Abstract
Previous studies have defined transcriptional control elements, in addition to the promoters, that both lie near individual human β-globin locus genes and have been implicated in their differential stage-specific regulation during development (i.e., are believed to directly participate in hemoglobin switching). We have reinvestigated the activities during erythropoiesis that might be conferred by two of the more intensively analyzed of these elements, the ɛ-globin gene 5′ silencer and the β-globin gene 3′ enhancer, by deleting them from a yeast artificial chromosome that spans the human β-globin locus, and then analyzing transgenic mice for expression of all of the human genes. These studies show that sequences within the ɛ-globin “silencer” are not only required for silencing but are also required for activation of ɛ-globin transcription; furthermore, deletion of the silencer simultaneously reduced γ-globin transcription during the yolk sac stage of erythroid development. Analysis of the adult β-globin gene 3′ enhancer deletion showed that its deletion affects only that gene.
Keywords: ɛ-globin silencer, β-globin 3′ enhancer
The five genes of the human β-globin locus are transcriptionally regulated by both gene-proximal and -distal control elements. The well characterized distal elements are the five 5′ DNase I hypersensitive sites (HS; refs. 1 and 2) comprising the β-globin locus control region (LCR; refs. 3 and 4); the LCR is generally acknowledged to be required for the regulation of all five genes of the locus during erythroid development. While the promoters of each gene play critical roles in their individual responses to LCR-mediated stimulation, other transcriptional control elements (lying in close physical proximity to the genes) have also been implicated, but their roles in developmental stage-specific control over gene expression in the locus are less clear.
The human β-globin LCR has been studied most extensively after dissection into its constituent components: the three central tissue-specific HS sites (HS2, HS3, and HS4) are thought to play a major role in elaborating the multiple activities attributed to the LCR, while HS5 and HS1 play currently less well defined roles. Early analysis of its activity represented the LCR as an integral organizational, and perhaps structural, unit (5, 6) that has since been referred to as a “holocomplex” (7, 8). A key experiment in the progressive dissection of LCR function showed that each of the three central HS elements was capable of conferring significant stage specificity to activation of the γ- and β-globin genes, and yet none of the individual HS elements was able to fully restore LCR activity (9). These and other studies have led to general acceptance of the hypothesis that the LCR functions as a single, cooperative unit for β-globin locus gene regulation.
Human β-globin locus transcriptional regulation has also been described as arising from either competitive or autonomous gene control, based principally on the chicken β/ɛ-globin gene enhancer/promoter regulatory paradigm (10). Autonomous control implies that a gene is not influenced by placing other transcriptional elements nearby, whereas competitive control implies that when other regulatory elements are placed in cis, or alternatively removed from a particular construct, normal tissue-specific or temporal control is lost (10, 11). Early studies indicated that while the human ɛ-globin gene is regulated autonomously (12) and that the fetal γ- and adult β-globin genes are regulated through competition (13), the mode of regulation for the γ-globin genes was disputed (14, 15). An important caveat in the interpretation of these experiments was that each of the transgenes was analyzed in the context of a DNA construction with arbitrary endpoints, where other participating transcriptional control elements may have been inadvertently omitted. Experiments analyzing gene regulatory activity in the murine locus first implicated the LCR as a direct participant in competition (16). Competitive regulatory mechanisms have more recently been proposed to explain regulation over allele-specific imprinting of the IGF2 and h19 genes (17–19), control over HOM-C complex temporal regulation (20) and control of allele-specific transcription of the olfactory receptor genes (21).
Studies on the transcriptional regulation of the human β-globin locus genes, and dissection of the regulatory mechanisms that are employed during this well characterized developmental program, have recently been advanced by incorporating large DNAs that span the human β-globin locus [linked cosmids and yeast artificial chromosomes (YACs); refs. 22 and 23] into the germ line of transgenic mice, as well as through gene targeting experiments in the murine LCR. Two well characterized YACs have been shown to contain all currently characterized distal and gene-proximal regulatory elements required for human β-globin locus gene control, and both of these, examined as transgenes in mice, have been shown to faithfully recapitulate the activation and developmental stage-specific inactivation of the human ɛ- and γ-globin genes at paralogous hematopoietic sites and developmental stages during murine embryogenesis to finally result in appropriate temporal activation of the adult β-globin gene (24, 25). The YACs can be precisely modified using homologous recombination in yeast, and analysis of the functional consequences of such mutations has led to novel insights into the role the LCR may play in hemoglobin switching. Based on the analysis of several LCR “core” element mutant YACs incorporated as transgenes in mice, we concluded that all of the human locus genes are controlled by competition between an integral functional complex (the LCR holocomplex) and other (as yet undefined) transcriptional control elements that probably lie near the genes, and further that the individual core elements could be substituted for one another to partially or fully restore mutant LCR function (26).
If the LCR is controlling human β-globin transcription by regulatory sequence competition, which and where are the competing elements that interact with the LCR? At least 11 distinct hypothetical or demonstrable transcriptional control sequences lie in close physical proximity to the five genes. These include all five gene promoters; an embryonic ɛ-globin gene silencer, required for suppression of ɛ-globin transcription during the fetal stage (27); 5′ γ-globin gene elements (28–32); a 3′ γ-globin gene enhancer (33); and both gene internal and 3′ β-globin gene enhancers (34, 35). Positive and negative regulation of ɛ-globin gene transcription has been the focus of particularly intensive study (36–44).
As the initial step in efforts to more precisely define elements that may contribute to temporally specific transcription in the human β-globin gene locus, we describe here the effects of deletion of two of these elements in a YAC, and their expression as mutant YAC transgenes: the embryonic ɛ-globin 5′ silencer and the adult β-globin 3′ enhancer. Analysis of transgenic lines bearing these mutants shows that the adult β-globin 3′ enhancer affects expression from only that gene, indicating that this element does not directly participate in competition for LCR-directed activity. Analysis of sequences that were originally characterized as the ɛ-globin gene “silencer” shows that mutation of this element confers unexpected properties: sequence elements within the silencer region are required for activation of both the embryonic ɛ- and fetal γ-globin genes.
MATERIALS AND METHODS
Generation of Element-Specific YAC Deletion Mutants by Homologous Recombination in Yeast.
To generate the targeting construct for deleting the ɛ-globin 5′ silencer using homologous recombination in yeast, a 645-bp fragment immediately flanking, and 5′ to, position −304 of the silencer was amplified from the parental β-globin locus YAC using PCR, where primers incorporated new ClaI and XhoI (italic lettering) sites on the 5′ and 3′ ends, respectively. The 5′ fragment primers were: E5FF (sense strand 5′- …GACTATCGATTCACTTTTTTCTCTGTTTGA) and E5FR (antisense strand 5′- …AGCTCTCGAGCCTCATATAAAGGAGCAAAT), where the underlined nucleotides correspond to positions 18,637 and 19,282, respectively, of the human β-globin locus sequence (web site: globin.cse.psu.edu). A 1900-bp 3′ flanking sequence (incorporating XhoI and SacI sites at the 5′ and 3′ ends) was also amplified by PCR to generate the 3′ flanking sequence to the silencer. The 3′ fragment primers were E3FF (sense strand 5′- …GTACCTCGAGGGATCCAGCACACATTATCA) and E3FR (antisense strand 5′-…GCATGAGCTCCTTAATTAACCATTTTCCCA), where the two underlined nucleotides correspond to positions 19,407 and 21,307, respectively, of the locus. These two PCR fragments were then digested with the appropriate restriction enzymes and ligated together into yeast shuttle vector pRS306, which had been previously digested with ClaI and SacI, to generate the final ɛ-silencer deletion targeting construct. The joining points of the two fragments (as well as ≈1 kbp of the ɛ-globin gene) were resequenced to verify that no mutations had been introduced during PCR.
A λ phage clone containing the 3′ end of the human adult β-globin gene was used to generate the plasmid employed for deleting the β-globin 3′ enhancer (26). A 4.65-kbp BamHI/SmaI adult β-globin gene fragment was isolated from a parental λ genomic clone (26) and subcloned into pGEM7. The 250-bp enhancer was removed by PstI digestion and the flanking sequences were religated. This fragment, now missing the enhancer, was then transferred into pRS306 to generate the final β-globin enhancer deletion targeting construct.
Most of the yeast methods used here were derived from a single source (45). Yeast integrative plasmid pRS306 contains the URA3 selectable marker used to monitor homologous integration into, and excision from, the YAC genome (46), as previously detailed (26). To take advantage of this selection, the YAC right vector arm in the original clone A201F4 (22) was modified to replace the resident URA3 gene with the LYS2 gene (called A201F4.2; ref. 26). The size and integrity of the two final mutated YACs examined in this study were assayed by restriction digestion of the yeast DNA and subsequent Southern blotting with probes spanning the locus (22).
YAC DNA Isolation/Transgenic Mice.
YAC DNA was isolated from pulse-field gels according to standard protocols (47, 48) with minor modifications (26). The DNA was injected into fertilized mouse eggs (CD1; Charles River Breeding Laboratory) before transfer to foster mothers (B6D2F1/J) as described (49). Tail DNAs from offspring were analyzed for the presence of the left and right YAC vector arms by PCR, and then for integrity of the transgenic β-globin locus by Southern blot hybridization of SfiI/SalI digests of DNA from transgenic organs (see below; ref. 22).
Transgene Copy Number and YAC Integrity.
To assess the integrated copy number of human β-globin YACs, genomic DNA recovered from transgenics was digested with PstI, which has two sites in the pYAC4-derived left vector arm. A 1.7-kbp EarI fragment spanning the 3′ PstI site was used for Southern blot hybridization, where a 1.6-kbp common band, as well as others that vary in size, would be detected, depending on the transgene copy number and YAC integration pattern(s).
To assess the integrity of the integrated YACs, hematopoietic cells from 0.5 g of transgenic spleen were suspended in 1 ml of cell suspension buffer (10 mM Tris·HCl, pH 7.5/20 mM NaCl/50 mM EDTA, pH 8.0). This solution was then diluted 1:10 in 0.4% agarose solution and poured into clamped homogeneous electric field (CHEF) plug molds. Solidified plugs were then treated at 50°C in Proteinase K buffer (100 mM EDTA, pH 8.0/0.2% sodium deoxycholate/1% Sarcosyl/10 mg/ml Proteinase K) for 48 h, followed by several 1-h washes with 10 mM Tris·HCl (pH 7.4) and 1 mM EDTA buffer. The plugs were digested with SfiI at 50°C for 12 h, and then with SalI for 12 h before pulse field gel electrophoresis and Southern blot hybridization. Two radiolabeled probes were employed (corresponding to HS4 or the Aγ-globin gene) that would be expected to hybridize to 10-kbp (SfiI/SfiI) or 52-kbp (SfiI/SalI) bands, respectively, in the event that the integrated human β-globin YAC transgenes were intact.
Semiquantitative Multiplex Reverse Transcriptase—PCR Assay.
The assay was developed and described in detail elsewhere (11, 26). A titration for cycle number was performed on each cDNA sample to verify that the amplification of all globin templates was within the linear range. An aliquot of each PCR reaction was electrophoresed on 8% polyacrylamide gels, dried, and subjected to autoradiography. Individual band intensities were quantified on a Molecular Dynamics PhosphorImager (11); expression per gene copy was calculated based on normalization to the four genomic copies of the murine α-globin gene (26). Multiple samples representing each developmental stage in erythropoiesis for each transgenic line were analyzed in like manner using different initial RNA preparations as well as different cDNA synthesis reactions.
RESULTS AND DISCUSSION
Generation of Gene-Proximal Regulatory Element huβYAC (human β-globin YAC) Mutants.
The human ɛ-globin gene 5′ silencer was originally identified as a 215-bp fragment (−392 to −177 relative to the ɛ-globin mRNA cap site; ref. 27) that was later found to possibly overlap a positive regulatory element (41). We therefore deleted only 125 bp of the ɛ-globin upstream sequence (from −304 to −179), containing one YY1 and two GATA factor binding sites that had previously been shown to participate in ɛ-globin negative transcriptional control (36, 44, 50). To generate a transplacement targeting construct for deleting this fragment from an otherwise intact human β-globin locus YAC, a 645-bp 5′ flanking sequence homology and the 1.9 kbp of 3′ flanking sequence were amplified from the parental A201F4 YAC clone DNA (Fig. 1A). Similarly, a 250-bp PstI fragment originally identified as the adult β-globin gene 3′ enhancer was removed by PstI digestion of a 4.65-kbp BamHI/SmaI genomic fragment of the β-globin gene (34). The flanking sequences were religated and the fragment, now missing the 250-bp enhancer sequence, was excised and subcloned into pRS306. These two yeast targeting subclones were then linearized and used to transform the (LYS2-retrofitted) A201F4.2 YAC (26).
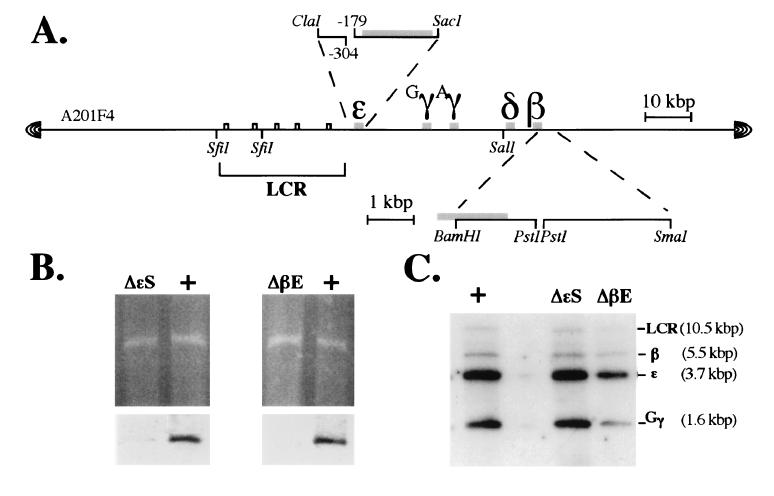
Figure 1.
Structure of the wild-type and mutant human β-globin YACs. (A) YAC clone A201F4 (22) was modified as described (26) to permit ease of manipulation for targeted mutagenesis in yeast. Expanded diagrams depicting the two targeting constructs generated here are shown above (the ɛ-globin silencer deletion mutant) or below (the β-globin enhancer deletion mutant) the diagram of the YAC clone (the position of the deleted elements is indicated by a break in the contiguity of the subclone). Note the position of the SfiI and SalI sites in the locus that were used in the mapping experiments described in Fig. 2. (B Upper) An ethidium bromide-stained pulse field gel of the wild-type, ɛ-silencer, and β-enhancer deletion mutant YACs. (B Lower) Autoradiographic exposure of the blots of the gels shown in the Upper panel after hybridization to probes corresponding to the ɛ-globin gene silencer (Left) or β-globin gene enhancer (Right). (C) Southern blots of both wild-type and mutant YAC DNAs depicting the size and integrity of the EcoRI fragments bearing the LCR and the ɛ-, γ-, and β-globin genes.
After transformation of yeast clone A201F4.2 with either of the two targeting plasmids, a majority of colonies growing on the selective medium (Trp−Lys−Ura−) were found to have plasmid integrated at the homologous human sequence. This initial targeting event creates a duplication of the homologous sequences separated by the vector, pRS306. Selective excision of the targeting plasmid was performed on medium containing uracil, followed by counterselection on 5-fluroorotic acid-containing plates (lethal to cells retaining the URA3 gene). Excision of pRS306 results in reversion to either the parental YAC structure or replacement of parental sequences by the mutant, now missing either the ɛ-globin silencer or the globin enhancer. The structures of the mutant and wild-type YACs were verified by Southern blot analysis (Fig. 1 B and C, respectively).
Transgenic Mice/Transgene Structure.
The mutated YAC DNAs were isolated from pulse-field gels and purified as described (see Materials and Methods and ref. 26). These DNAs were then microinjected into pronuclei of fertilized mouse ova. Founder tail DNAs were initially screened by PCR, using primers specific for both right and left YAC vector arms, and then verified by Southern blot hybridization. Southern blotting of each of the F1 or F2 lines, using probes that span the locus (22) showed that each contained restriction fragments of the expected size (e.g., Fig. 1C), and that no other bands (indicative of breakpoints in the YACs) were present. Fragmented YAC transgenes detected through this analysis were excluded from further consideration.
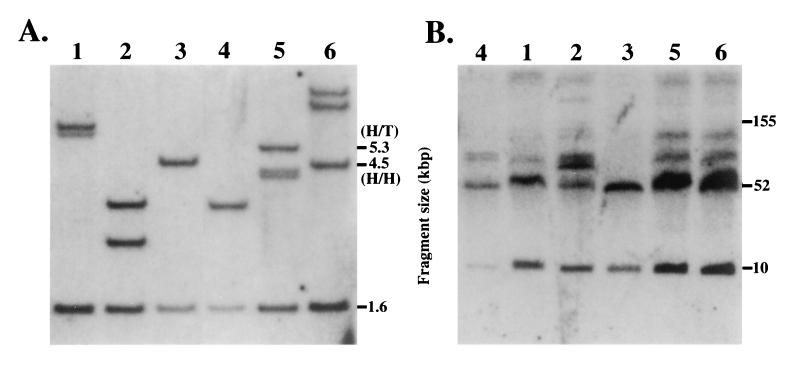
The YAC transgene copy number in the lines chosen for final expression analysis was determined using a YAC end assay, employing a probe that would detect a common 1.6-kbp band as well as other characteristic fragments in each digest. For example, a head-to-head tandem integration was predicted from the map of pYAC4 to yield an additional fragment 4.52 kbp in size (Fig. 2A, lanes 3 and 6), whereas a head-to-tail tandem integration would result in the generation of a fragment of 5.36 kbp (Fig. 2A, lane 5); if the YAC end was embedded in mouse chromosomal DNA (as either a single copy transgene or as the flanking copy of a multi-YAC integration), the size of the fragment would be dependent on the next random PstI site encountered in the mouse genome. From these data, combined with the data obtained from standard Southern blots and comparison of internal fragment intensities, we estimate that the three ɛ-silencer deletion lines contain three, two, and two transgene copies (Fig. 2A, lanes 1–3), respectively, whereas the two β-globin enhancer deletion lines both harbor four tandemly integrated YAC copies (Fig. 2A, lanes 5 and 6). Furthermore, the analysis shows that, as previously stated, the wild-type YAC transgenic line HS4321b (26) is single copy (Fig. 2A, lane 4).
Figure 2.
Copy number and integrity of YAC transgenes. Lanes: 1–3, ɛ-globin silencer deletion mutant transgenic lines 1–3, respectively; 4, wild type β-globin YAC transgenic line HS4321b (26); 5 and 6, β-globin enhancer deletion mutant lines 1 and 2, respectively (numbering at the top of each panel). (A) Tail DNA from F1 or F2 pups representing each of the transgenic lines was digested with PstI and electrophoresed and blotted normally. A 1.7-kbp EarI fragment isolated from plasmid vector pYAC4 was radiolabeled and then hybridized to the Southern blot, and finally exposed for autoradiography. Each lane displays a (common) internal fragment band from the pYAC4 vector (1.58 kbp) as well as other bands, depending on the transgene copy number and integration site. Head-to-head tandem integrations would be expected to yield a 4.5-kbp band, whereas head-to-tail tandem integrations are predicted to yield a 5.3-kbp band. (B) Spleen cells were isolated from each transgenic line and digested with SfiI plus SalI as described prior to electrophoresis on pulse field gels. The gel was then blotted to nylon and hybridized to probes corresponding to either LCR HS4 or the Gγ-globin gene.
The integrity of the YAC transgenes was assayed by examining spleen cell DNA that had been separated on pulse field gels for the presence of 10- and 52-kbp diagnostic bands on Southern blots after digestion with SfiI and SalI (Fig. 2B). The two β-globin locus fragments detected in this assay extend from beyond LCR HS5 at the 5′ end of the locus to a position near the start of the δ-globin gene (22). The data are shown in Fig. 2B, where the two expected diagnostic bands are clearly the most prominent in each of the transgenic lines; in particular, it should be noted that there are no bands detected below 60 kbp other than the two that are expected, thus adding credence to the contention that the YACs examined here are unbroken within the 62-kbp interval surveyed in this assay. It should also be noted that other bands of higher molecular weight are clearly detected in the same blot, but other experiments show that these are primarily due to partial digestion of the spleen cell plugs by SalI, a vexing problem we have not yet fully surmounted. We assume that this partial digestion is due to methylation of some SalI CpG sites.
Sequences Within the ɛ-Globin Gene Silencer Are Important for Embryonic Stage-Specific Activation of Both the ɛ- and γ-Globin Genes.
In transgenic mice, the human ɛ-globin gene is expressed in the embryonic yolk sac beginning between day 8.5 and 9.5 days postcoitus of gestation and switches off by day 12.5 (23–25); this repression is postulated to result from the binding of negative regulatory proteins to the ɛ-globin gene silencer. Multiple regulatory elements have been described in the ɛ-globin gene upstream region (41), and the most prominent of these is a silencer located from −304 to −179 relative to the ɛ-globin gene transcription initiation site (27). This silencer element contains consensus binding sites both for GATA factors and for YY1. If the silencer harbors only the expected negative regulatory activity, one would predict that deletion of this element from an otherwise intact human β-globin gene locus would result in the continued expression of the ɛ-globin gene into the fetal stage of erythropoiesis. We therefore directly tested the hypothesis that the silencer acts to restrict expression of the ɛ-globin gene to the embryonic yolk sac stage.
To our surprise, deletion of the silencer did not lead to high level ɛ-globin gene transcription in the fetal liver period, as anticipated, but rather caused a significant decline in ɛ-globin gene transcription in the yolk sac, thus demonstrating that this narrowly defined silencer sequence element of 125 bp still harbors cryptic activity that is required for stimulation of ɛ-globin RNA synthesis (Fig. 3). An even more unexpected observation was that deletion of the ɛ-globin gene silencer not only diminished ɛ-globin gene transcription, but also resulted in a significant decline in γ-globin RNA synthesis in the yolk sac (Fig. 3). Transcription of the γ- and β-globin genes during both the fetal liver and adult stages was not as significantly affected in these animals, thereby implying that these ɛ-globin 5′ sequences harbor a more general function in regulatory control over globin genes that are expressed in the embryonic yolk sac.
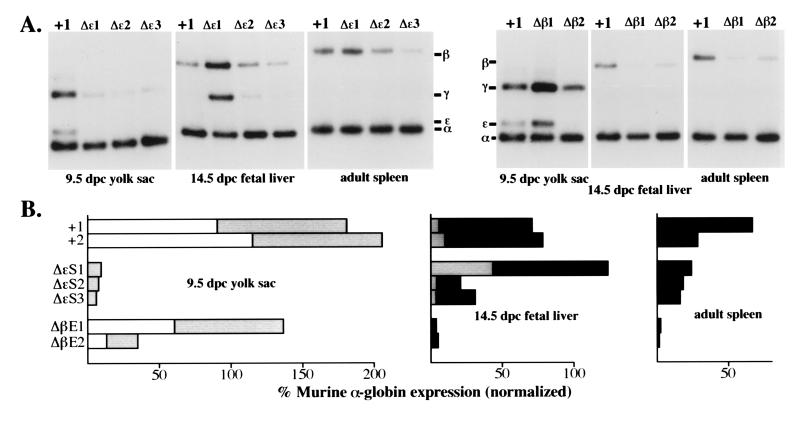
Figure 3.
Expression of human β-globin YAC mutants during erythropoiesis. (A Upper). Typical reverse transcriptase–PCR expression data for the human ɛ-, γ-, and β-globin genes as compared with the murine α-globin used as the internal control for each RNA sample. The transgenic line examined is abbreviated at the top of each lane, while the developmental stage and sites are shown below each panel. A +1 refers to RNA isolated from the single copy wild-type YAC transgenic line, HS4321b (26). (B) Quantification of the expression data shown in A after PhosphorImager analysis and correction for gene copy number (Fig. 2). Open rectangles represent ɛ-globin expression, shaded boxes represent γ-globin expression, and filled boxes represent β-globin expression levels.
We should note that the variation in expression levels is sporadically greater in all of the transgenic lines we have examined in this study than we had observed previously (26), where all the independent lines appeared to express the genes during different developmental stages with no greater than 10% variation. It may be that expression in our previous transgenic lines was fortuitously homogeneous for unknown reasons, or that the expression from single copy transgenes perturbs the erythroid cell transcriptional environment less than the presence of the two to four copies found in the animals under present analysis. Additionally, deletion of important regulatory DNA elements within the locus could (to some extent) affect the position-independent expression of the YAC (G. Li, J.B., and J.D.E., unpublished observations). For these reasons, we focused our interpretation and discussion on those results that display the most dramatic deviation(s) from the expression characteristics of the wild-type YAC transgenic lines and, more importantly, on those results that appeared to be consistent among the mutant lines.
It has been shown previously that binding sites for the ubiquitous transcription factor YY1 and for GATA factors contribute to the repressing activity of this element. However, Raich et al. (44) also demonstrated that two GATA sites immediately downstream of the YY1 site have a pronounced positive effect on ɛ-globin gene transcription. These results clearly demonstrate the presence of additional, functionally important positive regulatory elements even within this narrowly defined silencer region.
Because there are other positive regulatory elements both upstream and downstream of the silencer, it could be argued that the mutant is defective in activation because the missing sequence elements must cooperate with these flanking positive elements to confer efficient globin gene transcription (i.e., that the deleted sequences per se have no positive regulatory effect). Alternatively, changing the spacing between the more upstream and the more gene-proximal positive regulatory elements (after deleting the 125-bp silencer) might impair the formation of an active transcription complex on the ɛ-globin gene promoter. Finally, it is possible that the 3′ part of the deleted silencer is additionally the extreme 5′ end of an extended ɛ-globin gene promoter, thus affecting assembly of the basal apparatus (but clearly this cannot be the only effect; next paragraph). Further studies introducing specific mutations that do not alter the spacing within this region should distinguish between these several possibilities.
The more remarkable observation from analysis of the ɛ-silencer deletion mutant was that it also significantly reduced transcription of the γ-globin genes in erythroid cells of the embryonic yolk sac (Fig. 3). This effect was erythroid stage-specific, not significantly affecting expression of either the γ- or β-globin genes during either the fetal liver or adult stages. We interpret this observation to mean that deletion of sequences within the upstream region of the ɛ-globin gene generally affects embryonic stage-specific globin gene expression. This is the second such mutation in the locus that appears to quite specifically affect the activity of these same genes during the embryonic stage of murine erythropoiesis. We previously reported that duplication of HS4 of the LCR resulted in a decline in ɛ-, and a simultaneous, reciprocal increase in γ-, globin mRNA accumulation in the yolk sac (26). Taken together with the data presented here, both mutations point to an effect which may best be explained by assuming that the LCR as well as the ɛ- and γ-globin genes are in a different chromosomal domain in the yolk sac than in the fetal liver or adult spleen. Under the assumption that an LCR holocomplex must physically interact with other gene proximal protein/DNA complexes, these observations could be further interpreted to infer that deletion of the ɛ-globin silencer not only reduces the ability of the LCR to productively interact with the ɛ-globin gene promoter, but that these sequences may be involved in establishing this specific chromatin conformation that is required for proper γ-globin gene transcription in the murine yolk sac.
Previously, Wijgerde et al. (7) showed that two globin genes can be transcribed inside a single erythroid cell within only a brief time span. In terms of the competition model (10), these data were also interpreted to suggest that an LCR holocomplex (26) somehow distinguishes which gene to activate, presumably depending on the array and concentration of transcription factors present in an erythroid cell at different developmental stages. It has been shown experimentally that the human γ-globin genes are more abundantly expressed than the ɛ-globin gene in the murine embryonic yolk sac, thus implying that in a competitive regulatory environment, the human LCR interacts with the more distant human γ-globin gene promoters either more frequently or more stably than with the ɛ-globin gene promoter at this stage of murine erythropoiesis. However, the ɛ-globin upstream region might be important for recruiting or positioning the LCR, not only to enhance ɛ-globin gene transcription but also to allow efficient interaction with the γ-globin gene promoters by decreasing the distance between LCR and γ-globin genes. Deletion of such a hypothetical LCR “positioning” element would then result in a decline in expression of both the ɛ- and γ-globin genes.
Competition Between γ- and β-Globin Genes Is Not Mediated by the β-Globin 3′ Enhancer.
Previous work has shown that the human fetal γ- and adult β-globin genes are regulated by competition (13), but the participating cis-regulatory elements have not yet been defined. We therefore tested the hypothesis that the adult β-globin gene enhancer might be playing a role in this competition by deleting it from the huβYAC (human β-globin YAC) and then analyzing expression of the globin locus genes.
Deletion of the β-globin 3′ enhancer resulted in a significant loss of β-globin expression both in the fetal liver and in the adult spleen (Fig. 3), indicating, as expected, that this enhancer activity is required for adult β-globin gene transcription (34, 35). However, γ-globin synthesis was not elevated in the fetal liver, nor did its expression persist into the adult stage of erythropoiesis, even though β-globin synthesis had dramatically declined, thereby demonstrating that the adult β-globin gene 3′ enhancer does not play a role in the competition between the γ- and β-globin genes in either the fetus or in adult animals. Furthermore, deletion of the β-globin gene 3′ enhancer had a quite specific effect, solely on the expression of the adult β-globin gene.
Acknowledgments
We thank Jie Fan for outstanding technical assistance. We gratefully acknowledge fellowship support from the Cooley’s Anemia Foundation (Q.L.), the Deutsche Forschungsgemeinschaft (Grant A2:Bu 895/1-1), and the American Heart Association (J. B.), and research support from the National Institutes of Health (HL 24415; J.D.E.).
Footnotes
Abbreviations: YAC, yeast artificial chromosome; HS, hypersensitive sites; LCR, locus control region.
References
- 1.Tuan D, London I M. Proc Natl Acad Sci USA. 1984;81:2718–2722. doi: 10.1073/pnas.81.9.2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tuan D, Solomon W, Li Q, London I M. Proc Nat Acad Sci USA. 1985;82:6384–6388. doi: 10.1073/pnas.82.19.6384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Forrester W C, Takegawa S, Papayannopoulou T, Stamatoyannopoulos G, Groudine M. Nucleic Acids Res. 1987;15:10159–10177. doi: 10.1093/nar/15.24.10159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grosveld F, van Assendelft G B, Greaves D R, Kollias G. Cell. 1987;51:975–985. doi: 10.1016/0092-8674(87)90584-8. [DOI] [PubMed] [Google Scholar]
- 5.Hanscombe O, Whyatt D, Fraser P, Yannoutsos N, Greaves D, Dillon N, Grosveld F. Genes Dev. 1991;5:1387–1394. doi: 10.1101/gad.5.8.1387. [DOI] [PubMed] [Google Scholar]
- 6.Stamatoyannopoulos G. Science. 1991;252:383. doi: 10.1126/science.2017679. [DOI] [PubMed] [Google Scholar]
- 7.Wijgerde M, Grosveld F, Fraser P. Nature (London) 1995;377:209–213. doi: 10.1038/377209a0. [DOI] [PubMed] [Google Scholar]
- 8.Ellis J, Tan U K, Harper A, Michalovich D, Yannoutsos N, Philipsen S, Grosveld F. EMBO J. 1996;15:562–568. [PMC free article] [PubMed] [Google Scholar]
- 9.Fraser P, Pruzina S, Grosveld F. Genes Dev. 1993;7:106–113. doi: 10.1101/gad.7.1.106. [DOI] [PubMed] [Google Scholar]
- 10.Choi O-R, Engel J D. Cell. 1988;55:17–26. doi: 10.1016/0092-8674(88)90005-0. [DOI] [PubMed] [Google Scholar]
- 11.Foley K P, Engel J D. Genes Dev. 1992;6:730–744. doi: 10.1101/gad.6.5.730. [DOI] [PubMed] [Google Scholar]
- 12.Shih D, M, Wall R J, Shapiro S G. Nucleic Acids Res. 1990;18:5465–5472. doi: 10.1093/nar/18.18.5465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Enver T, Raich N, Ebens A J, Papayannopoulou T, Costantini F, Stamatoyannopoulos G. Nature (London) 1990;344:309–313. doi: 10.1038/344309a0. [DOI] [PubMed] [Google Scholar]
- 14.Behringer R R, Ryan T M, Palmiter R D, Brinster R L, Townes T M. Genes Dev. 1990;4:380–389. doi: 10.1101/gad.4.3.380. [DOI] [PubMed] [Google Scholar]
- 15.Dillon N, Grosveld F. Nature (London) 1991;350:252–254. doi: 10.1038/350252a0. [DOI] [PubMed] [Google Scholar]
- 16.Kim C G, Epner E M, Forrester W C, Groudine M. Genes Dev. 1992;6:928–938. doi: 10.1101/gad.6.6.928. [DOI] [PubMed] [Google Scholar]
- 17.Sasaki H, Jones P A, Chaillet J R, Ferguson-Smith A C, Barton S C, Reik W, Surani M A. Genes Dev. 1992;6:1843–1856. doi: 10.1101/gad.6.10.1843. [DOI] [PubMed] [Google Scholar]
- 18.Leighton P A, Ingram R S, Eggenschwiler J, Efstratiadis A, Tilghman S M. Nature (London) 1995;375:34–39. doi: 10.1038/375034a0. [DOI] [PubMed] [Google Scholar]
- 19.Leighton P A, Saam J R, Ingram R S, Stewart C L, Tilghman S M. Genes Dev. 1995;9:2079–2089. doi: 10.1101/gad.9.17.2079. [DOI] [PubMed] [Google Scholar]
- 20.Lewis E B. J Am Med Assoc. 1992;267:1524–1531. [Google Scholar]
- 21.Chess A, Simon I, Cedar H, Axel R. Cell. 1994;78:823–834. doi: 10.1016/s0092-8674(94)90562-2. [DOI] [PubMed] [Google Scholar]
- 22.Gaensler K M L, Burmeister M, Brownstein B H, Taillon-Miller P, Myers R M. Genomics. 1991;10:976–984. doi: 10.1016/0888-7543(91)90188-k. [DOI] [PubMed] [Google Scholar]
- 23.Strouboulis J, Dillon N, Grosveld F. Genes Dev. 1992;6:1857–1864. doi: 10.1101/gad.6.10.1857. [DOI] [PubMed] [Google Scholar]
- 24.Gaensler K M, Kitamura M, Kan Y W. Proc Natl Acad Sci USA. 1993;90:11381–11385. doi: 10.1073/pnas.90.23.11381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peterson K R, Clegg C H, Huxley C, Josephson B M, Haugen H S, Furukawa T, Stamatoyannopoulos G. Proc Natl Acad Sci USA. 1993;90:7593–7597. doi: 10.1073/pnas.90.16.7593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bungert J, Davé U, Lieuw K E, Lim K-C, Shavit J A, Liu Q, Engel J D. Genes Dev. 1995;9:3083–3096. doi: 10.1101/gad.9.24.3083. [DOI] [PubMed] [Google Scholar]
- 27.Cao S X, Gutman P D, Dave H P, Schechter A N. Proc Natl Acad Sci USA. 1989;86:5306–5309. doi: 10.1073/pnas.86.14.5306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mantovani R, Superti-Furga G, Gilman J, Ottolengi S. Nucleic Acids Res. 1989;17:6681–6691. doi: 10.1093/nar/17.16.6681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martin D I K, Tsai S-F, Orkin S H. Nature (London) 1989;338:435–438. doi: 10.1038/338435a0. [DOI] [PubMed] [Google Scholar]
- 30.Weatherall D J, Clegg J B, Higgs D R, Wood W G. The Hemoglobinopathies. 6th Ed. New York: McGraw-Hill; 1989. pp. 2281–2339. [Google Scholar]
- 31.Berry M, Grosveld F, Dillon N. Nature (London) 1992;358:499–502. doi: 10.1038/358499a0. [DOI] [PubMed] [Google Scholar]
- 32.Stamatoyannopoulos G, Josephson B, Zhang J W, Li Q. Mol Cell Biol. 1993;13:7636–7644. doi: 10.1128/mcb.13.12.7636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bodine D, Ley T. EMBO J. 1987;6:2997–3004. doi: 10.1002/j.1460-2075.1987.tb02605.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Behringer R R, Hammer R E, Brinster R L, Palmiter R D, Townes T M. Proc Natl Acad Sci USA. 1987;84:7056–7060. doi: 10.1073/pnas.84.20.7056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Antoniou M, deBoer E, Habets G, Grosveld F. EMBO J. 1988;7:377–384. doi: 10.1002/j.1460-2075.1988.tb02824.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gutman P D, Cao S X, Dave H P, Mittelman M, Schechter A N. Gene. 1992;110:197–203. doi: 10.1016/0378-1119(92)90648-9. [DOI] [PubMed] [Google Scholar]
- 37.Raich N, Papayannopoulou T, Stamatoyannopoulos G, Enver T. Blood. 1992;79:861–864. [PubMed] [Google Scholar]
- 38.Wada K Y, Peters B, Noguchi C T. J Biol Chem. 1992;267:11532–11538. [PubMed] [Google Scholar]
- 39.Gumucio D L, Shelton D A, Bailey W J, Slightom J L, Goodman M. Proc Natl Acad Sci USA. 1993;90:6018–6022. doi: 10.1073/pnas.90.13.6018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peters B, Merezhinskaya N, Diffley J F, Noguchi C T. J Biol Chem. 1993;268:3430–3437. [PubMed] [Google Scholar]
- 41.Trepicchio W L, Dyer M A, Baron M H. Mol Cell Biol. 1993;13:7457–7468. doi: 10.1128/mcb.13.12.7457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Watt P, Lamb P, Proudfoot N J. Gene Expression. 1993;3:61–75. [PMC free article] [PubMed] [Google Scholar]
- 43.Perez S C. J Biol Chem. 1994;269:33109–33115. [PubMed] [Google Scholar]
- 44.Raich N, Clegg C H, Grofti J, Romeo P H, Stamatoyannopoulos G. EMBO J. 1995;14:801–809. doi: 10.1002/j.1460-2075.1995.tb07058.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guthrie, C. & Fink, G. R. (1991) Methods Enzymol. 194. [DOI] [PubMed]
- 46.Sikorski R S, Hieter P. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Peterson K R, Zitnik G, Huxley C, Lowrey C H, Gnirke A, Leppig K A, Papayannopoulou T, Stamatoyannopoulos G. Proc Natl Acad Sci USA. 1993;90:11207–11211. doi: 10.1073/pnas.90.23.11207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schedl A, Larin Z, Montoliu L, Thies E, Kelsey G, Lehrach H, Schutz G. Nucleic Acids Res. 1993;21:4783–4787. doi: 10.1093/nar/21.20.4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hogan B, Costantini F, Lacy E. Manipulating the Mouse Embryo. Plainview, NY: Cold Spring Harbor Lab. Press; 1986. [Google Scholar]
- 50.Gong Q-H, Stern J, Dean A. Mol Cell Biol. 1991;11:2558–2566. doi: 10.1128/mcb.11.5.2558. [DOI] [PMC free article] [PubMed] [Google Scholar]