1740. [DOI] [PubMed] [Google Scholar]
- 21.Oliner J D, Andresen J M, Hansen S K, Zhou S, Tjian R. Genes Dev. 1996;10:2903–2911. doi: 10.1101/gad.10.22.2903. [DOI] [PubMed] [Google Scholar]
- 22.Perkins N D, Felzien L K, Betts J C, Leung K, Beach D H, Nabel G J. Science. 1997;275:523–527. doi: 10.1126/science.275.5299.523. [DOI] [PubMed] [Google Scholar]
- 23.Gerritsen M E, Williams A J, Neish A S, Moore S, Shi Y, Collins T. Proc Natl Acad Sci USA. 1997;94:2927–2932. doi: 10.1073/pnas.94.7.2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kamei Y, Xu L, Heinzel T, Torchia J, Kurokawa R, Gloss B, Lin S-C, Heyman R A, Rose D W, Glass C K, et al. Cell. 1996;85:403–414. doi: 10.1016/s0092-8674(00)81118-6. [DOI] [PubMed] [Google Scholar]
- 25.Chakravarti D, LaMorte V J, Nelson M C, Nakajima T, Schulman I G, Juguilon H, Montminy M, Evans R M. Nature (London) 1996;383:99–103. doi: 10.1038/383099a0. [DOI] [PubMed] [Google Scholar]
- 26.Smith C L, Oñate S A, Tsai M-J, O’Malley B W. Proc Natl Acad Sci USA. 1996;93:8884–8888. doi: 10.1073/pnas.93.17.8884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yao T-S, Ku G, Zhou N, Scully R, Livingston D M. Proc Natl Acad Sci USA. 1996;93:10626–10631. doi: 10.1073/pnas.93.20.10626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hanstein B, Eckner R, DiRenzo J, Halachmi S, Liu H, Searcy B, Kurokawa R, Brown M. Proc Natl Acad Sci USA. 1996;93:11540–11545. doi: 10.1073/pnas.93.21.11540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Palvimo J J, Kallio P J, Ikonen T, Mehto M, Jänne O A. Mol Endocrinol. 1993;7:1399–1407. doi: 10.1210/mend.7.11.8114755. [DOI] [PubMed] [Google Scholar]
- 30.Palvimo J J, Reinikainen P, Ikonen T, Kallio P J, Moilanen A, Jänne O A. J Biol Chem. 1996;271:24151–24156. doi: 10.1074/jbc.271.39.24151. [DOI] [PubMed] [Google Scholar]
- 31.Ikonen T, Palvimo J J, Jänne O A. J Biol Chem. 1997;272:29821–29828. doi: 10.1074/jbc.272.47.29821. [DOI] [PubMed] [Google Scholar]
- 32.Schmitz M L, Baeuerle P A. EMBO J. 1991;10:3805–3817. doi: 10.1002/j.1460-2075.1991.tb04950.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ikonen T, Palvimo J J, Kallio P J, Reinikainen P, Jänne O A. Endocrinology. 1994;135:1359–1366. doi: 10.1210/endo.135.4.7925097. [DOI] [PubMed] [Google Scholar]
- 34.Palvimo J J, Partanen M, Jänne O A. Biochem J. 1996;315:993–998. doi: 10.1042/bj3160993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reinikainen P, Palvimo J J, Jänne O A. Endocrinology. 1996;137:4351–4357. doi: 10.1210/endo.137.10.8828495. [DOI] [PubMed] [Google Scholar]
- 36.Akusjärvi G. Trends Microbiol. 1993;1:163–170. doi: 10.1016/0966-842x(93)90085-6. [DOI] [PubMed] [Google Scholar]
- 37.Kallio P J, Poukka H, Moilanen A, Jänne O A, Palvimo J J. Mol Endocrinol. 1995;9:1017–1028. doi: 10.1210/mend.9.8.7476976. [DOI] [PubMed] [Google Scholar]
- 38.Cavailles N, Dauvois S, Danielian P S, Parker M G. Proc Natl Acad Sci USA. 1994;91:10009–100013. doi: 10.1073/pnas.91.21.10009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oñate S A, Tsai S Y, Tsai M-J, O’Malley B W. Science. 1995;270:1354–1357. doi: 10.1126/science.270.5240.1354. [DOI] [PubMed] [Google Scholar]
- 40.Lee J W, Ryan F, Swaffield J C, Johnston S A, Moore D D. Nature (London) 1995;374:91–94. doi: 10.1038/374091a0. [DOI] [PubMed] [Google Scholar]
- 41.Voegel J J, Heine M J S, Zechel C, Chambon P, Gronemeyer H. EMBO J. 1996;15:3667–3675. [PMC free article] [PubMed] [Google Scholar]
- 42.Sakai D D, Helms S, Carlstedt-Duke J, Gustafsson J-Å, Rottman F M, Yamamoto K R. Genes Dev. 1988;2:1144–1154. doi: 10.1101/gad.2.9.1144. [DOI] [PubMed] [Google Scholar]
- 43.Drouin J, Trifiro M A, Plante R K, Nemer M, Eriksson P, Wrange Ö. Mol Cell Biol. 1989;9:1266–1272. doi: 10.1128/mcb.9.12.5305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Drouin J, Sun Y L, Chamberland M, Gauthier Y, De Léan A, Nemer M, Schmidt T J. EMBO J. 1993;12:145–156. doi: 10.1002/j.1460-2075.1993.tb05640.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cairns C, Cairns W, Okret S. DNA Cell Biol. 1993;12:695–702. doi: 10.1089/dna.1993.12.695. [DOI] [PubMed] [Google Scholar]
- 46.Heckert L L, Wilson E M, Nilson J H. Mol Endocrinol. 1993;11:1497–1506. doi: 10.1210/mend.11.10.9996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pfahl M. Endocr Rev. 1993;14:651–658. doi: 10.1210/edrv-14-5-651. [DOI] [PubMed] [Google Scholar]
- 48.Cato A B C, Wade E. BioEssays. 1996;18:371–378. doi: 10.1002/bies.950180507. [DOI] [PubMed] [Google Scholar]
- 49.McEwan I J, Wright A P H, Gustafsson J-Å. BioEssays. 1997;19:153–160. doi: 10.1002/bies.950190210. [DOI] [PubMed] [Google Scholar]
- 50.Nakajima T, Uchida C, Anderson S F, Parvin J D, Montminy M. Genes Dev. 1997;11:738–747. doi: 10.1101/gad.11.6.738. [DOI] [PubMed] [Google Scholar]
- 51.Tan J A, Joseph D R, Quarmby V E, Lubahn D B, Sar M, French F S, Wilson E M. Mol Endocrinol. 1988;2:1276–1285. doi: 10.1210/mend-2-12-1276. [DOI] [PubMed] [Google Scholar]
- 52.Sato N, Sadar M D, Bruchovsky N, Saatcioglu F, Rennie P S, Sato S, Lange P H, Gleave M E. J Biol Chem. 1997;272:17485–17494. doi: 10.1074/jbc.272.28.17485. [DOI] [PubMed] [Google Scholar]
- 53.Shemshedini L, Knauthe R, Sassone-Corsi P, Pornon A, Gronemeyer H. EMBO J. 1992;10:3839–3849. doi: 10.1002/j.1460-2075.1991.tb04953.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bubukya A, Wise S C, Shen X-Q, Burmeister L A, Shemshedini L. J Biol Chem. 1996;271:24583–24589. doi: 10.1074/jbc.271.40.24583. [DOI] [PubMed] [Google Scholar]
- 55.Ray A, Prefontaine K E. Proc Natl Acad Sci USA. 1994;91:752–756. doi: 10.1073/pnas.91.2.752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stein B, Yang M X. Mol Cell Biol. 1995;15:4971–4979. doi: 10.1128/mcb.15.9.4971. [DOI] [PMC free article] [PubMed] [Google Scholar]