Abstract
Horizontal gene transfer is thought to contribute to the wide distribution of group I introns among organisms. Integration of an intron into foreign RNA or DNA by reverse self-splicing, followed by reverse transcription and recombination, could lead to its transposition. Reverse self-splicing of group I introns has been demonstrated in vitro, but not in vivo. Here we report RNA-dependent integration of the Tetrahymena intron into the 23S rRNA in Escherichia coli. Analysis of products by Northern blot and reverse transcription–PCR amplification revealed precise intron insertion into a site homologous to the natural splice junction. Products are sensitive to treatment with RNase but not DNase and depend on the splicing activity of the intron. Partial reaction with 11 novel sites in the 23S RNA that are complementary to the guide sequence of the intron illustrates lower specificity than intron homing. Reverse splicing of the Tetrahymena intron in bacteria demonstrates the possibility of RNA-catalyzed transposition of group I introns in foreign hosts.
Group I introns are small genetic elements that are capable of catalyzing their splicing from primary transcripts (1). These introns are found in the genomes of bacteria and organelles, as well as the rDNA of lower eukaryotes (2). The broad distribution of group I introns has been attributed to genetic mobility (3, 4), but the mechanisms of dispersal remain in question (2). It is well established that group I introns can transpose site-specifically to intronless alleles of the same gene after cleavage of the target DNA by an intron-encoded DNA endonuclease (2, 5). This process, termed homing, is highly specific because of the large recognition sites (15–35 bp) of homing endonucleases (5).
Reverse splicing, coupled with reverse transcription and recombination, has been proposed as an alternative mechanism for intron mobility (6–8). A related mechanism after reverse splicing into DNA has been shown to operate during homing of group II introns in yeast mitochondria (9, 10). Self-splicing of the group I intron from Tetrahymena thermophila rDNA is fully reversible in vitro (8) and yields a product that resembles the precursor RNA. Substrates for reverse splicing are recognized by a short sequence (4–6 nt) that can base-pair with the internal guide sequence (IGS) of the intron (6). Therefore, reverse splicing is expected to be less sequence-specific than homing endonucleases and could result in transposition to new genes. Indeed, partial reverse splicing of a group I intron from Chlamydomonas reinhardtii chloroplast 23S rRNA into the cytoplasmic 5.8S rRNA in vitro has been reported (11). Here, we provide an example of reverse splicing of a group I intron in vivo by demonstrating that the Tetrahymena intron can integrate into 23S rRNA of E. coli.
MATERIALS AND METHODS
Growth of Cells and Plasmid Construction.
Plasmid pMALEC-IVS was prepared by ligating an EcoRI-HindIII fragment containing the Tetrahymena EC intron into pMAL-c2 (12) such that the lacZ′ translation frame is restored by splicing. The intron-containing fragment was prepared by PCR amplification of pTZEC (13). JM109 cells transformed with pMALEC-IVS were grown in Luria–Bertani medium plus 50 mg/liter ampicillin at 37°C until A600 = 0.4, before addition of 1 mM isopropyl β-d-thiogalactoside (IPTG). Chloramphenicol (100 mg/liter) was added immediately before harvest. pLKEC(438), which encodes a pre-RNA with short exons under control of the λ PL promoter (14), was induced by shifting cultures to 42°C. The sequences 8 nt preceding and the 29 nt following the intron are identical in pLKEC(438) and pMALEC-IVS.
RNA Isolation and Northern Blots.
Total RNA was isolated from 10 ml of culture after lysis with guanidine isothiocyanate (Perfect RNA kit, 5 Prime → 3 Prime). In control reactions, linear intron RNA was transcribed with T7 RNA polymerase and purified from 4% polyacrylamide gels, and 15 pmol was added to the guanidine isothiocyanate solution before lysis. For Northern blots, total RNA was denatured with glyoxal before electrophoresis in a 1.2% agarose gel, transferred to Nytran-S membrane (Schleicher & Schuell), and hybridized with a uniformly 32P-labeled intron-specific or malE-specific probe. Primer extensions in the presence of ddCTP were carried out as previously described (15) by using primer IP006–18, which is complementary to the 5′ end of the EC intron. Reactions contained 0.1 pmol 32P-labeled primer and 0.25 μg total RNA. Extension products specific for the cleaved or uncleaved 5′ splice site were quantitated as reported previously (15).
Membranes were quantitated by using a PhosphorImager (Molecular Dynamics) to determine the ratio of intron to reverse-splicing product. The moles of intron and product RNAs in 1 μg total RNA after 120 min of induction was estimated by comparison with a standard curve prepared with known amounts of intron RNA. The moles of 23S rRNA per μg total RNA was 0.48 and was determined by hybridization of dot blots with 23S probes (16).
Reverse Transcription (RT) and PCR Amplification.
For RT-PCR, total RNA was filtered through a Chromaspin TE-1000 gel filtration column (CLONTECH) to eliminate unreacted intron RNA. cDNA was synthesized in a 10-μl reaction containing 0.5 μg RNA, 5 pmol downstream primer, 25 mM Tris⋅HCl (pH 7.5), 60 mM NaCl, 10 mM DTT, 3 mM magnesium acetate, 0.5 mM deoxynucleotide triphosphates, and 2 units of AMV reverse transcriptase (Life Sciences) at 48°C for 45 min. Primer annealing was performed at 65°C for 3 min in the absence of magnesium, nucleotides, and enzyme. Reactions were terminated by incubating at 95°C for 2 min.
A 2-μl aliquot from reverse transcription was amplified by touchdown PCR (17). Fifty-microliter reactions contained 50 mM KCl, 10 mM Tris⋅HCl (pH 9), 0.1% Triton X-100, 5 μg gelatin, 0.2 mM each deoxynucleoside triphosphate, 1 mM MgCl2, 1 μM each downstream and upstream primers, and 2.5 units Taq DNA polymerase (Promega). Initial parameters: two cycles of 95°C for 1 min; 65°C for 1 min; and 72°C for 1 min. Subsequently, the annealing temperature of the reaction was decreased 1°C every second cycle from 65 to 55°C, at which temperature 10 cycles were carried out. Typically, 10 μl was analyzed on a 1.6% agarose gel and stained with ethidium bromide. Primers: DP-1286, AAACGGATCCTTTCACCCGCTTTATCGTTAC; UPI-94, DPI-336, UP-1666, DP-2180, and DP-2675 were described previously (13).
RESULTS
Reverse Splicing of Tetrahymena Intron in E. coli.
To investigate reverse splicing in vivo, the Tetrahymena intron was expressed in E. coli by inserting the intron sequences into an expression vector under the control of the IPTG-inducible Ptac promoter (Fig. 1A). Natural exon sequences were minimized (8 bp 5′ and 29 bp 3′) to reduce the possibility of homologous recombination between the plasmid and E. coli rRNA genes. The intron is spliced by the normal group I mechanism in E. coli (18) at a rate similar to that in Tetrahymena (15), suggesting that a species-specific protein cofactor is not required for splicing in vivo.
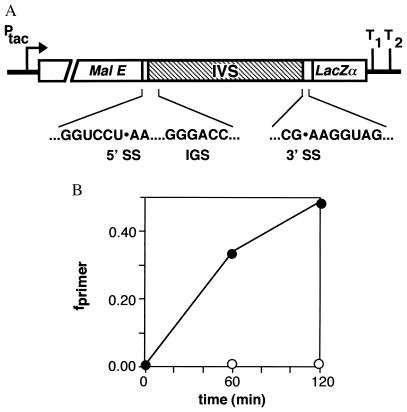
Figure 1.
Expression of the Tetrahymena group I intron in E. coli. (A) Diagram of pMALEC-IVS. An EcoRI-HindIII fragment containing the Tetrahymena intron (IVS) was ligated into pMAL-c2 (12). The 5′ exon contains 8 bp and the 3′ exon contains 29 bp of rRNA sequence. Sequences of 5′ and 3′ splice sites (SS) and the IGS are shown. (B) Induction of intron RNA vs. time after addition of IPTG. Fraction of primer extension product corresponding to spliced intron (closed) and pre-RNA (open) are plotted vs. time after addition of IPTG to cultures. The primer anneals to the 5′ end of the intron and distinguishes whether the 5′ splice site has been cleaved. As almost no pMALEC-IVS intron–3′ exon intermediate can be detected on Northern blots, this product corresponds closely to the amount of linear intron in the cell.
The high concentration of rRNA is expected to provide an abundant substrate for reverse splicing in vivo and could account for the preferential distribution of group I introns among rRNA genes (11, 19). Accordingly, the intron was targeted against the site in the E. coli 23S rRNA that is homologous to its natural splice junction by mutation of the IGS to 5′-GGGACC (Fig. 1A), such that the expected site of reaction is between U1926 and A1927. The mutated intron, which we designate “EC IVS,” splices efficiently when inserted in the rrnB operon at this position of the 23S coding sequence (15). We have also observed reverse splicing in vitro at this site with both 23S rRNA and 50S subunits as substrates (13).
Total RNA was isolated from cells transformed with pMALEC-IVS (Fig. 1A) 0–2 hr after induction with IPTG. Accumulation of intron RNA was monitored by primer extension that detects cleavage of the 5′ splice site (15). Only 0.2% of plasmid transcripts were unspliced after 2 hr of induction (Fig. 1B). To determine whether the intron had reacted with other transcripts, total RNA was separated on agarose gels and hybridized with an intron-specific probe (Fig. 2A). This revealed several large, intron-containing bands that increased in intensity with time after induction of the intron. These RNAs were not derived from the pMALEC-IVS precursor, which was identified by hybridization of the membrane with a malE probe (data not shown).
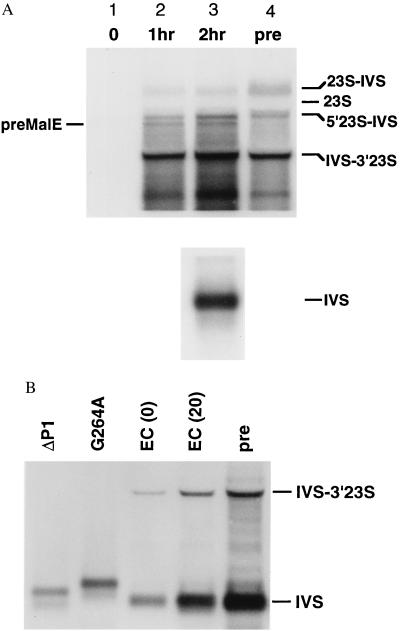
Figure 2.
Analysis of intron-containing RNAs. (A) Northern blot of intron-containing RNAs and hybridization with a probe specific for the Tetrahymena intron. Lanes 1–3, cells transformed with pMALEC-IVS and grown 0, 1 hr, or 2 hr in the presence of IPTG; lane 4 (pre), transformed with a plasmid containing the rrnB operon with the Tetrahymena intron at position 1926 of the 23S rRNA [pLK45-IVS:C26A (16)]. (Upper) Five micrograms total RNA, the bottom of the membrane containing the free intron was removed before hybridization. (Lower) Lane 3, 0.5 μg of total RNA. 23S-IVS, unspliced 23S rRNA plus intron; 23S, mature 23S rRNA; 5′23S-IVS, 5′ exon–intron; IVS-3′23S, intron–3′ exon intermediate; IVS, spliced intron; preMalE, initial transcript of pMALEC-IVS. Approximate sizes were determined by comparison with molecular weight standards (not shown) and to 23S and 16S rRNAs. (B) Northern blot of splicing-defective mutants, as in A. Cells were transformed with derivatives of pLKEC-IVS (14). ΔP1, deletion of P1; G264A, G-binding site; EC, active intron at 0 and 20 min; pre, pre-23S rRNA as in A. ΔP1 and G264A RNAs are inefficiently spliced and migrate more slowly than the linear intron. Bands represent ΔP1IVS-3′exon and G264A pre-RNA, respectively.
Several of these novel RNAs were found to comigrate with in vivo splicing products of an authentic pre-23S rRNA (Fig. 2A, lane 4). The pre-23S rRNA was obtained from a control strain transformed with a plasmid containing a poorly spliced variant of the EC intron (EC:C26A) inserted into the 23S gene after U1926 (16). The 1.6-kb RNA in lanes 2 and 3 (Fig. 2A) has the same mobility as the splicing intermediate (IVS-3′ 23S), in which the 3′ end of the intron is ligated to A1927 of the 23S rRNA. More exciting was to find that we could also detect a larger product that comigrated with the full-length pre-23S rRNA (pre, Fig. 2A). We postulated that these intron-containing products represented RNAs that had undergone the first step or both steps of reverse splicing with the 23S rRNA, respectively. The significant accumulation of the IVS-3′ 23S product (lane 3, Fig. 2A) suggested that reverse splicing was most likely to occur at U1926, the position targeted by the IGS mutations. We also detected a 2.4-kb RNA (5′23S-IVS) that is the size expected if the 5′ half of the 23S was ligated to the intron. Such a product is best explained by hydrolysis of the 3′ splice site of a complete “integrant.”
We next investigated whether these products depended on the self-splicing activity of the intron by introducing two splicing-deficient mutations. The EC-ΔP1 intron (14) is unable to undergo reverse splicing because of deletion of the P1 helix (nucleotides 1–30) that contains the IGS (16). The mutation G264 to A in the intron core damages the guanosine-binding site and significantly reduces its catalytic activity (20). None of the intron-containing products were observed when either of these splicing-defective introns were expressed in E. coli as short transcripts (Fig. 2B).
These short transcripts have the same 8- and 29-nt sequences flanking the intron [pLKEC(438)] (14, 16), but lack the upstream mal E sequences to avoid the instability of unspliced pMALEC-IVS transcripts. Active EC IVS expressed from pLKEC at 42°C yielded results comparable to those shown in Fig. 2A, although expression of the intron was 2- to 3-fold lower (16). The temperature of induction has only a modest effect on reverse splicing in vivo, as similar results were obtained at 30, 37, and 42°C from cells transformed with pMALEC-IVS (data not shown).
Amplification of Intron–23S rRNA Junctions.
To determine whether the products detected by Northern hybridization contain the sequences of the expected reverse-splicing products, 5′ and 3′ integration junctions in the 23S rRNA were amplified by RT-PCR (13). The 3′ integration junction at position 1926 was amplified with an upstream primer that anneals to the intron and a downstream primer complementary to 23S rRNA (Fig. 3A). The corresponding 5′ integration junction was amplified by using a downstream primer complementary to intron sequences and an upstream primer complementary to the 5′ exon (Fig. 3B). PCR products of the expected sizes for both 5′ and 3′ integration junctions were readily obtained, regardless of whether the intron was expressed from pMALEC-IVS (Fig. 3) or pLKEC (data not shown). Sequencing of PCR products confirmed that they contained precise junctions corresponding to insertion of the full-length intron between U1926 and A1927 of the 23S rRNA.
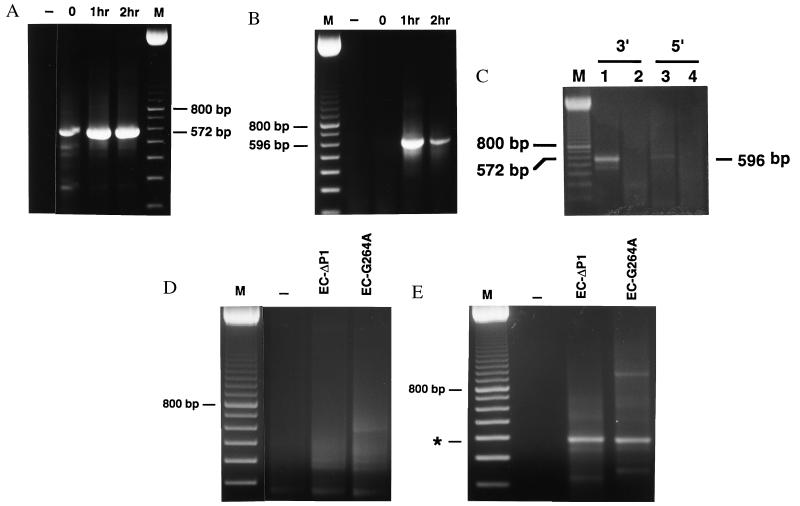
Figure 3.
RT-PCR amplification of intron–23S rRNA junctions. RNA was from cells transformed with pMALEC-IVS. Lanes: (–), no template; 0, 1, and 2 hr, time after IPTG induction; M, 100-bp ladder. (A) Amplification of 3′ integration junctions in 23S domain IV. Upstream primer (UPI-94) is intron-specific; downstream primer (DP-2180) anneals to 23S rRNA. The 572-bp product represents 3′ integration junctions at positions 1924, 1925, and 1926. (B) Amplification of 5′ integration junctions in 23S domain IV, with a 23S-specific upstream primer (UP-1666) and intron-specific downstream primer (DPI-336). The 596-bp fragment represents the 5′ integration junction at position 1926. (C) Lanes 1 and 3, total RNA from cells transformed with pMALEC-IVS after 2 hr of induction; lanes 2 and 4, in vitro transcribed intron RNA added during isolation of total RNA from cells transformed with pMAL-c2. RT-PCR as in A and B. (D) RT-PCR of splicing-defective mutants. Reactions as in A. RNA was from cells transformed with pLKEC-ΔP1 and pLKEC-G264A. (E) As in C, except using primers from B. ∗, 340-bp product of spurious single-primer amplification of intron RNA.
Integration junctions were not amplified (5′) or weakly amplified (3′) when IPTG was not added during cell growth, confirming that integration events are dependent on expression of the intron. PCR products were not obtained when a comparable amount of linear intron RNA was added to untransformed cells during lysis, showing that reverse splicing did not occur in vitro after RNA isolation (Fig. 3C). Neither 3′ nor 5′ integration junctions were amplified in RT-PCRs with RNA from the splicing-defective strains EC-ΔP1 and EC-G264A (Fig. 3 D and E). As the intron must be catalytically active for reverse splicing to occur, these controls confirmed that integration is the result of the reactivity of the intron and not recombination between the plasmid and rRNA genes or template switching during PCR amplification. Furthermore, 5′ and 3′ integration junctions were detected in samples treated with DNase, but not RNase (data not shown). This confirms the previous result and substantiates that the integration target is RNA, not DNA.
Reaction at Novel Sites.
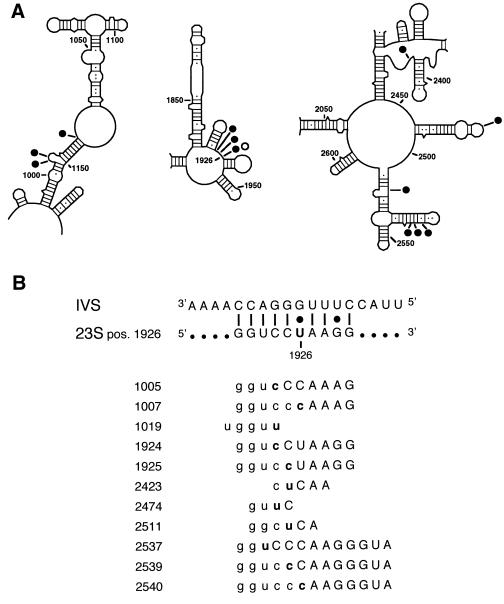
As reverse splicing only requires 4- to 6-nt recognition sequence in the substrate (8), long RNAs are expected to contain several possible integration targets. The E. coli 23S RNA contains five sites that are fully complementary to the EC IGS. To determine whether the intron reacts with other sites in the 23S rRNA in vivo, integration junctions were amplified by using the strategy above, but with primers specific for 23S rRNA domains II and V (Fig. 4). Although 5′ junctions were not detected at sites other than 1926, PCR products spanning putative 3′ integration junctions were cloned and sequenced. We identified a total of 12 reaction sites for EC-intron in the 23S rRNA from 39 positive clones (Fig. 5A). This probably represents a minimum number, as minor products may have been lost during amplification.
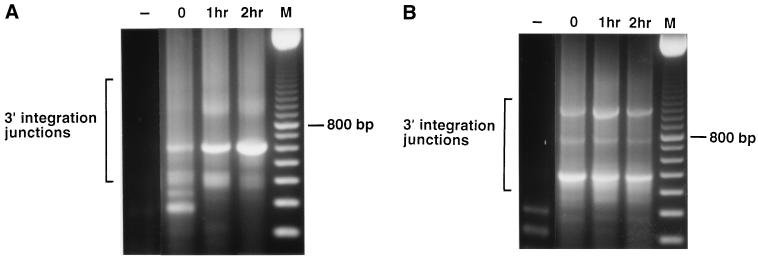
Figure 4.
RT-PCR of 3′ integration junctions in domains II and V of the 23S rRNA. Reactions as in Fig, 2, except that the PCR primers were specific for domains II and V. (A) 3′ integration junctions in domain II. Primers were UPI-94 and DP-1286. (B) 3′ integration junctions in domain V. Primers were UPI-94 and DP-2675. M, 100-bp ladder.
Figure 5.
Sites of intron integration in the 23S rRNA. (A) Reaction sites in domains II, IV, and V. Solid circle, 3′ junctions; open circle, 5′ junction. Position 1926 in domain IV is indicated. Secondary structure is from ref. 32. (B) Alignment of target sequences. Numbers on the left give the position preceding the intron insertion, and sequences on the right are complementary to the IGS, shown at the top. Uppercase, experimentally determined sequence; lowercase, inferred from 23S sequence; boldface, nucleotide 5′ of the integration site.
Sequences immediately upstream and downstream of the integration sites were analyzed for base complementarity with the IGS (Fig. 5B). All of the target sequences could form at least 4 bp with the IGS, and some as many as 13 bp. Several products resulted from attack at different positions within the same target sequence (e.g., 1924, 1925, 1926). Interestingly, the cleavage site was preceded by a C in half of the integration products (Fig. 5B). In most group I introns the 5′ exon ends in a U, which forms a U⋅G wobble pair that contributes to correct positioning of the 5′ splice site in the active site of the intron (21, 22). However, a C-G pair at the 5′ splice site does not inhibit the second step of splicing (23, 24) or reverse splicing in vitro (13). In vivo, we cannot distinguish whether the prevalence of 3′ junctions is due to inefficiency of the second step of reverse splicing or to rapid cleavage of the 5′ splice sites of novel integrants by G-addition.
Efficiency of Intron Integration in Vivo.
The extent of reverse splicing in vivo was estimated by quantitation of Northern blots as described in Methods and from the mole ratio of intron to 23S rRNA after 120 min induction, which was approximately 1:3. From these data, approximately 2% of the 23S rRNA formed a 3′ junction with the EC-IVS and as much as 0.1% underwent complete intron integration. Remarkably, this is only 10-fold less efficient than reverse splicing in vitro (8, 13). This is despite the fact that the ratio of intron to substrate is lower in vivo than in typical in vitro reactions and that some products of the reverse reaction in vivo are probably destroyed by forward splicing or by hydrolysis of the newly created 5′ or 3′ splice sites. As splicing of the Tetrahymena intron is rapid in E. coli (15), the integrants that we are able to detect may represent a stable subset of products that are unable to undergo forward splicing, perhaps because of a conformational change in the RNA (25).
CONCLUSION
This report presents evidence for complete reverse splicing of a group I intron in vivo. Expression of the Tetrahymena intron in E. coli resulted in integration of the intron into a site homologous to the natural splice junction in the bacterial 23S rRNA. The expression of splicing-defective mutants did not generate integration products, demonstrating that the process depends on the catalytic activity of the intron. Additional products, representing the first step of reverse splicing, arise from the attack of the intron at other locations in the 23S rRNA. Analysis of sequences flanking the reaction sites clearly shows that these novel sites are specified by complementarity with the IGS.
These experiments do not distinguish whether integration occurred in newly synthesized rRNA or into ribosomal complexes. Nucleotides near the integration site (U1926) interact with peptidyl tRNA and elongation factors and consequently are thought to be positioned at the surface of the 50S subunit (26). We have recently shown that the EC intron remains noncovalently bound to 50S complexes in E. coli and that this also involves base pairing with the 23S rRNA (16).
During homing of group II introns in yeast mitochondria, the template for target-primed reverse transcription is provided by direct integration of the intron into the DNA via partial or full reverse splicing (9, 10). Target recognition also appears to involve base pairing with the intron RNA (27), and this could lead to occasional integration at nonallelic sites (28, 29). Our data show that transient acquisition of a self-splicing intron by reverse splicing into an abundant transcript, such as rRNA, is quite feasible. Unlike group I and group II homing, this mechanism does not require intron-encoded proteins. However, stable transposition into the genome would presumably require reverse transcriptase activity in the host (2). This activity could be supplied by “indigenous” group II introns (30) or by retroelements prevalent in many cell types (31). Importantly, we find that reverse splicing is significantly less specific than homing and could expand the repertoire of intron-containing sites. These results affirm the hypothesis that reverse splicing contributes to the spread of intron sequences during evolution and suggest ways in which self-splicing introns could be used as simple genetic vectors.
Acknowledgments
This work was supported by the Pew Scholars Program in the Biomedical Sciences (S.W.) and a National Institutes of Health predoctoral fellowship (J.R.). We thank Mary Rubin and Tania Nikolcheva for their assistance and Stephen Mount for helpful comments on the manuscript.
ABBREVIATIONS
- IGS
internal guide sequence
- IPTG
isopropyl β-d-thiogalactoside
- RT
reverse transcription
References
- 1.Cech T R. Annu Rev Biochem. 1993;59:543–568. doi: 10.1146/annurev.bi.59.070190.002551. [DOI] [PubMed] [Google Scholar]
- 2.Belfort M, Perlman P S. J Biol Chem. 1995;270:30237–30240. doi: 10.1074/jbc.270.51.30237. [DOI] [PubMed] [Google Scholar]
- 3.Dujon B. Gene. 1989;82:91–114. doi: 10.1016/0378-1119(89)90034-6. [DOI] [PubMed] [Google Scholar]
- 4.Cavalier-Smith T. Trends Genet. 1991;7:145–148. [PubMed] [Google Scholar]
- 5.Byrk M, Mueller J E. In: Ribosomal RNA and Group I Introns. Green R, Schroeder R, editors. Austin, TX: R. G. Landes; 1996. pp. 221–241. [Google Scholar]
- 6.Cech T R. Int Rev Cytol. 1985;93:3–22. doi: 10.1016/s0074-7696(08)61370-4. [DOI] [PubMed] [Google Scholar]
- 7.Sharp P A. Cell. 1985;42:397–400. doi: 10.1016/0092-8674(85)90092-3. [DOI] [PubMed] [Google Scholar]
- 8.Woodson S A, Cech T R. Cell. 1989;57:335–345. doi: 10.1016/0092-8674(89)90971-9. [DOI] [PubMed] [Google Scholar]
- 9.Zimmerly S, Guo H, Perlman P S, Lambowitz A M. Cell. 1995;82:545–554. doi: 10.1016/0092-8674(95)90027-6. [DOI] [PubMed] [Google Scholar]
- 10.Yang J, Zimmerly S, Perlman P S, Lambowitz A M. Nature (London) 1996;381:332–334. doi: 10.1038/381332a0. [DOI] [PubMed] [Google Scholar]
- 11.Thompson A J, Herrin D L. J Mol Biol. 1994;236:455–468. doi: 10.1006/jmbi.1994.1157. [DOI] [PubMed] [Google Scholar]
- 12.Maina C V, Riggs P D, Grandea A G, III, Slatko B E, Moran L S, Tagliamonte J A, McReynolds L A, Guan C D. Gene. 1988;74:365–373. doi: 10.1016/0378-1119(88)90170-9. [DOI] [PubMed] [Google Scholar]
- 13.Roman J, Woodson S A. RNA. 1995;1:478–490. [PMC free article] [PubMed] [Google Scholar]
- 14.Roman J. Ph.D. thesis. College Park: University of Maryland; 1996. [Google Scholar]
- 15.Zhang F, Ramsey E S, Woodson S A. RNA. 1995;1:284–292. [PMC free article] [PubMed] [Google Scholar]
- 16.Nikolcheva T, Woodson S A. RNA. 1997;3:1016–1027. [PMC free article] [PubMed] [Google Scholar]
- 17.Don R H, Cox P T, Wainwright B J, Baker K, Mattick J S. Nucleic Acids Res. 1991;19:4008. doi: 10.1093/nar/19.14.4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Price J V, Cech T R. Science. 1985;228:719–722. doi: 10.1126/science.2986286. [DOI] [PubMed] [Google Scholar]
- 19.Damberger S H, Gutell R R. Nucleic Acids Res. 1994;22:3508–3510. doi: 10.1093/nar/22.17.3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Michel F, Hanna M, Green R, Bartel D P, Szostak J W. Nature (London) 1989;342:391–395. doi: 10.1038/342391a0. [DOI] [PubMed] [Google Scholar]
- 21.Knitt D S, Narlikar G J, Herschlag D. Biochemistry. 1994;33:13864–13879. doi: 10.1021/bi00250a041. [DOI] [PubMed] [Google Scholar]
- 22.Strobel S A, Cech T R. Science. 1995;267:675–679. doi: 10.1126/science.7839142. [DOI] [PubMed] [Google Scholar]
- 23.Barfod E T, Cech T R. Mol Cell Biol. 1989;9:3657–3666. doi: 10.1128/mcb.9.9.3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Doudna J A, Cormack B P, Szostak J W. Proc Natl Acad Sci USA. 1989;86:7402–7406. doi: 10.1073/pnas.86.19.7402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Woodson S A, Emerick V L. Mol Cell Biol. 1993;13:1137–1145. doi: 10.1128/mcb.13.2.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Noller H F. Annu Rev Biochem. 1991;60:191–227. doi: 10.1146/annurev.bi.60.070191.001203. [DOI] [PubMed] [Google Scholar]
- 27.Eskes R, Yang J, Lambowitz A M, Perlman P S. Cell. 1997;88:865–874. doi: 10.1016/s0092-8674(00)81932-7. [DOI] [PubMed] [Google Scholar]
- 28.Müller M W, Allmaier M, Eskes R, Schweyen R J. Nature (London) 1993;366:174–176. doi: 10.1038/366174a0. [DOI] [PubMed] [Google Scholar]
- 29.Sellem C H, Lecellier G, Belcour L. Nature (London) 1993;366:176–178. doi: 10.1038/366176a0. [DOI] [PubMed] [Google Scholar]
- 30.Kennell J C, Moran J V, Perlman P S, Butow R A, Lambowitz A M. Cell. 1993;73:133–146. doi: 10.1016/0092-8674(93)90166-n. [DOI] [PubMed] [Google Scholar]
- 31.Eickbush T H. In: Evolutionary Biology of Viruses. Morse S S, editor. New York: Raven; 1994. pp. 121–157. [Google Scholar]
- 32.Noller H F, Kop J A, Wheaton V, Brosius J, Gutell R R, Kopylov A M, Dohme F D, Herr W, Stahl D A, Gupta R, Woese C R. Nucleic Acids Res. 1981;9:6167–6189. doi: 10.1093/nar/9.22.6167. [DOI] [PMC free article] [PubMed] [Google Scholar]