SUMMARY
The synthesis of ribosomes in eukaryotic cells is a complex process involving many nonribosomal protein factors and snoRNAs. In general, the processes of rRNA transcription and ribosome assembly are treated as temporally or spatially distinct. Here, we describe the identification of a point mutation in the second largest subunit of RNA polymerase I near the active center of the enzyme that results in an elongation-defective enzyme in the yeast Saccharomyces cerevisiae. In vivo, this mutant shows significant defects in rRNA processing and ribosome assembly. Taken together, these data suggest that transcription of rRNA by RNA polymerase I is linked to rRNA processing and maturation. Thus, RNA polymerase I, elongation factors, and rRNA sequence elements appear to function together to optimize transcription elongation, coordinating cotranscriptional interactions of many factors/snoRNAs with pre-rRNA for correct rRNA processing and ribosome assembly.
INTRODUCTION
Biosynthesis of ribosomes in eukaryotic cells is extremely complex, as revealed from extensive studies using the yeast Saccharomyces cerevisiae (called “yeast” in this paper). Tandemly repeated chromosomal rRNA genes are first transcribed by RNA polymerase (Pol) I, to generate 35S pre-rRNA that undergoes a series of endo- and exonucleolytic cleavage steps, producing the mature 18S, 5.8S, and 25S rRNAs (Figure 1). Over 170 nonribosomal proteins including 18 known or presumptive RNA helicases and 70 small nucleolar RNAs (snoRNAs) participate in ribosome assembly in yeast (reviewed in Venema and Tollervey [1999], Tschochner and Hurt [2003], Fromont-Racine et al. [2003], and Granneman and Baserga [2005]).
Figure 1. Structure of 35S Pre-rRNA and the rRNA Processing Intermediates in the Major rRNA Processing Pathways in S. cerevisiae.
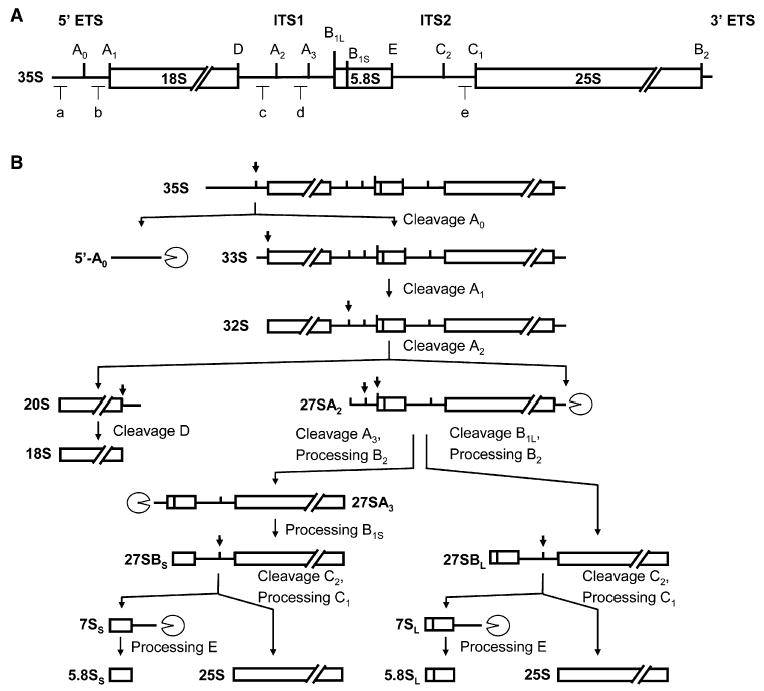
(A) Positions of two internal transcribed spacers (ITS1 and ITS2) and 5′ and 3′ external transcribed spacers (ETS) are shown. Pre-rRNA cleavage sites are indicated by upper-case letters. Lower-case letters indicate the location of oligonucleotide probes used for northern hybridization and primer extension.
(B) The major pre-rRNA processing pathways adapted from Venema and Tollervey (1999). The 27SA2 pre-rRNA is processed by two alternative pathways, giving rise to two forms of 5.8S rRNA, the major short form 5.8Ss, and a minor long form 5.8SL. Exonuclease activities are indicated by circles with cutouts.
For proper assembly and maturation of a ribosome, ~100 different positions in the rRNA must be accurately modified by snoRNA-protein complexes (snoRNPs; reviewed in Decatur and Fournier [2003] and Bertrand and Fournier [2004]), 78 ribosomal proteins (r-proteins) must properly associate with the rRNA, and the rRNAs themselves must adopt precise, intricate structures. One way to coordinate these complex reactions would be to couple transcription elongation with rRNA processing, that is, to use Pol I elongation rate (perhaps including specific pausing and release of pausing events) to regulate rRNA folding/maturation, similar to checkpoint functions studied in connection with mRNA capping and splicing in Pol II transcription systems (reviewed in Zorio and Bentley [2004] and Aguilera [2005]).
Recent studies using EM visualization of nascent rRNA transcripts demonstrated a cleavage at ITS1 that takes place cotranscriptionally in more than half (~55%) of the transcripts in exponentially growing yeast cells (Osheim et al., 2004). In addition, small 5′-terminal knobs seen on the transcripts by EM and known to contain U3 snoRNA and associated proteins (Mougey et al., 1993; Dragon et al., 2002) were found to become larger knobs cotranscriptionally before the cleavage at ITS1, indicating that rRNA folding, processing/modification, and r-protein addition might also take place cotranscriptionally (Osheim et al., 2004). Thus, processing of rRNA in yeast appears to begin during Pol I transcription, setting up a scenario in which changes in Pol I transcription elongation could affect rRNA processing and vice versa, i.e., these two processes previously thought to be temporally distinct might be functionally coupled to one another.
We have recently demonstrated that Pol II elongation factors Spt4 and Spt5 participate in Pol I transcription elongation, and that deletion of the SPT4 gene causes clear defects in rRNA processing even at permissive temperatures (Schneider et al., 2006). The yeast transcription factor consisting of Spt4 and Spt5 (Spt4/5 complex; the mammalian homolog is called DSIF) is known to be involved in the checkpoint function that ensures recruitment of mRNA capping enzymes to a Pol II elongation complex. Together with other factors, Spt4/5 pauses Pol II proximal to the promoter until the mRNA is properly capped, at which point Spt5 is modified and the pause is released (for reviews, see Zorio and Bentley [2004] and Aguilera [2005]). Thus, our previous observations are consistent with the conclusion that the Spt4/5 complex plays roles in coupling rRNA transcription to rRNA processing and ribosome assembly, similar to its role with Pol II. However, since Spt4 and Spt5 are involved in Pol II transcription, it is difficult to differentiate direct from indirect effects of the spt4 deletion on rRNA processing. To test whether Pol I transcription is functionally coupled to rRNA processing/maturation, we isolated mutants of Pol I that impaired elongation and looked for defects in rRNA processing and ribosome assembly. In this paper, we describe isolation of such mutants and demonstrate direct evidence for functional coupling between Pol I transcription and rRNA processing/ribosome assembly.
RESULTS
Isolation of an rpa135 Mutant with Defects in Transcription Elongation
It was previously shown that 6-azauracil (6AU) inhibits growth of yeast cells by reducing cellular UTP and GTP levels (Exinger and LaCroute, 1992), and that decreased nucleotide concentrations increase RNA polymerase pausing and elongation arrest in vitro (Uptain et al., 1997). 6AU-hypersensitive (6AUS) yeast mutants with alterations in Pol II subunits were also isolated that show defects in Pol II elongation in vivo (Archambault et al., 1992; Langelier et al., 2005). To isolate elongation-defective mutations in Pol I, we introduced PCR-mutagenized copies of RPA135 (gene for second largest Pol I subunit) to a yeast strain carrying an rpa135Δ mutation (NOY975), which grows by virtue of Pol II transcription of the 35S rRNA coding region fused to the GAL7 promoter (“GAL7-35S rDNA”) on a plasmid (see Table 1). Several independent 6AUS mutants (as judged by their growth on glucose with and without 6AU; cf. Figure 2A) were isolated. One of these mutants contained a missense mutation resulting in a glycine at amino acid position 784 rather than an aspartate. The acidic residue at this position is conserved among most or all known multisubunit RNA polymerases, including yeast Pol I, yeast Pol II, human Pol II, and E. coli RNA polymerase (Figure 2B). In Pol II this residue is located close to the catalytic center and might play a role in loading substrate NTP at the active site during Pol II transcription (Cramer et al., 2001; Langelier et al., 2005).
Table 1.
Yeast Strains and Plasmids Used in This Study
| Strain or Plasmid | Description |
|---|---|
| Strains | |
| NOY388 | MATa ade2-1 ura3-1 trp1-1 leu2-3,112 his3-11 can1-100 |
| NOY975 | MATa ade2-1 ura3-1 trp1-1 leu2-3,112 his3-11 can1-100 rpa135Δ::LEU2 carrying pNOY199 |
| NOY1075 | MATa ade2-1 ura3-1 trp1-1 leu2-3,112 his3-11 can1-100 rrn3(S213P) (see Claypool et al. [2004]) |
| NOY2169 | MATa ade2-1 ura3-1 trp1-1 leu2-3,112 his3-11 can1-100 rpa135Δ::LEU2 carrying pNOY742 |
| NOY2171 | MATa ade2-1 ura3-1 trp1-1 leu2-3,112 his3-11 can1-100 rpa135Δ::LEU2 carrying pNOY743 |
| NOY2172 | MATa ade2-1 ura3-1 trp1-1 leu2-3,112 his3-11 can1-100 rpa135(D784G) |
| NOY2173 | MATa ade2-1 ura3-1 trp1-1 leu2-3,112 his3-11 can1-100 rpa135Δ::LEU2 carrying pNOY744 |
| NOY2174 | MATa ade2-1 ura3-1 trp1-1 leu2-3,112 his3-11 can1-100 rpa135Δ::LEU2 carrying pNOY745 |
| NOY2175 | MATa ade2-1 ura3-1 trp1-1 leu2-3,112 his3-11 can1-100 rrp6Δ::HIS3 |
| NOY2176 | MATa ade2-1 ura3-1 trp1-1 leu2-3,112 his3-11 can1-100 rpa135(Δ784G) rrp6Δ::HIS3 |
| Plasmids | |
| pNOY199 | High copy number plasmid carrying GAL7-35S rDNA, 2 mm, TRP1 |
| pRS316 | pBluescript, CEN6, ARSH4, URA3 (see Sikorski and Hieter [1989]) |
| pNOY742 | pRS316 (pBluescript, CEN6, ARSH4, URA3) derivative containing RPA135 |
| pNOY743 | pRS316 (pBluescript, CEN6, ARSH4, URA3) derivative containing rpa135(D784G) |
| pNOY744 | pRS316 (pBluescript, CEN6, ARSH4, URA3) derivative containing His6-(HA)3-RPA135 |
| pNOY745 | pRS316 (pBluescript, CEN6, ARSH4, URA3) derivative containing His6-(HA)3-rpa135(D784G) |
| pNOY746 | Derivative of high copy number plasmid pNOY373 (Wai et al., 2000) containing rDNA with promoter, but with C mutated to G between +1 and +56 relative to transcription start site |
Figure 2. Mutation of Aspartate 784 to Glycine in A135 Impairs Pol I Elongation Rate.
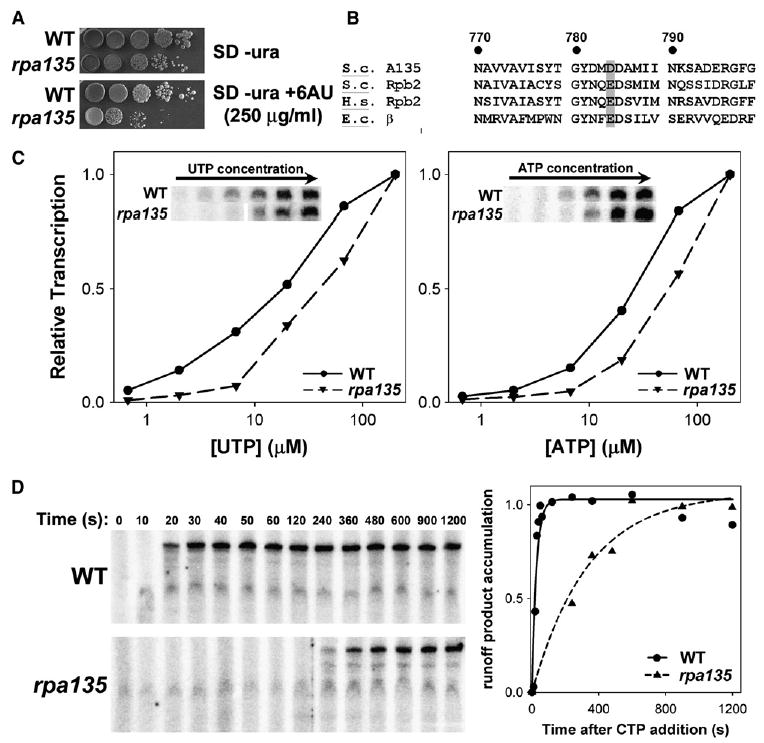
(A) Ten-fold dilutions of yeast cells with WT A135 (NOY388) and rpa135(D784G) (NOY2172) carrying pRS316 were plated on SD −Ura medium and SD −Ura +250 μg/ml 6AU.
(B) Protein sequences flanking D784 of S. cerevisiae (S.c.) A135 aligned with S.c. Rpb2, Homo sapiens (H.s.) Rpb2, and Escherichia coli (E.c.) β. Numbering is relative to S.c. A135, and conserved acidic residues equivalent to D784 are shaded in gray.
(C) Multiround in vitro transcription assays varying the concentration of UTP (left) or ATP (right) using purified WT or rpa135(D784G) Pol I. Runoff product accumulation was normalized to the value at the highest NTP concentration (the values were as follows: for UTP, WT 6983 and rpa135 6893; for ATP, WT 5328 and rpa135 8254 in arbitrary units) and plotted versus the varied NTP concentration. Raw data are inset in each plot.
(D) Elongation rate assay measuring runoff product (763 nt) accumulation versus time after release of elongation complexes arrested at +56 (relative to transcription start site) using WT and rpa135(D784G) mutant Pol I. Raw data are shown on the left, with quantification of the amount of runoff product normalized to maximum and plotted versus time on the right.
In Vitro Analyses of Pol I Carrying the rpa135(D784G) Mutation
We constructed strains expressing His6-(HA)3-tagged versions of WT (NOY2173) and mutant (NOY2174) A135. We purified Pol I from these strains and observed no difference in Pol I subunit composition or stability (as assessed by SDS-PAGE, data not shown).
Using a standard multiround Pol I transcription assay system with all purified initiation factors (Keener et al., 1998), we found that when one of four NTP substrates was reduced (UTP and ATP analyzed), transcription by the mutant Pol I was reduced relative to the WT Pol I, and that the difference in the activities between the two polymerases was less at higher NTP concentrations (Figure 2C). This observation is consistent with the selected phenotype of the mutant, i.e., 6AU sensitivity. However, since these multiround assays do not differentiate defects in elongation from other kinetic steps (i.e., initiation, pausing, arrest, or termination), we developed an assay for Pol I elongation rate.
We constructed a linear rDNA template (−247 to +763) in which all 6 CG base pairs encoding C between +1 to +56 are replaced by GC base pairs (eliminating CTP requirement for transcription up to +56) to compare elongation rates in vitro. Pol I was first incubated with the template in the presence of purified factors, UAF, CF, TBP, and Rrn3, followed by the addition of ATP, UTP, GTP, and [α-32P]GTP but without CTP. Then heparin was added to prevent reinitiation of transcription. Finally, CTP was added to allow the paused Pol I (presumed to be at +56, which precedes the first base pair encoding C) to continue elongation. Accumulation of the full-length runoff product (763 nt) was then monitored over time (Figure 2D). From these data we calculated that the WT elongation rate at room temperature (~20°C) was ~30 nt/s, whereas the mutant enzyme was ~10-fold slower. These data clearly demonstrate that the rpa135(D784G) mutation impaired transcription elongation compared to WT. (For technical reasons, we were not able to clearly observe the stalled +56 complex on our gel; however, because equal amounts of WT and mutant Pol I yield approximately equal amounts of product, the mutant Pol I does not arrest or terminate prematurely.)
Defects in Ribosome Assembly Caused by the rpa135(D784G) Mutation
To measure effects of the rpa135 mutation in vivo, the mutation was introduced into the chromosomal RPA135 gene in our standard strain (NOY388; “WT”), yielding the mutant strain NOY2172. Both the mutant and the WT strains were grown exponentially in synthetic glucose complete (SD) medium at 30°C. The growth rate of the mutant was about 55% of the growth rate of the WT strain. In sucrose gradient sedimentation analysis, the mutant extract showed a significant decrease in the amount of free 60S relative to free 40S subunit and an appearance of “halfmers” which represent 80S monosomes or polysomes containing an additional 40S subunit (Helser et al., 1981; Figure 3A). This pattern suggested a preferential reduction in 60S relative to 40S subunits (Rotenberg et al., 1988). To confirm that this anomalous ribosome profile was not a consequence of a general decrease in Pol I transcription rate, we examined the ribosome profile of a strain in which Pol I transcription initiation was specifically impaired by mutation of the transcription initiation factor Rrn3 (rrn3[S213P]; Claypool et al., 2004). When grown at the semipermissive temperature (30°C), this strain grew slightly slower than the rpa135 strain; however, the ribosome profile appeared normal (Figure 3A). Thus, only when Pol I transcription elongation is impaired do we observe defects in ribosome assembly.
Figure 3. rRNA Accumulation and Ribosome Assembly Are Impaired in the rpa135(D784G) Mutant Compared to WT.
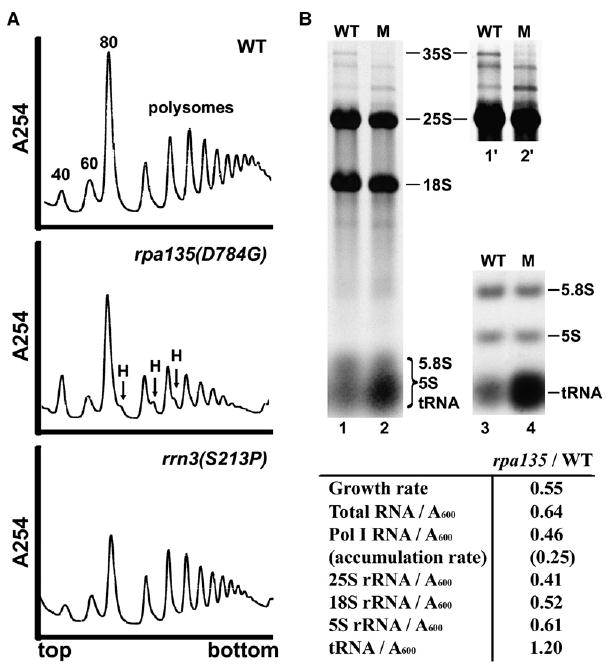
(A) Sucrose gradient analysis of ribosome profiles in WT (NOY388), rpa135(D784G) (NOY2172), and rrn3(S213P) (NOY1075). Cells were grown in YEPD at 30°C, and extracts containing approximately equal amounts of RNA were analyzed by sucrose gradient centrifugation. Positions of 40S, 60S, 80S, polysomes, and halfmers (H) are indicated.
(B) RNA was purified from WT and rpa135(D784G) (lanes labeled “M”) cells that were grown in SD −Ura containing [14C]uracil (1 μCi/ml; 5 μg/ml). RNA containing equal amounts of 14C radioactivity was analyzed on formaldehyde-agarose gels (lanes 1 and 2) or agarose:acrylamide composite gels (lanes 3 and 4; only a lower part shown). Lanes 1′ and 2′ are 3-fold longer exposures of the upper part of lanes 1 and 2, for visualization of low abundance RNAs. The locations of various RNA species are indicated. RNA species were excised from the gels after autoradiography, and amounts of individual species were quantified. The amounts of individual RNA species in the mutant cells were normalized to WT and are summarized below.
To quantify effects of the rpa135 mutation on individual RNA species, we grew cells in SD −Ura medium containing [14C]uracil for many generations and measured total RNA (cold-TCA-precipitable 14C-labeled RNA) as well as individual species excised after gel electrophoresis. The results are summarized in Figure 3B. Calculation showed that even though cell growth rate in the mutant was significantly reduced due to a strong (~4.0-fold) decrease in the rate of production of mature rRNA species by Pol I, no decrease in the amount of tRNA (per cell mass) was observed. In contrast to tRNA, the amount of Pol III-derived 5S rRNA in the rpa135 mutant was significantly lower than in WT cells (per cell mass), similar to the levels of Pol I-derived rRNA species. It is likely that 5S rRNA is synthesized normally by Pol III in the rpa135 mutant, but because fewer functional ribosomes are assembled due to impaired Pol I elongation, excess 5S rRNA is degraded.
We also observed a small but reproducible decrease (~20%) in the amount of 25S rRNA relative to 18S rRNA in the mutant strain compared to WT (Figure 3B). This observation is consistent with the ribosome profiles in Figure 3A. Finally, there was a reduction in the amount of 35S pre-rRNA in the mutant cells compared to WT, consistent with experiments described below. All of these data support the conclusion that impairment of Pol I elongation affects ribosome assembly.
Defects in rRNA Processing Caused by the rpa135(D784G) Mutation
To examine defects in specific rRNA processing steps, we analyzed the abundance of various pre-rRNA species in the WT and rpa135 cells by northern blot and primer extension. Because it has been shown previously that polyadenylated forms of normal and aberrant rRNA processing intermediates accumulate when the function of the nuclear exosome is disrupted (Kuai et al., 2004; Fang et al., 2004; Houseley et al., 2006), we included two additional strains in these studies. One strain contained a deletion of the gene for the nuclear-specific exosome subunit Rrp6 in our WT strain background (NOY2175). The other strain was a double mutant: rrp6 rpa135 (NOY2176).
We observed abnormal processing of 35S pre-rRNA leading to mature 18S, 5.8S, and 25S rRNAs in the rpa135 mutants. First, there was a large reduction in the amount of the 35S pre-rRNA in the two strains carrying the rpa135 mutation compared to control strains (Figure 4A, compare even-numbered lanes with odd-numbered lanes). This result is consistent with the results obtained by direct analysis of cellular RNAs fully labeled with [14C]uracil mentioned above (see Figure 3B).
Figure 4. The rpa135(D784G) Mutation Causes Defects in rRNA Processing and Accumulation of Aberrant Pre-rRNA Species.
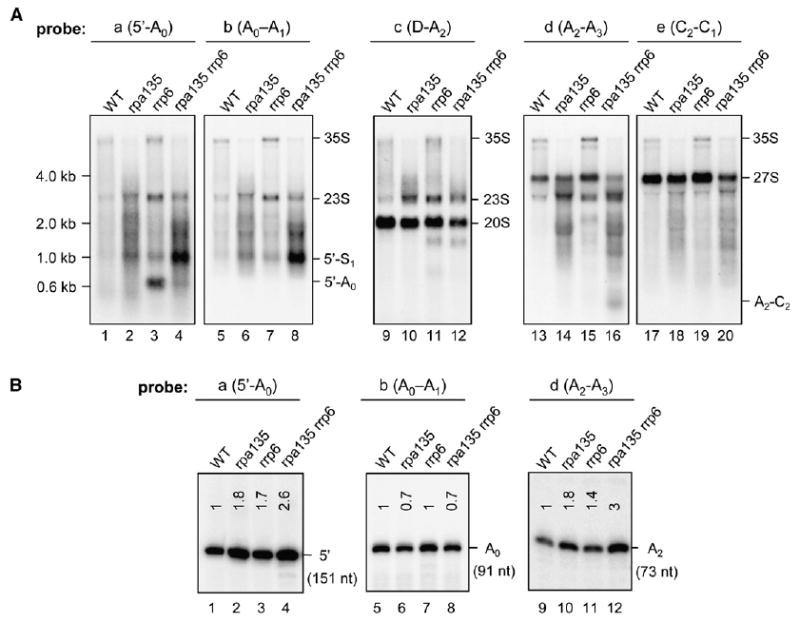
(A) Total RNA was prepared from strains WT (NOY388), rpa135 (NOY2172), rrp6 (NOY2175), and rpa135 rrp6 (NOY2176) and analyzed by northern hybridization with oligonucleotide probes indicated on the top of each panel (see Figure 1A for locations). Positions of size markers are shown on the left.
(B) The amounts of RNA with the 5′ end at +1 (5′), A0, and A2 in the preparations used for northern hybridization were analyzed by primer extension using primers “a,” “b,” and “d,” respectively. Length of the extended products is shown in parentheses. The amount of extended products in the mutant strains relative to WT was quantified by phosphorimager analysis. The average values normalized to WT calculated from four experiments are indicated in each lane.
Processing in the 5′ ETS region of 35S pre-rRNA is abnormal in the rpa135 strains. Normally, cleavage at A0 releases an ~0.6 kb fragment that is degraded by the TRAMP [Trf4/Air2/Mtr4 polyadenylation complex]-exosome complex (de la Cruz et al., 1998; Allmang et al., 2000; Zanchin and Goldfarb, 1999; for TRAMP, see Kadaba et al. [2004], LaCava et al. [2005], Vanacova et al. [2005], Wyers et al. [2005], and Houseley et al. [2006]). We observed accumulation of this 5′-A0 fragment in the rrp6 strain, consistent with this model (Figure 4A, lane 3). However, this fragment is diminished in the rrp6 rpa135 double mutant (Figure 4A, lane 4) demonstrating that impaired Pol I elongation resulted in either a large decrease in normal cleavage at A0 or additional abnormal cleavage events. These data are supported by primer extension analyses which showed that rpa135 cells contain more RNAs with intact 5′ ends (Figure 4B, lanes 1–4) but have less properly processed A0 ends (Figure 4B, lanes 5–8) when quantified per total RNA. Thus, processing at A0 is impaired in the rpa135 mutant strains.
Processing of the 18S rRNA is further impaired in the rpa135 mutant cells. With both probes “a” (for 5′-A0) and “b” (for A0–A1), we observed increased intensities of a smear from ~0.8 kb to ~2 kb in the strains carrying the rpa135 mutation. Within this smear, several bands were observed and an ~1 kb band was especially strong in the rrp6 rpa135 strain (Figure 4A, lanes 4 and 8; indicated as “5′-S1”). These fragments result from endonucleolytic cleavage(s) within the 18S region of pre-rRNA. They contain most (or all) of the 5′ ETS as well as proximal 18S rRNA (i.e., they were not cleaved at either A0 or A1). The accumulation of these aberrant cleavage products explains why the production of mature 18S rRNA (per total RNA) (Figure 3B) and its normal precursors 20S rRNA (Figure 4A, lanes 9–12) and 35S rRNA (Figures 3B and 4A) is reduced in the rpa135 mutants even though we detect higher amounts of the 5′ end (i.e., +1) of pre-rRNA in these mutants than in WT by primer extension (Figure 4B, lanes 1–4). For convenience in discussion, we call the major aberrant cleavage site (or sites, see below) responsible for ~1 kb RNA fragment “S1” (small subunit rRNA cleavage site 1; Figure 4A). We note that a fragment corresponding to 5′-S1 can be detected weakly in WT cells. Thus, it appears that this aberrant cleavage occurs at a very low frequency normally, the rpa135 mutation causes a large increase in this frequency, and the TRAMP-exosome complex plays a role in eliminating this aberrant product.
Processing events leading to formation of 5.8S rRNA were also found to be aberrant in the rpa135 strains. With probe “d” (for A2–A3), an ~0.4 kb band was clearly observed in the rpa135 rrp6 strain (Figure 4A, lane 16, “A2–C2”), but only weakly in the rpa135 and rrp6 strains and not in the WT strain (Figure 4A, lanes 13–15). The same signal was observed with a probe for the A3–B1 region (data not shown), but not with probe “e” (for C2–C1) (Figure 4A, lane 20) in the rpa135 rrp6 strain. This band likely corresponds to the A2–C2 fragment, which was previously observed in mutants defective in processing factor Ssf1 (Fatica et al., 2002) or Npa1 (Dez et al., 2004), in cells depleted of exosome components (Allmang et al., 2000) or those treated with 5-fluorouracil (Fang et al., 2004). This fragment is apparently produced by premature cleavage at C2 prior to the cleavage at A3 (Fatica et al., 2002).
Another feature we observed for the rpa135 strain is an increase of the ~3 kb RNA relative to WT detected by probes a, b, c, and d (but not by e), which was also increased by the rrp6 mutation (Figure 4A; indicated as 23S). This RNA must include most of the 5′ ETS through A3 region and may correspond to the 23S pre-rRNA that is known to be produced by the cleavage at A3 as an alternative pathway when cleavages at A0, A1, and A2 are inefficient (Venema and Tollervey, 1999).
Finally, we observed degradation of precursors for 25S rRNA. With the A2–A3 and C2–C1 probes (d and e), all of which detected the presence of 27S pre-rRNA (~3.9 kb), we observed increased intensities of several bands and smears ranging from ~0.7 kb to ~2 kb in both the rpa135 and rpa135 rrp6 strains relative to controls (Figure 4A, lanes 13–20). These fragments likely contain the A2 site as their 5′ end, with various sites within 25S rRNA as their 3′ end, since we detect more RNA with an A2 5′ end in the rpa135 mutant cells by primer extension (Figure 4B, lanes 9–12) but equal or less 27S pre-rRNA in the rpa135 mutants than in control strains (Figure 4A, lanes 13–20). These results indicate that the rpa135 mutation impairs the normal processing pathways for production of the 25S rRNA via 27S pre-rRNA by causing cleavages (and degradation) within the region corresponding to the mature 25S rRNA.
Aberrant RNA Fragments Are Polyadenylated
Preliminary experiments suggested that the aberrant RNA species generated in the rpa135 mutant were polyadenylated, indicating a potential role for the TRAMP complex, which is involved in rRNA quality control (LaCava et al., 2005; Houseley et al., 2006). We confirmed this prediction for the aberrant 5′-S1 RNA. As shown in Figure 5, poly(A) tailed RNAs were detected first by reverse transcription using an oligo(dT)-adaptor primer, followed by PCR amplification of the products using a primer with the 5′ end at +674 combined with the adaptor portion of the primer used for the reverse transcription (Figure 5B, lanes 1–4). From the size of the band (~300 bp), the major cleavage site (S1) was calculated to be ~270 nt from the 5′ end of the mature 18S rRNA. PCR products obtained in these experiments were cloned and sequenced to identify cleavage/polyadenylation sites (Figures 5C and 5D). Although there is an indication of a “hot spot” around position 262, other cleavage/polyadenylation sites were also identified. We conclude that cleavage or cleavages followed by polyadenylation take place within the 18S region during processing of precursor rRNA; these RNAs are degraded by the nuclear exosome, and the frequency of such cleavage/degradation of precursor rRNA is increased in the rpa135 mutant strains. Similar cleavage/polyadenylation of precursor rRNA followed by degradation also takes place within the 25S region, but we have not characterized these events in detail.
Figure 5. The 5′-S1 Fragment Is Polyadenylated.
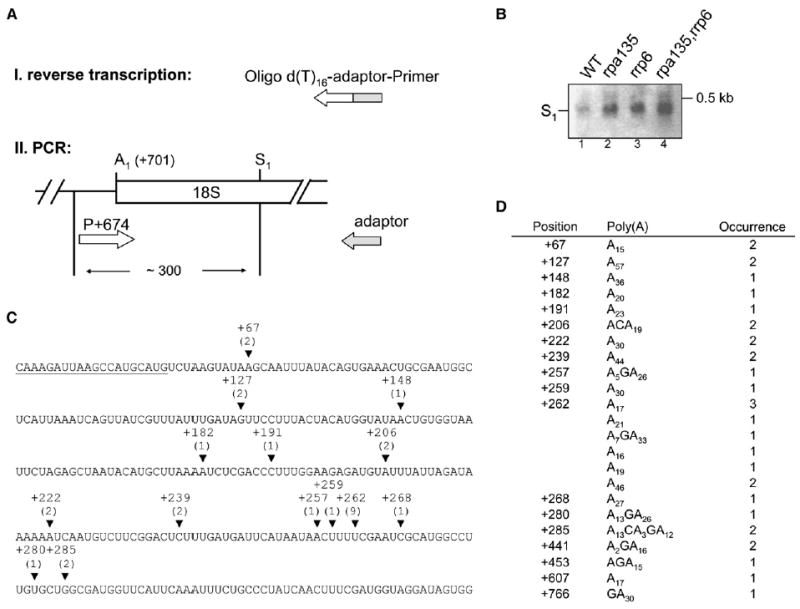
Total RNA from the four strains analyzed in Figure 4 was reverse transcribed with an oligo(dT)-adaptor primer. cDNA was then PCR amplified with the adaptor primer in combination with the forward primer (position +674 relative to the 5′ end of 35S pre-rRNA). PCR products were cloned and sequenced.
(A) Schematic diagram of the procedure and the approximate size of PCR products are indicated.
(B) EtBr-stained gel obtained after agarose gel electrophoresis of PCR products. S1 indicates PCR products derived from RNA formed by cleavage/polyadenylation at S1.
(C) Sequence of 18S rRNA is shown from position +37 to +337 with respect to the 5′ end of mature 18S rRNA. Position of forward primer for amplification of sequencing product is underlined. Polyadenylation sites identified between +441 and +766 are not shown (see table in [D]). Arrowheads indicate the position of polyadenylation sites (from strain NOY2176). If adenosines in the 18S rRNA sequence precede a poly(A) stretch the last nucleotide is indicated. The number of sequenced transformants are given in parentheses.
(D) The table shows a list of transformants analyzed, position of polyadenylation sites, length and composition of poly(A) tails, and number of sequenced transformants. We note that the sequences of poly(A) tails occasionally contained bases other than A. It is not clear whether this use of non-A in poly(A) tails was actually carried out by TRAMP in vivo. Similar observations were recently made for polyadenylation of human rRNA (Slomovic et al., 2006).
The rpa135 Mutant Polymerase Is as Processive as WT
To confirm that the aberrant RNA transcripts observed (Figures 3–5) resulted from abnormal cleavage/processing of the rRNA rather than premature termination of transcription by the mutant polymerase, we measured the processivity of Pol I using two methods: (a) biochemical analysis of Pol I processivity using a chromatin immunoprecipitation (ChIP)-based assay and (b) direct visualization of transcribing Pol I molecules by EM analysis of Miller chromatin spreads.
Our ChIP assay for processivity compares Pol I association with the 35S rRNA gene near the 5′ end to that near the 3′ end (Mason and Struhl, 2005; Schneider et al., 2006). Pol I (and crosslinked DNA) was immunoprecipitated with a polyclonal antibody against A190 followed by extensive washing and reversal of the crosslinks. The ratio of ChIP signal (IP divided by input) at the 3′ end relative to the 5′ end can be used to estimate the “relative processivity” of Pol I in a given strain (Figure 6A). When quantified and normalized to WT, it is clear that there is no decrease in the relative processivity of Pol I in the rpa135 mutant cells (Figure 6A, right panel). These data indicate that defects in rRNA processing/ribosome assembly cannot be explained by defects in processivity, in vivo.
Figure 6. Processivity of Pol I Is Not Affected by rpa135 Mutation.
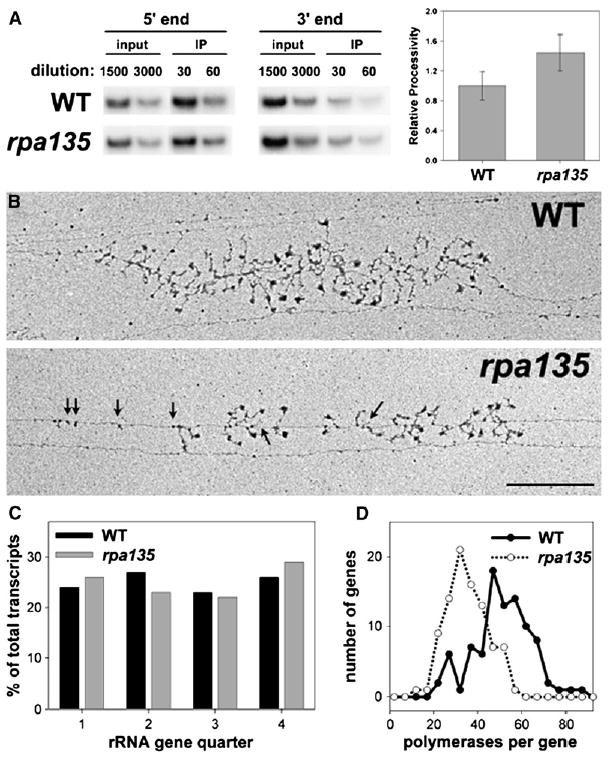
(A) ChIP analysis of Pol I association with the 5′ and 3′ ends of the rDNA in WT and rpa135 cells. PCR products shown are as follows: 5′ end, +67 to +388; 3′ end, +6067 to +6388. Dilutions of input and IP DNA are indicated. To measure relative processivity, IP/input signal at the 3′ end was divided by that at the 5′ end. This value for both strains was normalized to WT (WT = 1). Averages of values from three independent cultures are shown; error bars equal ± 1 standard deviation.
(B) EM analysis of Miller chromatin spreads. One representative rDNA gene of WT and rpa135 mutant cells is shown. Bar, 0.5 μm. Polymerase density in each example is close to average for the population. Shorter than expected RNA molecules in the rpa135 mutant are indicated (arrows).
(C) Polymerase density within the four quarters of each rDNA repeat was quantified for rpa135 and WT cells, and values are expressed as a percentage of total polymerases on the gene. Only genes whose 5′ and 3′ ends were unambiguously identified were analyzed. Number of individual genes analyzed in this way was 21 for WT and 35 for rpa135, corresponding to 1143 and 1259 transcripts, respectively. (D) Individual rDNA repeats were analyzed for total number of polymerases per gene (N = 90 for both WT and rpa135). Number of polymerases per gene was then plotted versus the number of genes with that value. The average polymerase densities for the populations were 49 ± 13 polymerases per gene for WT and 33 ± 9 polymerases per gene for rpa135.
EM Miller chromatin spread analysis of the WT and rpa135 mutant cells further confirmed that there are no processivity defects in the mutant strain. If the rpa135 mutant polymerase were more prone to premature termination, then EM analysis would detect differences in the polymerase density near the 5′ end of the gene versus the 3′ end compared to WT. As seen in both the representative EM pictures (Figure 6B) and the quantification of Pol I occupancy within the four quarters of the gene for a larger sampling of genes (Figure 6C), there is no evidence for increased premature termination or release of Pol I in the mutant compared to WT. These data support the interpretation that elongation defects of the mutant polymerase cause defects in rRNA processing and ribosome assembly.
In addition to confirming the absence of polymerase processivity defects, EM analysis revealed several other features of rRNA transcription in the mutant. First, by quantification of the polymerase density of a large number of genes, we observed an ~30% decrease in the average polymerase density per gene in the rpa135 mutant compared to WT (33 Pols/gene compared to 49 Pols/gene in WT; Figure 6D). Thus, the rpa135 mutation decreases the rate of initiation ~30% more than the overall rate of elongation in vivo. However, because defects in initiation in the rrn3 mutant do not cause defects in rRNA processing/assembly (Figure 3A), the defect in initiation caused by the rpa135(D784G) mutation cannot be responsible for the observed defects in rRNA processing/assembly observed in this mutant.
The micrographs also showed that transcripts attached to Pol I seen in the rpa135 mutant are often short or missing relative to those at similar positions within a gene in the WT strain (examples shown by arrows in Figure 6B), suggesting that the anomalous pre-rRNA cleavages/degradation observed for the mutant (Figures 4 and 5) take place cotranscriptionally. Such cotranscriptional defects are consistent with the conclusion that efficient rRNA processing/ribosome assembly requires proper transcription elongation. This cotranscriptional rRNA degradation is being investigated further.
DISCUSSION
Proper Transcription of rRNA Genes by Pol I Is Important for rRNA Processing and Ribosome Assembly
We have demonstrated that a single amino acid alteration in the A135 subunit of Pol I located near the active center decreases transcription elongation rate of the enzyme measured in vitro and causes significant defects in rRNA processing and assembly in vivo, leading to unbalanced production of the two ribosomal subunits and an overall reduction in the rate of ribosome synthesis by ~4-fold. This decrease in ribosome synthesis rate is not due to a decrease in the processivity of Pol I. EM Miller chromatin spread analysis demonstrated that the rate of Pol I initiation decreased slightly more than the rate of elongation in the mutant strain compared to WT. Although we have not extensively studied the effects of the rpa135 mutation on transcription initiation in vitro, it is likely that the “initiation step,” i.e., productive recruitment of Pol I and/or promoter escape requires an efficient binding of NTP to Pol I, and thus it is not surprising that this step is also sensitive to the mutation in the A135 subunit. Although the initiation step appears to further limit the overall Pol I transcription rate in the rpa135 mutant strain in vivo, it is unlikely that the initiation defect is responsible for the observed processing defects. First, decreasing the initiation step alone by mutating the essential initiation factor Rrn3 does not cause a defect in rRNA processing/ribosome assembly (Figure 3A), even though this rrn3 mutation caused a larger decrease in the overall rate of Pol I transcription compared to the rpa135 mutation. Second, in the only reported case of coupling between Pol I initiation and rRNA processing, the processing defects are very different than those observed here and result in accumulation of more 35S rRNA and long uncleaved nascent transcripts (Gallagher et al., 2004). Therefore, we conclude that the observed rRNA processing defects are more consistent with the postulate that Pol I transcription at post-Pol I recruitment step(s) is specifically coupled with rRNA processing and ribosome assembly, and that the mutation in Pol I that slows down elongation rate in vitro interferes with this coupling in vivo.
As an additional control, we also examined ribosome synthesis in the rpa135 mutant and WT strains carrying the GAL7-35S rDNA fusion gene on a multicopy plasmid. The presence of this plasmid improved the growth rate of the mutant, but not WT, in galactose. In addition, by analyzing the production of the tagged 18S and 25S rRNAs from the GAL7 promoter, we found no negative effects of the rpa135 mutation on the Pol II-driven rRNA synthesis (our unpublished data). Therefore, the negative effects of the rpa135 mutation on rRNA processing are limited to rRNA molecules produced by the mutant Pol I; rRNA molecules produced by Pol II in the same nucleus are not affected. Thus, we conclude that the proper elongation of Pol I transcription itself is important for cotranscriptional as well as posttranscriptional rRNA processing events to produce mature rRNAs and ribosomes.
The possibility of coupling of Pol I elongation with rRNA processing/ribosome assembly was suggested recently in connection with the discovery of regulation of Pol I elongation (via UBF) by growth factor signaling in mammalian cells (Stefanovsky et al., 2006). The present work provides direct support of this suggestion. Coupling between Pol I initiation and rRNA processing has also been reported (Gallagher et al., 2004). A subset of the small ribosomal subunit (SSU) processome proteins known as t-UTPs apparently participates in Pol I initiation step(s), suggesting coordinate regulation of Pol I transcription and rRNA processing. However, as noted above, this type of coupling is different from the mechanism(s) described here.
If Pol I transcription elongation is functionally coupled to rRNA processing, then why is rRNA produced by Pol II processed relatively normally (as discussed above and shown previously; e.g., Nogi et al. [1991] and Sharma and Tollervey [1999])? One likely explanation is that there is a kinetic balance between the rate of rRNA synthesis (i.e., Pol I transcription elongation rate) and the rates of rRNA folding and maturation. Perhaps Pol II transcription elongation rate through rDNA is fortuitously similar to that of Pol I, explaining the relatively normal ribosome assembly observed from GAL7-35S rDNA-derived rRNA or from Pol II-derived rRNA in polymerase switched strains (our unpublished data; for polymerase switched strains, see Vu et al. [1999] and Oakes et al. [1999]).
Although drastic defects in rRNA assembly are not detected when rRNA is produced by Pol II, it seems clear that Pol II transcription of rRNA does not perfectly mimic transcription elongation by Pol I. Cbf5 is one of four core proteins of the H/ACA snoRNPs and is almost certainly the catalytic component responsible for pseudouridylation of pre-rRNA (Zebarjadian et al., 1999; Lafontaine et al., 1998). Carbon, Fournier, and coworkers constructed several cbf5-ts mutants and observed that transcription of 35S pre-rRNA from the GAL7-35S rDNA gene by Pol II improved the growth rate of some mutants, showed no effect on other mutants, and interestingly, made one mutant inviable on galactose at the permissive temperature, i.e., caused an allele-specific dominant synthetic lethality (Zebarjadian et al., 1999). Thus, mutant alleles of CBF5 produced different phenotypes depending on whether the rRNA gene was transcribed by Pol I or Pol II. Similar observations were also made for Nop2, which is a factor required for 60S subunit synthesis. Two out of six ts alleles of NOP2 showed synthetic lethality when the GAL7-35S rDNA gene was expressed at the permissive temperature (cited and discussed in Hong et al. [2001]). These observations suggest specific, possibly direct interaction(s) between rRNA processing factors and Pol I and provide additional support for functional coupling of rRNA transcription to rRNA processing.
Possible Effects of Pol I Mutations on Cotranscriptional Recruitment of Ribosome Assembly Factors
Cotranscriptional recruitment of the mRNA capping machinery, splicing factors, and the 3′ end processing/polyadenylation machinery has been extensively studied in regard to Pol II transcription of mRNAs (reviewed in Sims et al. [2004], Zorio and Bentley [2004], Aguilera [2005], and Buratowski [2005]). The primary mechanism for regulating the coupling of Pol II elongation to mRNA processing seems to be the modulation of the phosphorylation state of the C-terminal domain (CTD) of the largest subunit of Pol II (Rpb1). Although Pol I has no discernible structure similar to the CTD of Pol II, several factors involved in modulating Pol II elongation via phosphorylation of the CTD have been implicated in Pol I transcription (kinases Kin28 and Ctk1 [Iben et al., 2002; Bouchoux et al., 2004], and the phosphatase Fcp1 [Fath et al., 2004]). Perhaps these and other factors can modulate cotranscriptional rRNA processing events by reversible modification of Pol I and some other factors, such as Spt5p. It has been shown that the Spt4/5 complex (which plays a crucial role in coupling Pol II transcription to mRNA capping) physically interacts with both Pol I and rDNA and appears to play a role in coupling Pol I transcription to rRNA processing (Schneider et al., 2006).
Several aberrant processing events were observed in the rpa135 mutant and some of them resemble those observed in cells depleted of known ribosome assembly factors or cells carrying a mutation in such factors. For example, the appearance of the aberrantly processed A2–C2 fragment was previously observed in cells depleted of Ssf1 (Fatica et al., 2002) or Npa1 (Dez et al., 2004). It was suggested that recruitment of such factors prevents premature cleavage at C2 prior to cleavage at A3 and that ordered recruitment of various factors to preribosomal particles combined with their proper release from the particles ensures a correct order or orders of cleavage reactions for efficient ribosome assembly (Fatica et al., 2002). Thus, the appearance of the aberrant A2–C2 fragment in the rpa135 mutant in the present study might imply disordered or incomplete recruitment of processing factors, such as Ssf1 and Npa1 to the rRNA.
Quality Control of Ribosome Synthesis
By combining the rpa135 mutation with deletion of RRP6, we have demonstrated that the rpa135 mutation leads to accumulation of many aberrant rRNA fragments carrying poly(A) tails (this article and our unpublished data), indicating a robust participation of the quality-control system including the known TRAMP-exosome pathway. Participation of TRAMP in pre-rRNA degradation in the rpa135 mutant is also supported by a severe synthetic growth defect produced by combination of the rpa135 mutation with deletion of TRF4 encoding the poly(A) polymerase component of TRAMP (our unpublished data). In contrast, combination of the rpa135 mutation with the deletion of RRP6 showed much weaker synthetic growth defects. Thus, degradation of defective pre-rRNA might also use a 5′-3′ exonuclease, e.g., Rat1, as suggested previously (Fang et al., 2005).
The signal in the rRNA that induces the TRAMP-exosome system in the rpa135 mutant remains unknown. Previous studies on tRNA synthesis have demonstrated that TRAMP-exosome recognizes and degrades tRNA molecules with incorrect conformation rather than incorrect sequence (Vanacova et al., 2005). Therefore, it seems that impaired Pol I elongation leads to increased production of rRNA molecules with incorrect conformations. How rRNA molecules with incorrect conformations are distinguished from those with correct conformation is an interesting question for future research.
It was recently reported that the exosome and TRAMP components are concentrated in a subnucleolar focus called “No-body,” and preribosome surveillance was suggested to take place in that focus (Dez et al., 2006). On the other hand, rRNA modification and folding appear to take place cotranscriptionally (Osheim et al., 2004) and thus surveillance might also take place cotranscriptionally. Our EM Miller chromatin spread analysis supports the presence of such cotranscriptional surveillance. This subject is under current investigation.
In conclusion, we have demonstrated that proper transcription of rRNA by Pol I is required for efficient rRNA processing/ribosome assembly: i.e., they are functionally coupled. Thus, Pol I, elongation factors, and rRNA sequence elements must function together to optimize Pol I elongation for regulation of the complex but highly efficient ribosome assembly processes.
EXPERIMENTAL PROCEDURES
Media and Strains
Yeast strains and plasmids are listed in Table 1. Yeast extract/peptone with glucose (YEPD) and SD media were described previously (Claypool et al., 2004). Cells were grown at 30°C. The rpa135(D784G) mutant was isolated using standard methods (for details, see the Supplemental Data available with this article online).
In Vitro Transcription
RPA135- and rpa135(D784G)-containing Pol I was prepared as described previously (Keener et al., 1998) from cells carrying His6-(HA)3 epitope tags on the N terminus of A135 (NOY2173 and NOY2174). Multiround transcription with all pure components was performed as described at room temperature for 30 min (Keener et al., 1998) except that the concentration of a single nucleotide (either UTP or ATP) was varied while keeping the concentrations of the other three nucleotides constant.
The analysis of elongation rate in vitro is described in the Supplemental Data.
Sucrose Gradient Analysis
Polysome profiles were analyzed as described previously (Schneider et al., 2006).
Other Biochemical Analyses
Steady-state amounts of various RNA species (shown in Figure 3) were measured by growing cells for many generations in an exponentially growing state in the presence of [14C]uracil, followed by isolation of RNA and its separation into individual RNA species by gel electrophoresis. The details are described in the Supplemental Data. Analysis of polyadenylated RNA by reverse transcriptase-PCR (RT-PCR) was carried out as described (Salles et al., 1999). Northern analysis of RNA, primer extension analysis of 5′ ends of RNA, and PCR reactions were done using standard methods as described previously (Claypool et al., 2004). Oligonucleotide probes and primers used in these analyses are listed in the Supplemental Data. ChIP was performed as described previously (Schneider et al., 2006).
EM Analysis
Miller chromatin spreads and analysis by EM were performed as described after growth of cells in SD media (French et al., 2003). For NOY2172 (rpa135), polymerase number per gene was determined by counting RNA polymerases or nascent transcripts on all rRNA genes that could be unambiguously traced as individual genes (N = 90). Ninety representative control genes (NOY388) were similarly analyzed. To more carefully map the position of transcripts along the gene, amenable genes were selected (N = 21 for WT and 35 for rpa135). Gene lengths were normalized to 100 units, and each transcript was positioned accordingly and final values were plotted for the four quarters of the rDNA gene.
Supplementary Material
Supplemental Data include Supplemental Experimental Procedures, Supplemental References, and one table and can be found with this article online at http://www.molecule.org/cgi/content/full/26/2/217/DC1/.
Acknowledgments
We thank Dr. Suzanne Sandmeyer for critical reading of the manuscript. The work was supported by Public Health Service grants GM-35949 (to M.N.) and GM-63952 (to A.L.B.) and a postdoctoral fellowship from the Jane Coffin Child Memorial Fund for Medical Research (to D.A.S.).
References
- Aguilera A. Cotranscriptional mRNP assembly: from the DNA to the nuclear pore. Curr Opin Cell Biol. 2005;17:242–250. doi: 10.1016/j.ceb.2005.03.001. [DOI] [PubMed] [Google Scholar]
- Allmang C, Mitchell P, Petfalski E, Tollervey D. Degradation of ribosomal RNA precursors by the exosome. Nucleic Acids Res. 2000;28:1684–1691. doi: 10.1093/nar/28.8.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archambault J, LaCroute F, Ruet A, Friesen JD. Genetic interaction between transcription elongation factor TFIIS and RNA polymerase II. Mol Cell Biol. 1992;12:4142–4152. doi: 10.1128/mcb.12.9.4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand E, Fournier MJ. The snoRNPs and related machines: ancient devices that mediate maturation of rRNA and other RNAs. In: Olson MOJ, editor. The Nucleolus. Austin, TX: R.G. Landes & Co; 2004. pp. 223–257. [Google Scholar]
- Bouchoux C, Hautbergue G, Grenetier S, Carles C, Riva M, Goguel V. CTD kinase I is involved in RNA polymerase I transcription. Nucleic Acids Res. 2004;32:5851–5860. doi: 10.1093/nar/gkh927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratowski S. Connections between mRNA 3′ end processing and transcription termination. Curr Opin Cell Biol. 2005;17:257–261. doi: 10.1016/j.ceb.2005.04.003. [DOI] [PubMed] [Google Scholar]
- Claypool JA, French SL, Johzuka K, Eliason K, Vu L, Dodd JA, Beyer AL, Nomura M. Tor pathway regulates Rrn3p-dependent recruitment of yeast RNA polymerase I to the promoter but does not participate in alteration of the number of active genes. Mol Biol Cell. 2004;15:946–956. doi: 10.1091/mbc.E03-08-0594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer P, Bushnell DA, Kornberg RD. Structural basis of transcription: RNA polymerase II at 2.8 angstrom resolution. Science. 2001;292:1863–1876. [Google Scholar]
- Decatur WA, Fournier MJ. RNA-guided nucleotide modification of ribosomal and other RNAs. J Biol Chem. 2003;278:695–698. doi: 10.1074/jbc.R200023200. [DOI] [PubMed] [Google Scholar]
- de la Cruz J, Kressler D, Tollervey D, Linder P. Dob1p (Mtr4p) is a putative ATP-dependent RNA helicase required for the 3′ end formation of 5.8S rRNA in Saccharomyces cerevisiae. EMBO J. 1998;17:1128–1140. doi: 10.1093/emboj/17.4.1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dez C, Froment C, Noaillac-Depeyre J, Monsarrat B, Caizergues-Ferrer M, Henry Y. Npa1p, a component of very early pre-60S ribosomal particles, associates with a subset of small nucleolar RNPs required for peptidyl transferase center modification. Mol Cell Biol. 2004;24:6324–6337. doi: 10.1128/MCB.24.14.6324-6337.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dez C, Houseley J, Tollervey D. Surveillance of nuclear-restricted pre-ribosomes within a subnucleolar region of Saccharomyces cerevisiae. EMBO J. 2006;25:1534–1546. doi: 10.1038/sj.emboj.7601035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragon F, Gallagher JE, Compagnone-Post PA, Mitchell BM, Porwancher KA, Wehner KA, Wormsley S, Settlage RE, Shabanowitz J, Osheim Y, et al. A large nucleolar U3 ribonucleo-protein required for 18S ribosomal RNA biogenesis. Nature. 2002;417:967–970. doi: 10.1038/nature00769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Exinger F, LaCroute F. 6-Azauracil inhibition of GTP biosynthesis in Saccharomyces cerevisiae. Curr Genet. 1992;22:9–11. doi: 10.1007/BF00351735. [DOI] [PubMed] [Google Scholar]
- Fang F, Hoskins J, Butler JS. 5-fluorouracil enhances exosome-dependent accumulation of polyadenylated rRNAs. Mol Cell Biol. 2004;24:10766–10776. doi: 10.1128/MCB.24.24.10766-10776.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang F, Phillips S, Butler JS. Rat1p and Rai1p function with the nuclear exosome in the processing and degradation of rRNA precursors. RNA. 2005;11:1571–1578. doi: 10.1261/rna.2900205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fath S, Kobor MS, Philippi A, Greenblatt J, Tschochner H. Dephosphorylation of RNA polymerase I by Fcp1p is required for efficient rRNA synthesis. J Biol Chem. 2004;279:25251–25259. doi: 10.1074/jbc.M401867200. [DOI] [PubMed] [Google Scholar]
- Fatica A, Cronshaw AD, Dlakic M, Tollervey D. Ssf1p prevents premature processing of an early pre-60S ribosomal particle. Mol Cell. 2002;9:341–351. doi: 10.1016/s1097-2765(02)00458-6. [DOI] [PubMed] [Google Scholar]
- French SL, Osheim YN, Cioci F, Nomura M, Beyer AL. In exponentially growing Saccharomyces cerevisiae cells, rRNA synthesis is determined by the summed RNA polymerase I loading rate rather than by the number of active genes. Mol Cell Biol. 2003;23:1558–1568. doi: 10.1128/MCB.23.5.1558-1568.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromont-Racine M, Senger B, Saveanu C, Fasiolo F. Ribosome assembly in eukaryotes. Gene. 2003;313:17–42. doi: 10.1016/s0378-1119(03)00629-2. [DOI] [PubMed] [Google Scholar]
- Gallagher JE, Dunbar DA, Granneman S, Mitchell BM, Osheim Y, Beyer AL, Baserga SJ. RNA polymerase I transcription and pre-rRNA processing are linked by specific SSU processome components. Genes Dev. 2004;18:2506–2517. doi: 10.1101/gad.1226604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granneman S, Baserga SJ. Crosstalk in gene expression: coupling and co-regulation of rDNA transcription, pre-ribosome assembly and pre-rRNA processing. Curr Opin Cell Biol. 2005;17:281–286. doi: 10.1016/j.ceb.2005.04.001. [DOI] [PubMed] [Google Scholar]
- Helser TL, Baan RA, Dahlberg AE. Characterization of a 40S ribosomal subunit complex in polyribosomes of Saccharomyces cerevisiae treated with cycloheximide. Mol Cell Biol. 1981;1:51–57. doi: 10.1128/mcb.1.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong B, Wu K, Brockenbrough JS, Wu P, Aris JP. Temperature sensitive nop2 alleles defective in synthesis of 25S rRNA and large ribosomal subunits in Saccharomyces cerevisiae. Nucleic Acids Res. 2001;29:2927–2937. doi: 10.1093/nar/29.14.2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseley J, LaCava J, Tollervey D. RNA-quality control by the exosome. Nat Rev Mol Cell Biol. 2006;7:529–539. doi: 10.1038/nrm1964. [DOI] [PubMed] [Google Scholar]
- Iben S, Tschochner H, Bier M, Hoogstraten D, Hozak P, Egly JM, Grummt I. TFIIH plays an essential role in RNA polymerase I transcription. Cell. 2002;109:297–306. doi: 10.1016/s0092-8674(02)00729-8. [DOI] [PubMed] [Google Scholar]
- Kadaba S, Krueger A, Trice T, Krecic AM, Hinnebusch AG, Anderson J. Nuclear surveillance and degradation of hypomodified initiator tRNAMet in S. cerevisiae. Genes Dev. 2004;18:1227–1240. doi: 10.1101/gad.1183804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keener J, Josaitis CA, Dodd JA, Nomura M. Reconstitution of yeast RNA polymerase I transcription in vitro from purified components. TATA-binding protein is not required for basal transcription. J Biol Chem. 1998;273:33795–33802. doi: 10.1074/jbc.273.50.33795. [DOI] [PubMed] [Google Scholar]
- Kuai L, Fang F, Butler JS, Sherman F. Polyadenylation of rRNA in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2004;101:8581–8586. doi: 10.1073/pnas.0402888101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaCava J, Houseley J, Saveanu C, Petfalski E, Thompson E, Jacquier A, Tollervey D. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell. 2005;121:713–724. doi: 10.1016/j.cell.2005.04.029. [DOI] [PubMed] [Google Scholar]
- Lafontaine DL, Bousquet-Antonelli C, Henry Y, Caizergues-Ferrer M, Tollervey D. The box H + ACA snoRNAs carry Cbf5p, the putative rRNA pseudouridine synthase. Genes Dev. 1998;12:527–537. doi: 10.1101/gad.12.4.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langelier MF, Baali D, Trinh V, Greenblatt J, Archambault J, Coulombe B. The highly conserved glutamic acid 791 of Rpb2 is involved in the binding of NTP and Mg(B) in the active center of human RNA polymerase II. Nucleic Acids Res. 2005;33:2629–2639. doi: 10.1093/nar/gki570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason PB, Struhl K. Distinction and relationship between elongation rate and processivity of RNA polymerase II in vivo. Mol Cell. 2005;17:831–840. doi: 10.1016/j.molcel.2005.02.017. [DOI] [PubMed] [Google Scholar]
- Mougey EB, O’Reilly M, Osheim Y, Miller OL, Jr, Beyer A, Sollner-Webb B. The terminal balls characteristic of eukaryotic rRNA transcription units in chromatin spreads are rRNA processing complexes. Genes Dev. 1993;7:1609–1619. doi: 10.1101/gad.7.8.1609. [DOI] [PubMed] [Google Scholar]
- Nogi Y, Yano R, Nomura M. Synthesis of large rRNAs by RNA polymerase II in mutants of Saccharomyces cerevisiae defective in RNA polymerase I. Proc Natl Acad Sci USA. 1991;88:3962–3966. doi: 10.1073/pnas.88.9.3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakes M, Siddiqi I, Vu L, Aris J, Nomura M. Transcription factor UAF, expansion and contraction of ribosomal DNA (rDNA) repeats, and RNA polymerase switch in transcription of yeast rDNA. Mol Cell Biol. 1999;19:8559–8569. doi: 10.1128/mcb.19.12.8559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osheim YN, French SL, Keck KM, Champion EA, Spasov K, Dragon F, Baserga SJ, Beyer AL. Pre-18S ribosomal RNA is structurally compacted into the SSU processome prior to being cleaved from nascent transcripts in Saccharomyces cerevisiae. Mol Cell. 2004;16:943–954. doi: 10.1016/j.molcel.2004.11.031. [DOI] [PubMed] [Google Scholar]
- Rotenberg MO, Moritz M, Woolford JL., Jr Depletion of Saccharomyces cerevisiae ribosomal protein L16 causes a decrease in 60S ribosomal subunits and formation of half-mer polyribosomes. Genes Dev. 1988;2:160–172. doi: 10.1101/gad.2.2.160. [DOI] [PubMed] [Google Scholar]
- Salles FJ, Richards WG, Strickland S. Assaying the polyadenylation state of mRNAs. Methods. 1999;17:38–45. doi: 10.1006/meth.1998.0705. [DOI] [PubMed] [Google Scholar]
- Schneider DA, French SL, Osheim YN, Bailey AO, Vu L, Dodd J, Yates JR, Beyer AL, Nomura M. RNA polymerase II elongation factors Spt4p and Spt5p play roles in transcription elongation by RNA polymerase I and rRNA processing. Proc Natl Acad Sci USA. 2006;103:12707–12712. doi: 10.1073/pnas.0605686103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma K, Tollervey D. Base pairing between U3 small nucleolar RNA and the 5′ end of 18S rRNA is required for pre-rRNA processing. Mol Cell Biol. 1999;19:6012–6019. doi: 10.1128/mcb.19.9.6012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims RJ, III, Belotserkovskaya R, Reinberg D. Elongation by RNA polymerase II: the short and long of it. Genes Dev. 2004;18:2437–2468. doi: 10.1101/gad.1235904. [DOI] [PubMed] [Google Scholar]
- Slomovic S, Laufer D, Geiger D, Schuster G. Polyadenylation of ribosomal RNA in human cells. Nucleic Acids Res. 2006;34:2966–2975. doi: 10.1093/nar/gkl357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanovsky V, Langlois F, Gagnon-Kugler T, Rothblum LI, Moss T. Growth factor signaling regulates elongation of RNA polymerase I transcription in mammals via UBF phosphorylation and r-chromatin remodeling. Mol Cell. 2006;21:629–639. doi: 10.1016/j.molcel.2006.01.023. [DOI] [PubMed] [Google Scholar]
- Tschochner H, Hurt E. Pre-ribosomes on the road from the nucleolus to the cytoplasm. Trends Cell Biol. 2003;13:255–263. doi: 10.1016/s0962-8924(03)00054-0. [DOI] [PubMed] [Google Scholar]
- Uptain SM, Kane CM, Chamberlin MJ. Basic mechanisms of transcript elongation and its regulation. Annu Rev Biochem. 1997;66:117–172. doi: 10.1146/annurev.biochem.66.1.117. [DOI] [PubMed] [Google Scholar]
- Vanacova S, Wolf J, Martin G, Blank D, Dettwiler S, Friedlein A, Langen H, Keith G, Keller W. A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS Biol. 2005;3:e189. doi: 10.1371/journal.pbio.0030189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venema J, Tollervey D. Ribosome synthesis in Saccharomyces cerevisiae. Annu Rev Genet. 1999;33:261–311. doi: 10.1146/annurev.genet.33.1.261. [DOI] [PubMed] [Google Scholar]
- Vu L, Siddiqi I, Lee BS, Josaitis CA, Nomura M. RNA polymerase switch in transcription of yeast rDNA: role of transcription factor UAF (upstream activation factor) in silencing rDNA transcription by RNA polymerase II. Proc Natl Acad Sci USA. 1999;96:4390–4395. doi: 10.1073/pnas.96.8.4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wai HH, Vu L, Oakes M, Nomura M. Complete deletion of yeast chromosomal rDNA repeats and integration of a new rDNA repeat: use of rDNA deletion strains for functional analysis of rDNA promoter elements in vivo. Nucleic Acids Res. 2000;28:3524–3534. doi: 10.1093/nar/28.18.3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyers F, Rougemaille M, Badis G, Rousselle JC, Dufour ME, Boulay J, Regnault B, Devaux F, Namane A, Seraphin B, et al. Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell. 2005;121:725–737. doi: 10.1016/j.cell.2005.04.030. [DOI] [PubMed] [Google Scholar]
- Zanchin NI, Goldfarb DS. The exosome subunit Rrp43p is required for the efficient maturation of 5.8S, 18S and 25S rRNA. Nucleic Acids Res. 1999;27:1283–1288. doi: 10.1093/nar/27.5.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zebarjadian Y, King T, Fournier MJ, Clarke L, Carbon J. Point mutations in yeast CBF5 can abolish in vivo pseudouridylation of rRNA. Mol Cell Biol. 1999;19:7461–7472. doi: 10.1128/mcb.19.11.7461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorio DA, Bentley DL. The link between mRNA processing and transcription: communication works both ways. Exp Cell Res. 2004;296:91–97. doi: 10.1016/j.yexcr.2004.03.019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Data include Supplemental Experimental Procedures, Supplemental References, and one table and can be found with this article online at http://www.molecule.org/cgi/content/full/26/2/217/DC1/.


