Abstract
Replication of HIV-1 requires the viral Tat protein, which increases the extent of transcription elongation by RNA polymerase II after activation at the single viral long terminal repeat (LTR) promoter. This effect of Tat on transcription requires Tat interactions with a 5′ region (TAR) in nascent transcripts as well as Tat-specific cofactors. The present study identifies a cellular protein, TIP30, that interacts with Tat and with an SRB-containing RNA polymerase II complex both in vivo and in vitro. Coexpression of TIP30 specifically enhances transactivation by Tat in transfected cells, and immunodepletion of TIP30 from nuclear extracts abolishes Tat-activated transcription without affecting Tat-independent transcription. These results implicate TIP30 as a specific coactivator that may enhance formation of a Tat–RNA polymerase II holoenzyme complex.
The HIV-1 genome encodes a regulatory protein, Tat, that functions primarily to increase the level of transcription from the single promoter in the viral long terminal repeat (LTR) during viral growth (reviewed in ref. 1). Unlike most transcriptional activators, Tat binds to an RNA element, termed the Tat-responsive (TAR) element, near the 5′ end of the nascent transcript and acts mainly to enhance transcriptional elongation (1). Tat contains a TAR-binding domain and a nuclear localization signal, which lie in the carboxyl-terminal residues 49–58, and an activation domain, which lies within the amino-terminal 48 aa (1). Genetic and biochemical studies have suggested that at least two cellular cofactors are required for transactivation by Tat in human cells: one that binds to the loop of TAR and facilitates binding of Tat to TAR and another that interacts with the amino-terminal portion of Tat (1). Although several cellular proteins have been found to interact with TAR RNA in vitro, only the 185-kDa protein termed TRP-185 appears to be a prominent candidate for the primary TAR-binding cofactor (2, 3). Binding of TRP-185 to TAR depends on the loop sequences of TAR and on TRP-185-associated factors. Therefore, TRP-185 and its associated factors may represent the TAR loop-binding cofactor of Tat.
Evidence for the existence of a Tat-interacting cofactor activity in human cells first emerged from studies demonstrating that Tat mutants lacking the basic domain were able to inhibit transactivation by Tat (4, 5). Indirect studies also suggested that this cofactor activity can interact with the activation domain of HIV-1 Tat, but not with the activation domain of the herpes simplex virus protein VP16 (ref. 4). Earlier-reported candidates for Tat-interacting cofactors include TBP1 (6), TAP (7), Tip60 (8), and HT2A (9). Although these proteins have some of the expected characteristics of a Tat cofactor, none have been determined to participate directly as cofactors in the action of Tat. More recent studies using cell-free transcription assays have reported the partial purification of a novel Tat cofactor called Tat-SF (10). One component (Tat-SF1) of Tat-SF was identified as a 140-kDa phosphorylated protein, and a cognate cDNA has been cloned (11). The importance of Tat-SF1 for Tat-dependent transcription was suggested by combined immunodepletion and in vitro transcription assays and by cotransfection assays. Because the in vitro immunodepletion assays were performed with a Tat-SF fraction assayed in a partially purified reconstituted system rather than with a Tat-responsive nuclear extract, it is not clear that Tat-SF1 is essential for Tat-activated transcription. In addition, coexpression of Tat-SF1 and Tat did not increase Tat-activated transcription in vivo. Rather, Tat relieved an inhibition of transcription that was caused by overexpression of Tat-SF1 (11).
Other studies have suggested a model in which Tat increases transcriptional elongation by enhancing phosphorylation of the carboxyl-terminal domain (CTD) of the largest subunit of RNA polymerase II. This model was first indicated by the observation (12) that Tat-activated transcription in vitro is inhibited by dichlororibofuranosylbenzimidazole (DRB), a purine analog known to inhibit both RNA polymerase II elongation (13) and CTD phosphorylation (reviewed in ref. 13). Further support for this model was provided by the finding that the CTD is required for Tat-mediated transactivation in vitro (14–16) and in vivo (15). DRB may exert its effect by direct inhibition of a CTD kinase activity, and two DRB-sensitive cellular kinases, the CAK component of TFIIH (14, 17–19) and Tat-associated kinase (TAK) (20), have been found to interact with the activation domain of Tat. Consistent with these observations, the CTD kinase activity of TFIIH can be stimulated by Tat (14, 18, 19), and a pseudosubstrate inhibitor of CDK7 that blocks CAK-mediated phosphorylation significantly inhibits Tat-mediated synthesis of long HIV transcripts without affecting synthesis of short (attenuated), Tat-independent HIV transcripts (19). In addition, TAK recently was identified as the previously reported elongation factor P-TEFb (21–23).
We report here the identification of a cellular factor, TIP30, that interacts with the activation domain of Tat both in vivo and in vitro. We demonstrate that TIP30 is selectively required for Tat-mediated transactivation both by anti-TIP30 immunodepletion and recombinant TIP30 complementation assays in vitro and by transient transfection assays involving ectopic TIP30 expression in vivo. Our results suggest that, like previously characterized general transcription factors, TIP30 may also serve as a direct target for Tat.
MATERIALS AND METHODS
Plasmids.
The plasmid pGEX-3X-Tat(1–48) was described previously (18). The pGEX-3x-Tat(1–48K41T) and pGEX-3X-Tat(1–48F38A) plasmids used for expression of mutant GST-Tat proteins were constructed by digesting the plasmids pGEX-3X-TatK41T and pGEX-3X-TatF38A with EagI and EcoRI, filling in the ends, and recircularizing. To construct the series of mutant GST-Tat plasmids, the Tat-coding sequence (amino acids 1–72) was first cloned into pBluescript (+) (Stratagene), and mutations were then introduced at positions 41 and 38 by oligonucleotide-directed mutagenesis (Amersham). Subsequently, the Tat-coding sequences containing mutations were cloned into pGEX-3X in-frame with the glutathione S-transferase (GST) gene. For expressing TIP30 in bacteria and for generating polyclonal antibodies against TIP30, a PCR-based method was used to generate a TIP30 cDNA containing an NdeI site at the N-terminal end and a BamHI site at the C-terminal end after the stop codon. The resulting PCR product was subcloned into the plasmid 6HisT-pRSET (24) to generate 6HisT-pRSETTIP30. For production of TIP30 by in vitro transcription/translation, the plasmid pET11aTIP30 was used to produce TIP30. This plasmid was constructed by insertion of a 730-bp DNA fragment, which was isolated by digestion of 6HisT-pRSETTIP30 with NdeI and BamHI, into pET11a (Novagen). The plasmid (pRSETTat) used for producing Tat was constructed by inserting a BglII Tat cDNA fragment (encoding amino acids 1–72) into the BamHI site of pRSET-C (Invitrogen).
Affinity Chromatography and Protein Purification.
HeLa whole cell extract was prepared as described previously (25). To purify Tat-binding proteins on a large scale, 100 ml of HeLa whole cell extract (5 mg/ml protein) in ACB buffer (10 mM Hepes, pH 7.9/1 mM EDTA/1 mM DTT/10% glycerol) containing 100 mM NaCl was chromatographed on a 5-ml GST-Tat (1–48) affinity column. The bound proteins were eluted with 20 ml of 1 M NaCl in ACB and dialyzed against 0.1 M NaCl in ACB (25). Fifteen milliliters of the dialyzed bound proteins was loaded onto a 0.5-ml phosphocellulose column and step-eluted with 2.5 ml each of 0.3, 0.5, and 0.85 M NaCl in ACB. Twenty-microliter aliquots of the eluted fractions (0.5 ml) were first analyzed by SDS/PAGE followed by silver staining. The 30-kDa protein was present in the 0.3 M NaCl fractions, which were pooled, precipitated with trichloroacetic acid, and analyzed by SDS/PAGE. The 30-kDa band was visualized by staining with Coomassie blue, excised from the gel, and digested with endoproteinase C. The resulting peptides were isolated by HPLC and subjected to microsequencing. For antibody production, His-tagged TIP30 protein expressed in bacteria was purified and then used for immunization of rabbits as described previously (26). To purify endogenous TIP30, 10 ml of HeLa nuclear extract in BC buffer (20 mM Tris⋅HCl, pH 7.9/0.2 mM EDTA/1 mM DTT/20% glycerol) containing 100 mM KCl (BC100) was prepared as described (27) and chromatographed on a GST-Tat(1–48) affinity column (0.5 ml). Bound proteins were eluted with 2 ml of BC buffer containing 1 M KCl (BC1000) and then applied to a Sephacryl S-400 column (Pharmacia). The column was washed with BC100 containing 0.02% Nonidet P-40 (100 ml), and aliquots of the column fractions were subjected to SDS/PAGE followed by silver staining and immunoblotting with anti-TIP30 antibody.
Coimmunoprecipitation and Immunodepletion Assays.
A HeLa cell line (HeLa-HATat) stably expressing hemagglutinin (HA)-Tagged Tat was obtained by introduction of pCIN4-HA-Tagged Tat(1–72) into HeLa S3 and selection in G418-containing medium (ref. 28; details will be provided on request). For coimmunoprecipitation, 1 ml of nuclear extract prepared from the HeLa-HATat cells was mixed with 10 μl of anti-HA epitope immunoaffinity matrix (Babco, Richmond, CA) in BC100 containing 0.1% Nonidet P-40. The matrix then was washed extensively with BC100 plus 0.1% Nonidet P-40 and eluted with 20 μl of 1 mg/ml HA peptide (YPYDYPDYA) in BC100. As a control, nuclear extract made from parental HeLa S3 cells was subjected to the same procedure. Eluates were immunoblotted with polyclonal anti-Tat antibody (Agmed, Bedford, CA), antigen-affinity-purified anti-TIP30, anti-SRB7 (29), anti-RPC53 (30), and 8WG16 (31) antibodies. For immunodepletion of TIP30, anti-TIP30 antisera were purified by passage through a column containing immobilized His-tagged TBP protein followed by passage through a column containing His-tagged TIP30 protein. The anti-TIP30 antibodies were eluted with 200 mM glycine, pH 2.5. The antibodies were further purified by binding to protein A-Sepharose (Pharmacia) and then cross-linked to protein A with dimethylpimilidate (26). Four hundred microliters of HeLa nuclear extract in BC100 adjusted to 0.5 M NaCl were passed five times through 100 μl crossed-linked anti-TIP30-protein A-Sepharose or protein A-Sepharose columns, and the flow-through fractions were used for Western blotting and in vitro transcription analyses.
Transient Transfection Assays.
For transfection assays, TIP30 cDNA was subcloned into the mammalian expression vector pcDNA3.1 (Novagen) to create pcDNA3.1TIP30. Lipofectamine was used for transfection according to the manufacturer’s instructions (GIBCO/BRL). The same amounts of DNA were used for each dish. Cell lysates were first assayed for β-galactosidase activity (32). Normalized quantities of cell extracts were then analyzed by thin-layer chloramphenicol acetyltransferase (CAT) assays (32). Acetylated and nonacetylated forms of [14C]chloramphenicol were quantified by PhosphorImager (Molecular Dynamics Storm 840). Percent conversions of [14C]chloramphenicol to acetylated forms were determined. The fold stimulation by TIP30 was determined from an average of at least three independent experiments.
In Vitro Transcription Assays.
In vitro transcription assays with Gal-VP16 and Gal-Sp1 were performed in HeLa nuclear extracts as described previously (27). Gal-VP16 and Gal-Sp1 were purified as described previously (33, 34). In vitro transcription assays with Tat were performed as described previously (11) with modifications. Supercoiled pHIV+TAR-G400 (100 ng) and pHIVDTAR-G100 (100 ng) templates were used in each reaction (25 μl). The reactions were further incubated for 30 min and then stopped by addition of 1 μl of 250 mM EDTA. Each reaction was then supplemented with 2 μl of RNase T1 (5 units/μl) and incubated at 37°C for 10 min.
RESULTS
Identification of Tat-Interacting Proteins.
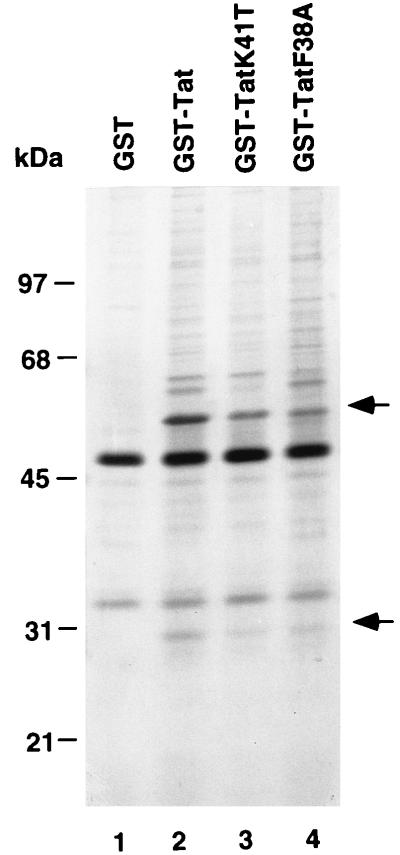
To identify cellular proteins that interact with the activation domain of Tat, we employed a GST fusion protein [GST-Tat(1–48)] containing the amino-terminal 48 aa of HIV-1 Tat for protein affinity chromatography. The bacterially expressed fusion protein was bound to glutathione-Sepharose and used as a ligand (17). As controls, two GST-Tat(1–48) fusion proteins containing amino acid substitutions K41T and F38A within the activation domain were also used as ligands. Both amino acid substitutions significantly weaken transcriptional activation by GAL4-Tat(1–48) (35), indicating that they might affect the interaction between Tat and its cofactor(s). To carry out affinity chromatography, a HeLa whole cell extract was loaded onto the columns at 100 mM NaCl, and the columns were then washed at 100 mM NaCl. Bound proteins were eluted with buffer containing 1M NaCl, resolved by SDS/PAGE, and stained with silver. Two polypeptides with apparent molecular masses of 30 kDa and 56 kDa selectively bound to the GST-Tat(1–48) column (Fig. 1, lane 2), but not to the GST column (lane 1). These two polypeptides bound somewhat less efficiently to the GST-Tat mutant columns (compare lane 2 with lanes 3 and 4). Because Tat is likely to interact with multiple cellular factors that are involved in stimulation of transcription, abrogation of Tat-mediated transactivation by these mutations may result from the cumulative effect of weakening several protein–protein interactions between Tat and cellular factors. Therefore, although they were not completely abolished by the mutations, the interactions between the activation domain of Tat and these two proteins may still be biologically important. We also have observed binding of the 30- and 56-kDa proteins when HeLa nuclear extracts, rather than whole cell extracts, were subjected to Tat affinity chromatography (data not shown). The 30-kDa Tat-interacting protein is designated TIP30, and the 56-kDa Tat-interacting protein is designated TIP56. Proteins similar in size to subunits of P-TEFb (21–23) were not apparent in the high-salt GST-Tat column eluates, although it is possible that they remained bound to the column.
Figure 1.
The activation domain of Tat binds specifically to a 30- and a 56-kDa protein. Aliquots of HeLa whole cell extract were chromatographed on affinity columns containing either GST or GST-Tat(1–48) as described previously (17). Bound proteins were subjected to SDS/PAGE on a 10% gel and stained with silver. Arrows indicate the 30- and 56-kDa proteins.
Identification of a Cognate cDNA and Characterization of the 30-kDa Tat-Interacting Protein (TIP30).
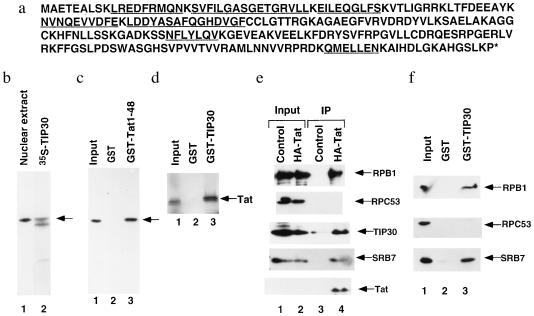
For further characterization, a large preparation of affinity-purified TIP30 was subjected to further purification and microsequencing. Of the seven peptide sequences obtained from TIP30, five matched amino acid sequences encoded by a randomly cloned human cDNA in the EST database. Our subsequent sequence analysis of a 1.2-kb insertion revealed an ORF encoding a 242-aa protein with a calculated mass of 27.3 kDa and an estimated isoelectric point of 9.01 (Fig. 2a). All seven of the original peptide sequences were found within this ORF (Fig. 2a, underlined sequences), which appears to encode full-length TIP30 on the basis of the following. First, there is a good match to the consensus Kozak sequence (CCA/GCCATGG) at the first AUG (36); the nucleotide at the −3 position is a G and the nucleotide at position +4 is G. Second, a search of the database with the entire TIP30 sequence resulted in the identification of 15 related cDNA clones derived from several libraries. An in-frame stop codon located 30-bp upstream of the first ATG codon was found in a cDNA clone derived from a pancreatic islet cell library. Third, as shown in Fig. 2b, the recombinant protein (lane 2) expressed from the cloned cDNA is indistinguishable in size from the endogenous TIP30 (lane 1) in HeLa nuclear extract. The smaller 35S-labeled polypeptide in lane 2 may be either a truncated TIP30 resulting from an internal translation start site or a proteolytic product. Finally, as shown below, the recombinant TIP30 has transcriptional activity.
Figure 2.
Sequence analysis and characterization of the cloned TIP30 cDNA. (a) Deduced amino acid sequences. The sequence for TIP30 is derived from the EST clone 82204, and the peptide sequences obtained by microsequencing of purified TIP30 fragments are underlined. (b) The cDNA encodes full-length TIP30. Two microliters of HeLa nuclear extract (lane 1) and 1 μl of in vitro translation mixture containing [35S]methionine-labeled recombinant TIP30 (lane 2) were resolved by SDS/PAGE (10%) and transferred onto a nitrocellulose membrane. The endogenous TIP30 was visualized by immunoblotting with antibodies raised against bacterially expressed TIP30, and recombinant TIP30 was visualized by autoradiography. (c) Anti-TIP30 antibody recognizes a 30-kDa nuclear extract protein that binds to the activation domain of Tat. Two hundred microliters of HeLa nuclear extract in BC100 was chromatographed on 40 μl GST and GST-Tat(1–48) columns. The columns were washed with BC100 and eluted with 160 ml of BC1000. Proteins eluted from the columns were separated by SDS/PAGE and immunoblotted with anti-TIP30 antibody. Lanes: 1, 10% of the input HeLa nuclear extract; 2, 25% of the GST column eluate; 3, 25% of the GST-Tat column eluate. (d) Binding of Tat to GST-TIP30. The binding assay was performed in BC100 as described (26). Lanes: 1, 10% of the input containing [35S]methionine-labeled Tat (1–72) expressed in a reticulocyte lysate translation/transcription system; 2, 25% of the eluate from the GST beads; 3, 25% of the eluate from the GST-TIP30 beads. (e) Coimmunoprecipitation of Tat and TIP30. Anti-HA monoclonal antibody matrix was used to immunoprecipitate HA-tagged Tat from nuclear extracts. Immunoprecipitates (lanes 3 and 4) as well as input of control HeLa nuclear extract (lane 1) and HeLa nuclear extract containing HA-Tat (lane 2) were immunoblotted with antibodies against Tat, TIP30, SRB7, RPB1 (the largest subunit of RNA polymerase II), and RPC53 (a subunit of RNA polymerase III). (f) Interactions of TIP30 with RNA polymerase II and SRB7. Affinity chromatography with GST-TIP30 was performed as described in c. Proteins eluted from the columns were immunoblotted with antibodies against SRB7, RPB1, and RPC53. Lanes: 1, 2.5% of the input HeLa nuclear extract; 2, 25% of the GST column eluate; 3, 25% of the GST-TIP30 column eluate.
Interaction Between TIP30 and the Activation Domain of Tat.
To further ascertain that the cDNA encodes authentic TIP30, the eluates from GST and GST-Tat(1–48) columns were immunoblotted with anti-TIP30 antibody (Fig. 2c). An immunoreactive polypeptide, identical in size to TIP30 in the nuclear extract (lane 1), was detected in the eluate from the affinity column containing the GST-Tat activation domain fusion protein (lane 3) but not in the eluate from the control column containing GST alone (lane 2). Moreover, in vitro translated Tat labeled with [35S]methionine was bound to a GST-TIP30 fusion protein (Fig. 2d) but not to GST alone. Bacterially expressed TIP30 was found to bind weakly to GST-Tat(1–48) fusion protein but not to GST in an in vitro binding assay (data not shown), suggesting that TIP30 may be able to contact directly the activation domain of Tat. However, the stronger binding of natural TIP30 from nuclear extracts or recombinant TIP30 from reticulocyte lysates leaves open the possibility that other proteins may facilitate interactions between Tat and TIP30 by acting as intermediates or by covalent modification (e.g., phosphorylation) of TIP30.
To test for interactions between Tat and TIP30 in vivo, nuclear extracts from control HeLa cells and from HeLa cells that stably express an HA-tagged Tat protein were immunoprecipitated by anti-HA antibodies. In this analysis (Fig. 2e), TIP30 was detected in the immunoprecipitate from a nuclear extract containing Tat protein (lane 4) but not in that from a control extract lacking Tat protein (lane 3). Consistent with an earlier report (37) that Tat can associate with an RNA polymerase II holoenzyme complex (29, 38), RNA polymerase II (RPB1 subunit) and SRB7 (a component diagnostic of yeast and mammalian holoenzyme; ref. 29), but not RNA polymerase III (RPC53 subunit), were also coimmunoprecipitated with Tat (Fig. 2e). A protein affinity chromatography assay (Fig. 2f) also demonstrated selective binding of RNA polymerase II and SRB7 in nuclear extract to GST-TIP30 relative to GST, whereas no binding of RNA polymerase III was found. Therefore, TIP30 may enhance formation of a Tat-containing RNA polymerase II holoenzyme complex via interactions with both Tat and components of an RNA polymerase II complex.
Selective Involvement of TIP30 in Tat-Mediated Transcription in Vitro.
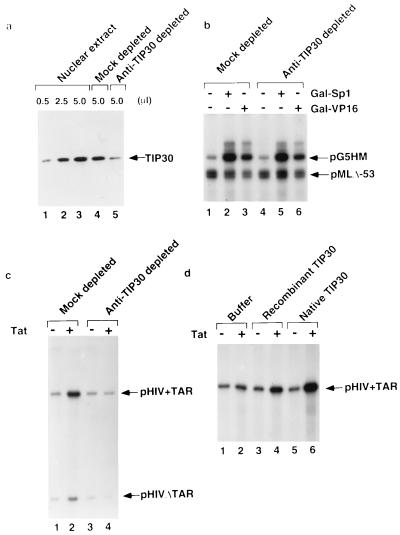
To directly assess whether TIP30 is required for both basal and activated transcription, we used antigen-purified anti-TIP30 antibody to immunodeplete TIP30 from HeLa nuclear extracts. A quantitative Western blot analysis with anti-TIP30 showed that this treatment removed approximately 90% of TIP30 from the extract, whereas a mock-depleted extract showed little loss of TIP30 (Fig. 3A). Mock-depleted and anti-TIP30-depleted nuclear extracts were first assayed for transcription from both the adenovirus major late core promoter and a promoter containing the HIV TATA box and five Gal4-binding sites upstream of the TATA box (27). As shown in Fig. 3B, there were no significant effects of TIP30 depletion on either basal transcription or on transcription activated by the DNA-binding activators Gal-VP16 and Gal-Sp1.
Figure 3.
TIP30 is required specifically for transactivation by Tat. (A) Immunodepletion of endogenous TIP30 from HeLa nuclear extract. Five microliters of HeLa (lane 3), mock-depleted (lane 4), and TIP30-depleted (lane 5) nuclear extract was immunoblotted with anti-TIP30 antibody. Lanes 1 and 2, 0.5 μl and 2.5 μl of HeLa nuclear extract, respectively. The arrowhead indicates the position of TIP30. (B) Effect of TIP30 on transcriptional activation by Gal-VP16 and Gal-Sp1. Five microliters of mock-depleted or TIP30-depleted nuclear extracts was assayed for transcription in the absence (lanes 1 and 4) or presence of 10 ng of Gal-VP16 (lanes 3 and 6) or 20 ng of Gal-Sp1 (lanes 2 and 5), as indicated. (C) Effect of TIP30 on transcriptional activation by Tat. Five microliters of mock-depleted or TIP30-depleted nuclear extract was assayed for transcription in the absence (lanes 1 and 3) or presence (lanes 2 and 4) of 200 ng of Tat, as indicated. The arrowheads indicate the transcripts from the pHIV+TAR-G400 and pHIVDTAR-G100 templates. (D) Restoration of Tat activation by recombinant TIP30 and endogenous TIP30. The TIP30-depleted extracts supplemented with compensating buffer (lanes 1 and 2), 100 ng of recombinant TIP30 (lanes 3 and 4), or 25 ng of endogenous TIP30 (lanes 5 and 6) were assayed for their ability to support Tat-activated transcription, as indicated.
We next tested whether TIP30 is required for in vitro activation of the HIV-1 promoter by Tat. As demonstrated in Fig. 3C, Tat strongly activated transcription in the mock-depleted extract (compare lanes 1 and 2). A 2-fold increase in the activity of a template with a mutant TAR site in response to Tat is consistent with previous reports by others (11, 14, 39) and may reflect the ability of a sufficiently high concentration of Tat to moderately suppress the effects of TAR mutations in vitro. An analogous DNA-binding-independent transcriptional activation by high levels of a DNA-binding activator has also been observed in prokaryotic systems (40). No significant effect on Tat-independent transcription from the HIV-1 promoter was observed in TIP30-depleted extract (Fig. 3C, lane 1 vs. lane 3). In striking contrast, the TIP30-depleted extract failed to support Tat-dependent transcription from either the highly responsive intact template or the weakly responsive TAR-deficient template (Fig. 3C, lane 2 vs. lane 4).
When added to the TIP30-depleted nuclear extract, the endogenous TIP30 preparation isolated from HeLa nuclear extract (about 10–20% pure judged by silver stain) fully restored Tat-activated transcription (Fig. 3D, lane 6 vs. lane 5), whereas bacterially expressed recombinant TIP30 only partially restored this activity (lane 4 vs. lane 3). In contrast, neither the endogenous TIP30 nor recombinant TIP30 increased transcription from the HIV-1 promoter in the absence of Tat (lane 1 vs. lanes 3 and 5). As expected, the endogenous TIP30 and recombinant TIP30 also restored some activity of the weakly responsive TAR-deficient templates in the presence of Tat (data not shown). This result strongly suggests that, among the activators tested, TIP30 is required specifically for transactivation by Tat. The observation that recombinant TIP30 is less active than the natural TIP30 indicates that the full activity of TIP30 may require either posttranslational modifications or an associated factor(s) that can be partially depleted by anti-TIP30 antibodies.
Potentiation of Tat-Mediated Transactivation by TIP30 in Vivo.
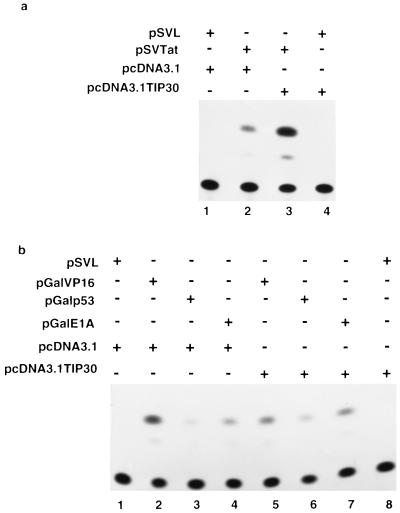
To examine the role of TIP30 in transcriptional activation by Tat in vivo, HeLa cells were cotransfected with a plasmid (pcDNA3.1TIP30) that contains the TIP30 cDNA under the control of a CMV promoter, a plasmid p167 (ref. 41) that contains the CAT gene under the control of the HIV LTR, and a plasmid (pSVtat) that expresses HIV-1 Tat (42). No significant effect on expression of the CAT gene was observed when TIP30 was expressed in the absence of Tat (Fig. 4a, lane 4 vs. lane 1). However, overexpression of TIP30 increased Tat-activated transcription by approximately 9-fold (8.9 ± 3, means ± SEM, n = 4) (lane 3 vs. lane 2).
Figure 4.
Effects of TIP30 on transcription in vivo. (a) Potentiation of Tat-activated gene expression by TIP30. HeLa cells were cotransfected with HIV-1 LTR-CAT reporter plasmid p167 (2 μg) and plasmids expressing Tat (20 ng) and TIP30 (4 μg), as indicated. (b) Effects of TIP30 on transcription controlled by other activators. HeLa cells were cotransfected with CAT reporter plasmid pG6–33HIVLTRDTAR (2 μg) and other plasmids (20 ng), as indicated.
We also tested whether TIP30 could increase activation by other regulatory factors. The pcDNA3.1TIP30 was cotransfected with vectors expressing Gal-VP16 (ref. 35), Gal-p53 (ref. 43), or Gal-E1A (ref. 35) and an HIV-LTR CAT reporter containing Gal4-binding sites. A representative experiment is shown in Fig. 4b, which revealed little (less than 2-fold) to no effect of TIP30 on activation by Gal-VP16 (lane 5 vs. lane 2), Gal-p53 (lane 6 vs. lane 3), or Gal-E1A (lane 7 vs. lane 4). When the levels of the CAT expression in three independent experiments were quantitated and averaged, there were no significant effects of TIP30 expression on activation by Gal-VP16, Gal-p53, or Gal-E1A (data not shown). These data are consistent with the results of the in vitro assays (Fig. 3b).
DISCUSSION
Despite considerable evidence that the ubiquitous TFIIH and TAK serve as direct targets for Tat (Introduction), Tat is unable to activate transcription in transfected rodent cells (1) or in cell-free systems reconstituted with partially purified protein fractions containing all general transcription factors (including TFIIH) and TAK (22). These observations clearly indicate that other cellular factors are required for Tat-mediated activation. We have identified and obtained a cognate cDNA clone for a Tat-interacting protein designated TIP30. TIP30 shares no significant homology with any of the putative Tat cofactors, suggesting that it is a novel Tat-interacting factor. In contrast to previous studies of other Tat-interacting proteins that include TBP1, Tip60, HT2A, and Tat-SF1, we have employed both recombinant protein and cognate antibodies to show that TIP30 is required specifically for Tat-activated transcription in vitro. Furthermore, unlike the Tat-SF1 that was isolated as a component of a Tat stimulator fraction in vitro (11), overexpression of TIP30 also potentiated transactivation by Tat in vivo. Therefore, it is likely that TIP30 plays a role that is distinct from that of Tat-SF1 in mediating transactivation by Tat.
Our results suggest that TIP30 interacts with Tat in nuclear extract. This conclusion is based on the observations that TIP30 in nuclear extract binds to GST-Tat, that in vitro translated Tat binds to GST-TIP30, and that Tat can be coimmunoprecipitated with TIP30 from nuclear extract (Fig. 2). However, it is not clear that TIP30 binds directly to the activation domain of Tat in vivo because bacterially expressed TIP30 bound only weakly to GST-Tat. Therefore, it remains possible that other cellular factors mediate interactions between Tat and TIP30.
Our analyses also showed that an SRB-containing RNA polymerase II complex, as well as TIP30, could be coimmunoprecipitated with Tat (Fig. 2e), and that TIP30 can interact with an SRB-containing RNA polymerase II complex in the absence of Tat (Fig. 2f). These results are consistent with an earlier report claiming that Tat is associated with an RNA polymerase II holoenzyme (37). However, in contrast to SRB proteins (29, 38) and CA150, another protein that has been reported to associate with RNA polymerase II and to be involved in Tat-mediated transcription (44), TIP30 is not tightly associated with RNA polymerase II and SRB proteins and can be separated from them by phosphocellulose chromatography. It also has been shown that the RNA polymerase II holoenzyme purified by phosphocellulose and other chromatographic steps does not support Tat-activated transcription in vitro, whereas a Tat binding fraction containing RNA polymerase II holoenzyme complex can do so (37). Hence, it is conceivable that a cellular Tat cofactor(s) separated from holoenzyme during conventional purification is necessary for Tat-mediated transactivation. Our demonstrations that TIP30 is associated with RNA polymerase II in nuclear extracts, that it can interact with Tat, and that it can be coimmunoprecipitated with Tat and RNA polymerase II raise the possibility that TIP30 is a cellular cofactor involved in Tat recruitment to an RNA polymerase II–holoenzyme complex.
The mechanisms of Tat-mediated activation may involve regulation of phosphorylation of the CTD of RNA polymerase II, because this event is correlated with the transition from initiation to productive elongation (reviewed in ref. 13) and because kinase inhibitors (DRB, H8) that block phosphorylation also block Tat-stimulated transcriptional elongation by RNA polymerase II (12, 14). Many cellular kinases have been shown to phosphorylate the CTD in vitro. Two of them, general transcription factor TFIIH (14, 18–20) and elongation factor P-TEFb/TAK (21–23), were found to interact with Tat; in the case of TFIIH, Tat was shown to enhance phosphorylation of the CTD (14, 18, 19). Thus, Tat may increase transcriptional elongation by increasing the CTD kinase activity of TFIIH, and this could be effected at initiation and/or promoter-clearance steps. Other kinases, such as TAK, might function in conjunction with Tat during elongation to keep RNA polymerase II in a hyperphosphorylated form, because P-TEFb (TAK) can stimulate elongation by RNA polymerase II after initiation and promoter clearance are completed (45).
Another possibility is that Tat may trigger a cascade of kinase reactions to increase phosphorylation of the CTD at a particular step. Indeed, it was suggested that higher levels of CTD phosphorylation (hyperphosphorylation) may require additional factors in addition to TFIIH (14). Such a scenario obviously predicts that cyclic phosphorylation/dephosphorylation events mediated by different components regulate Tat-activated transcription. Because TIP30 interacts with both Tat and an SRB-containing RNA polymerase II complex, it is possible that the proposed TIP30-facilitated recruitment of Tat into an RNA polymerase holoenzyme may serve to regulate phosphorylation of the RNA polymerase II CTD or of another component involved in elongation.
TIP30, unlike other Tat-interacting proteins such as TFIIH, TAK, and TFIID (46), does not increase the level of HIV transcription in the absence of Tat, suggesting that TIP30 is neither a general transcription factor nor a general elongation factor. Moreover, that TIP30 was not found to have any significant effect on transactivation by several other DNA-binding activators suggests that it might be specifically required for Tat-mediated transcription and, presumably, for certain other cellular activators. Whether there are such presumptive activators, whether they reflect human counterparts of Tat, or whether they might, like Tat, affect the elongation potential of RNA polymerase II are interesting questions that remain to be explored.
Acknowledgments
We thank R. Kobayashi (Cold Spring Harbor Laboratory) for peptide sequencing, and M. Green, Q. Zhou, and P. A. Sharp for providing plasmids. We are also grateful to our colleagues for discussions. This work was supported by grants to R.G.R. from the National Institutes of Health (AI37327) and the Tebil Foundation and by grants to J.G. from the Medical Research Council of Canada and the National Cancer Institute of Canada. J.G. is an International Research Scholar of the Howard Hughes Medical Institute. H.X. was supported in part by a fellowship from the National Institutes of Health.
ABBREVIATIONS
- GST
glutathione S-transferase
- CTD
carboxyl-terminal domain
- TAK
Tat-associated kinase
- HA
hemagglutinin
- CAT
chloramphenicol acetyltransferase
- LTR
long terminal repeat
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF039103).
References
- 1.Jones K A, Peterlin B M. Annu Rev Biochem. 1994;63:717–743. doi: 10.1146/annurev.bi.63.070194.003441. [DOI] [PubMed] [Google Scholar]
- 2.Wu F, Garcia J, Sigman D, Gaynor R. Genes Dev. 1991;5:2128–2140. doi: 10.1101/gad.5.11.2128. [DOI] [PubMed] [Google Scholar]
- 3.Sheline C T, Milocco L H, Jones K A. Genes Dev. 1991;5:2508–2520. doi: 10.1101/gad.5.12b.2508. [DOI] [PubMed] [Google Scholar]
- 4.Madore S J, Cullen B R. J Virol. 1993;67:3703–3711. doi: 10.1128/jvi.67.7.3703-3711.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Song C Z, Loewenstein P M, Green M. Proc Natl Acad Sci USA. 1994;91:9357–9361. doi: 10.1073/pnas.91.20.9357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nelbock P, Dillon P J, Perkins A, Rosen C A. Science. 1990;248:1650–1653. doi: 10.1126/science.2194290. [DOI] [PubMed] [Google Scholar]
- 7.Yu L, Zhang Z, Loewenstein P M, Desai K, Tang Q, Mao D, Symington J S, Green M. Virology. 1995;69:3007–3016. doi: 10.1128/jvi.69.5.3007-3016.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kamine J, Elangovan B, Subramanian T, Coleman D, Chinnadurai G. Virology. 1996;216:357–366. doi: 10.1006/viro.1996.0071. [DOI] [PubMed] [Google Scholar]
- 9.Fridell R A, Harding H, Bogerd P, Cullen B R. Virology. 1995;209:347–357. doi: 10.1006/viro.1995.1266. [DOI] [PubMed] [Google Scholar]
- 10.Zhou Q, Sharp P A. EMBO J. 1995;14:321–328. doi: 10.1002/j.1460-2075.1995.tb07006.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou Q, Sharp P A. Science. 1996;274:605–610. doi: 10.1126/science.274.5287.605. [DOI] [PubMed] [Google Scholar]
- 12.Marciniak R A, Sharp P A. EMBO J. 1991;10:4189–4196. doi: 10.1002/j.1460-2075.1991.tb04997.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bentley D L. Curr Opin Genet Dev. 1995;5:210–216. doi: 10.1016/0959-437x(95)80010-7. [DOI] [PubMed] [Google Scholar]
- 14.Parada C A, Roeder R G. Nature (London) 1996;384:375–378. doi: 10.1038/384375a0. [DOI] [PubMed] [Google Scholar]
- 15.Chun R F, Jeang K T. J Biol Chem. 1996;271:27888–27894. doi: 10.1074/jbc.271.44.27888. [DOI] [PubMed] [Google Scholar]
- 16.Yang X, Herrmann C H, Rice A P. J Virol. 1996;70:4576–4584. doi: 10.1128/jvi.70.7.4576-4584.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blau J, Xiao H, McCraken S, O’Hare P, Greenblatt J, Bentley D. Mol Cell Biol. 1996;16:2044–2055. doi: 10.1128/mcb.16.5.2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garcia-Martinez L F, Mavankal G, Neveu J M, Lane W S, Ivanov D, Gaynor R B. EMBO J. 1997;16:2836–2850. doi: 10.1093/emboj/16.10.2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cujec T P, Okamoto H, Fujinaga K, Meyer J, Chamberlin H, Morgan D O, Peterlin B M. Genes Dev. 1997;11:2645–2657. doi: 10.1101/gad.11.20.2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Herrmann C H, Rice A P. Virology. 1993;197:601–608. doi: 10.1006/viro.1993.1634. [DOI] [PubMed] [Google Scholar]
- 21.Zhu Y, Pe’ery T, Peng J, Ramannathan Y, Marshall N, Marshall T, Amendt B, Mathews M, Price D. Genes Dev. 1997;11:2622–2632. doi: 10.1101/gad.11.20.2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mancebo, H. S. Y., Lee, G., Flygare, J., Tomasssini, J., Luu, P., Zhu, Y., Peng, J., Blau, D., Price, D. & Flores, O. Genes Dev. 11, 2633–2656. [DOI] [PMC free article] [PubMed]
- 23.Yang, X. Gold, M. O., Tang, D. N., Lewis, D. E., Aguilar-Cordova, E., Roce, A. P. & Herrmann, C. H. Proc. Natl. Acad. Sci. USA 94, 12331–12336. [DOI] [PMC free article] [PubMed]
- 24.Hoffmann A, Roeder R G. J Biol Chem. 1996;271:18194–18202. doi: 10.1074/jbc.271.30.18194. [DOI] [PubMed] [Google Scholar]
- 25.Xiao H, Pearson A, Coulombe B, Truant R, Zhang S, Regier J L, Triezenberg S J, Reinberg D, Flores O, Ingles C J, Greenblatt J. Mol Cell Biol. 1994;14:7013–7024. doi: 10.1128/mcb.14.10.7013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tao Y, Guermah M, Martinez E, Oegelschlager T, Hasegawa S, Takada R, Yamamoto T, Horikoshi M, Roeder R G. J Biol Chem. 1997;272:6714–6721. doi: 10.1074/jbc.272.10.6714. [DOI] [PubMed] [Google Scholar]
- 27.Chiang C-M, Ge H, Wang Z, Hoffman A, Roeder R G. EMBO J. 1993;12:2749–2762. doi: 10.1002/j.1460-2075.1993.tb05936.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rees S, Coote J, Stables J, Goodson S, Harris S, Lee M G. Biotechniques. 1996;20:102–104. doi: 10.2144/96201st05. [DOI] [PubMed] [Google Scholar]
- 29.Chao D M, Gadbois E L, Murray P J, Anderson S F, Sonu M S, Parvin J D, Young R A. Nature (London) 1996;380:82–85. doi: 10.1038/380082a0. [DOI] [PubMed] [Google Scholar]
- 30.Wang Z, Roeder R G. Genes Dev. 1997;11:1315–1326. doi: 10.1101/gad.11.10.1315. [DOI] [PubMed] [Google Scholar]
- 31.Thompson N E, Steinberg T H, Aronson D B, Burgess R R. J Biol Chem. 1989;264:11511–11520. [PubMed] [Google Scholar]
- 32.Suen T-C, Hung M-C. Mol Cell Biol. 1991;11:354–362. doi: 10.1128/mcb.11.1.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chasman D I, Leatherwood J, Carey M, Ptashne M, Kornberg R. Mol Cell Biol. 1989;9:4746–4749. doi: 10.1128/mcb.9.11.4746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Emili A, Greenblatt J, Ingles C J. Mol Cell Biol. 1994;14:1582–1593. doi: 10.1128/mcb.14.3.1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Southgate C D, Green M R. Genes Dev. 1991;5:2496–2507. doi: 10.1101/gad.5.12b.2496. [DOI] [PubMed] [Google Scholar]
- 36.Kozak M. Cell. 1986;44:283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- 37.Cujec T P, Cho H, Maldonado E, Meyer J, Reinberg D, Peterlin B M. Mol Cell Biol. 1997;17:1817–1823. doi: 10.1128/mcb.17.4.1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maldonado E, Shiekhattar R, Sheldon M, Cho H, Drapkin R, Rickert P, Lees E, Anderson C W, Linn S, Reinberg D. Nature (London) 1996;381:86–89. doi: 10.1038/381086a0. [DOI] [PubMed] [Google Scholar]
- 39.Marciniak R A, Calnan B J, Frankel A D, Sharp P A. Cell. 1990;63:791–802. doi: 10.1016/0092-8674(90)90145-5. [DOI] [PubMed] [Google Scholar]
- 40.Ptashne M, Gann A. Nature (London) 1997;386:569–577. doi: 10.1038/386569a0. [DOI] [PubMed] [Google Scholar]
- 41.Rosen C A, Sodroski J G, Haseltine W A. Cell. 1985;41:813–823. doi: 10.1016/s0092-8674(85)80062-3. [DOI] [PubMed] [Google Scholar]
- 42.Emerman M, Guyader M, Montagnier L, Baltimore D, Muesing M A. EMBO J. 1987;6:3755–3768. doi: 10.1002/j.1460-2075.1987.tb02710.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fields S, Jang S K. Science. 1990;249:1046–1049. doi: 10.1126/science.2144363. [DOI] [PubMed] [Google Scholar]
- 44.Sune C, Hayyashi T, Liu Y, Lane W S, Young R A, Garcia-Blanco M A. Mol Cell Biol. 1997;17:6029–6039. doi: 10.1128/mcb.17.10.6029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marshall N F, Peng J, Xie Z, Price D H. J Biol Chem. 1996;271:27176–27183. doi: 10.1074/jbc.271.43.27176. [DOI] [PubMed] [Google Scholar]
- 46.Kashanchi F, Piras G, Radonovich M F, Duvall J F, Fattaey A, Chiang C M, Roeder R G, Brady J N. Nature (London) 1994;367:295–299. doi: 10.1038/367295a0. [DOI] [PubMed] [Google Scholar]