Abstract
Analysis of the function of a particular gene product typically involves determining the expression profile of the gene, the subcellular location of the protein, and the phenotype of a null strain lacking the protein. Conditional alleles of the gene are often created as an additional tool. We have developed a multifunctional, transposon-based system that simultaneously generates constructs for all the above analyses and is suitable for mutagenesis of any given Saccharomyces cerevisiae gene. Depending on the transposon used, the yeast gene is fused to a coding region for β-galactosidase or green fluorescent protein. Gene expression can therefore be monitored by chemical or fluorescence assays. The transposons create insertion mutations in the target gene, allowing phenotypic analysis. The transposon can be reduced by cre–lox site-specific recombination to a smaller element that leaves an epitope tag inserted in the encoded protein. In addition to its utility for a variety of immunodetection purposes, the epitope tag element also has the potential to create conditional alleles of the target gene. We demonstrate these features of the transposons by mutagenesis of the SPA2, ARP100, SER1, and BDF1 genes.
The yeast Saccharomyces cerevisiae has proved of great importance in characterizing basic biological processes. This utility can only become more marked now that the sequence of the entire yeast genome has been obtained, and additional homologs of yeast genes are identified in other organisms (1). Determining the function of a particular gene product usually entails characterization of the gene’s expression pattern, phenotypic analysis of a disruption mutant, and investigating the subcellular distribution of the encoded protein. Often, conditional alleles of the gene are generated as a means of gaining additional information about the in vivo function of a protein. Typically, these procedures require multiple independent manipulations.
Many techniques have been devised to simplify particular aspects of gene characterization. For example, mutagenesis of cloned yeast genes using mini-transposon (mTn) constructs can circumvent several steps. By a few simple manipulations of Escherichia coli strains, mTns containing an expression reporter construct can be inserted at multiple independent sites in a cloned gene (2, 3). The mutagenized genes may then be reintroduced into the yeast genome by homologous recombination. Yeast strains resulting from such transposon mutagenesis have been used to analyze coding regions and disruption phenotypes, structure–function relationships, differential gene expression, and protein localization (for examples, see refs. 2–7). Epitope-tagging is another useful and time-saving technique. By engineering an epitope recognized by a commercially available antibody into a protein of interest, the time and expense of generating specific antibodies and associated reagents is avoided (8). Finally, various mutagenesis procedures have been employed to generate conditional mutants (e.g., ref. 9).
We have created a new set of mTns that incorporate all of the features described above. With a single cloning step followed by a simple mutagenesis procedure using one of these mTns, it is possible to generate multiple constructs that allow rapid analysis of expression pattern, disruption phenotype and protein localization for a given gene. The mTns contain the coding region for either β-galactosidase (lacZ; β-gal) or the Aequorea victoria green fluorescent protein (GFP; refs. 10 and 11). In-frame fusions between a yeast coding region and the transposon can be identified by β-gal activity or fluorescence. The transposon insertion creates a truncation of the mutagenized gene, providing disruption alleles for phenotypic analysis. The inserted transposon can then be reduced by Cre-mediated site-specific recombination to an element of less than 300 bp that encodes several tandem copies of an epitope (12) that can be used for immunodetection. In addition, while a protein containing the epitope tag is often functional, conditional mutations may also be generated by this method. The construction and use of these transposons on several yeast genes are described below.
MATERIALS AND METHODS
Strains, Reagents, and Standard Procedures.
Yeast and bacterial strains used are listed in Table 1. Manipulation of DNA and yeast was according to standard procedures (14, 15). Methods for identification of strains with productive lacZ fusions and for performing immunocytology on yeast cells were as described by Burns et al. (16). The plasmid B227, containing LEU2 and the cre gene under the control of the GAL1 promoter (GAL-cre), was provided by F. Heffron and is a derivative of pBS49 (17). The 12CA5 mAb against the influenza virus hemagglutinin epitope (HA) was obtained from Babco (Richmond, CA). Fluorescence of GFP was viewed with a Leitz system 13 filter using either untreated cells or cells that were fixed and spheroplasted as described (16).
Table 1.
Yeast and bacterial strains
| Strain | Genotype | Use (source) |
|---|---|---|
| Yeast | ||
| Y800 | MATa leu2-Δ98 cry1R ade2-101 HIS3 | Mutagenesis of ARP100 |
| MATα leu2-Δ98 CRY1 ade2-101 his3-Δ200 | ||
| ura3-52 can1R lys2-801 CYH2 trp1-1 | ||
| ura3-52 CAN1 lys2-801 cyh2R TRP1 Ciro | ||
| Y800-T1a | MATa ade2-1 leu2-Δ98 ura3-52 lys2-801 trp1-1 his3-Δ200 Ciro | Mutagenesis of SPA2 |
| W303-1B | MATα ura3-1 ade2-1 trp1-1 his3-11,15 leu2-3,112 can1-100 | Mutagenesis of SER1 (R. Rothstein, Columbia University) |
| Y1279 | MATa leu2-Δ98 ade2-101 his3-Δ200 | Mutagenesis of BDF1 (B. Santos, Yale University) |
| MATα LEU2 ade2-101 HIS3 | ||
| ura3-52 trp1-901 lys2-801 | ||
| ura3-52 trp1-901 lys2-801 | ||
| Bacteria | ||
| B224/RDP146 | F−recA1 (Δlac-pro) rpsE | Recipient for F factor in mating (2) |
| B211 | B224 plus pLB101 (pACYC184 with tnpA) | Constitutively expresses Tn3 transposase (2) |
| B425 | NG135 (K12 recA56 gal-ΔS165 strA) plus pNG54 (pACYC184 with tnpR) | Constitutively expresses Tn3 resolvase (13) |
| B426 | B224 plus pOX38::mTn-4xHA/lacZ | Transposon mutagenesis |
| B427 | B224 plus pOX38::mTn-3xHA/lacZ | Transposon mutagenesis |
| B428 | B224 plus pOX38::mTn-3xHA/GFP | Transposon mutagenesis |
| DH5α | endA1 hsdR17 supE44 thi-1 recA1 gyrA relA1 Δ(lacZYA-argF) U169 deoR [φ80 dlacΔ(lacZ)M15] | Cloning, plasmid preparation |
Construction of Transposons.
Transposons mTn-3xHA/lacZ and mTn-4xHA/lacZ (see Fig. 1) were constructed using plasmids derived from pEIE, kindly provided by F. Heffron (18). The following changes were made. By cloning of a PCR product, the loxP site upstream of lacZ was replaced with a modified loxR site, 5′-ATCGCTTCGGATAACTCCTGCTATACGAAGTTAT-3′, that contains a single base deletion in the central region to maintain the same reading frame as loxP. In this process, an MluI site was introduced between loxR and lacZ. The loxP site adjacent to the tet gene in pEIE was found to contain an insertion mutation and was replaced with the consensus loxP, introducing an XbaI site between loxP and tet in the process. A PCR product containing the 115-bp Tn3 res element flanked by SpeI and XbaI sites was generated from plasmid pNG54 [kindly provided by Nigel Grindley (13)]. This product was cloned at the XbaI site adjacent to tet. A PCR product containing three copies of the HA epitope flanked by EagI sites was generated from a plasmid kindly provided by Bruce Futcher (12). This product was cloned at the NotI site adjacent to loxP.
Figure 1.
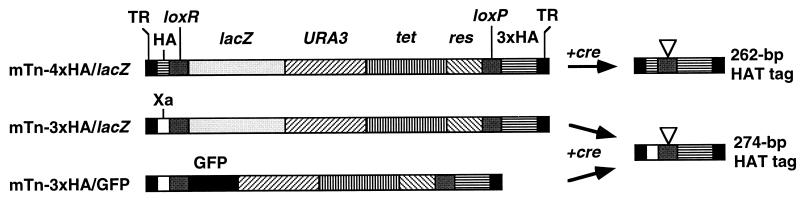
Schematic representation of the mTns constructed and the derived HAT tag elements. Each mTn contains the coding regions for the tetracycline efflux (Tet) and Ura3 proteins, and the res element from Tn3. mTn-4xHA/lacZ and mTn-3xHA/lacZ contain a truncated lacZ gene. mTn-3xHA/GFP contains the coding region for GFP mutant p11 (11). In each case, these are flanked by lox sites and Tn3 terminal repeats (TR). Between loxP and the right TR, each transposon contains a sequence encoding three tandem copies of the HA epitope. Between the left TR and loxR is a sequence encoding either an additional copy of the HA epitope (mTn-4xHA/lacZ) or the factor Xa protease cleavage site (mTn-3xHA/lacZ, mTn-3xHA/GFP). Exposure of these transposons to Cre recombinase catalyzes the formation of a smaller element encoding the HAT tag, shown to the right. The site of the lox recombination junction is indicated by a triangle. (Not drawn to scale.)
In the mTn-4xHA derivative, the complementary oligonucleotides 5′-GGCCTACCCATACGACGTCCCAGACTACGCGTT-3′ and 5′-GGCCAACGCGTAGTCTGGGACGTCGTATGGGTA-3′, encoding a single copy of the HA epitope, were annealed and cloned at the NotI site adjacent to loxR. In the mTn-3xHA derivatives, the complementary oligonucleotides 5′-GGCCATTGAAGGTAGAAGAGAAAATTTGTACTTCCAAAGAAAGAA-3′ and 5′-GGCCTTCTTTCTTTGGAAGTACAAATTTTCTCTTCTACCTTCAAT-3′, encoding a peptide that contains the factor Xa protease cleavage site (Ile-Glu-Gly-Arg; ref. 19), were annealed and cloned at the NotI site adjacent to loxR. To create mTn-3xHA/GFP (see Fig. 1), a plasmid containing mTn-3xHA/lacZ was digested with MluI, removing most of the lacZ coding region. This was replaced with a PCR fragment containing the coding region for GFP mutant p11 flanked by MluI sites, generated from a plasmid kindly provided by Roger Tsien (11). The three transposon constructs were then moved onto the F plasmid derivative pOX38 (2) by transposition, generating strains B426, B427, and B428 (Table 1).
Shuttle Mutagenesis of Yeast Genes.
Restriction fragments from the SPA2 (20), ARP100, SER1 (21), and BDF1 (6, 22) genes were cloned into the vector pHSS6 (2). The corresponding regions of the encoded proteins are indicated in the text or in the diagram in Fig. 2. Plasmids were transformed into strain B211, and transformants were mated to strains B426, B427, or B428 to initiate transposition. Products of this mating (transconjugants) were selected by their resistance to kanamycin, chloramphenicol, and tetracycline, and mated to strain B425 to resolve cointegrate structures. Transconjugants were then selected by their resistance to kanamycin, tetracycline, streptomycin, and chloramphenicol. Plasmid DNA was recovered from these transconjugants and transformed into strain DH5α. Plasmid DNA prepared from a pool of all transformants was digested with NotI and transformed into yeast, selecting for the URA3 marker.
Figure 2.
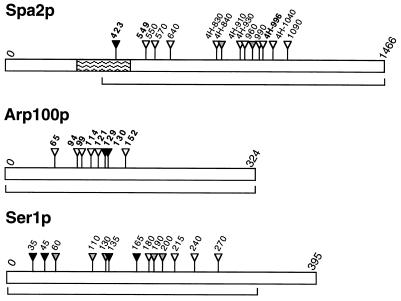
Map showing amino acid positions of HAT tag insertions in the yeast proteins Spa2, Arp100, and Ser1. Regions mutagenized are indicated by brackets. Insertion positions determined by sequencing are indicated in bold. The prefix “4H” indicates HAT tags derived from mTn-4xHA/lacZ; other tags are derived from mTn-3xHA/lacZ. For Spa2p, the coiled-coil region is indicated by wavy lines, an HAT tag that causes both a spa2 null phenotype and mislocalization of the protein is indicated by a black triangle, and two HAT tags that cause a partial mutant phenotype are indicated by grey triangles. For Arp100p, HAT tags that cause a null phenotype are indicated by black triangles. For Ser1p, HAT tags that cause a ser1 null phenotype are indicated by black triangles, and HAT tags that cause wild-type or temperature-sensitive phenotypes are indicated by white and grey triangles, respectively. Proteins are not drawn to the same scale.
Analysis of Transposon Insertion Sites and Recombinant loxP Sites.
PCR was performed on yeast genomic DNA samples from individual transformants, using a primer complementary to sequences encoding the HA epitope and a primer complementary to sequences in the appropriate gene. Sequence data were obtained from these PCR products using a primer complementary to sequences encoding the HA epitope.
Analysis of Cre-Mediated Excision.
Strains containing the transposon and pB227 were grown to saturation in synthetic medium lacking uracil and leucine (SC-Ura-Leu) with raffinose as carbon source. Cultures were then diluted 1/100 into SC-Leu medium with either glucose or galactose as carbon source, and grown to saturation. Cells were plated onto SC medium, and the resulting colonies were scored for loss of the URA3 marker by replica plating to SC-Ura medium. To obtain strains carrying the HA/transposon tag (HAT tag) routinely, the SC-Leu-raffinose culture was spotted onto medium containing 5-fluoroorotic acid to select Ura− cells.
RESULTS
New Transposons for Generating Insertions in Yeast Genes.
Three new Tn3-related transposons suitable for mutagenesis of yeast genes have been constructed: mTn-4xHA/lacZ, mTn-3xHA/lacZ, and mTn-3xHA/GFP (Fig. 1). These transposons are flanked by the Tn3 38-bp terminal repeats, which direct transposition, and contain the Tn3 res site for resolution of transposition cointegrates. Tn3-encoded enzymes catalyzing transposition and resolution are provided in trans. All three transposons contain the URA3 and tet genes for selection in S. cerevisiae and E. coli, respectively. Transposons mTn-3xHA/lacZ and mTn-4xHA/lacZ contain a lacZ gene that lacks an initiator methionine, while transposon mTn-3xHA/GFP contains the entire coding region for a mutant derivative of GFP that shows enhanced fluorescence (10, 11). These elements allow identification of in-frame fusions between a transposon and a yeast coding region by use of assays for either β-gal or fluorescence activity. Levels of both activities can be measured quantitatively and have been shown to provide indices of gene expression (e.g., refs. 4, 23, and 24).
A loxR element lies at one end of the transposons and a loxP element lies at the other end. These target sites for the Cre recombinase are divergent from one another and undergo low levels of spontaneous recombination. The lox sites are internal to sequences encoding multiple copies of an epitope from the influenza virus hemagglutinin protein (the HA epitope; ref. 25). The mTn-3xHA transposons also contain a factor Xa protease cleavage site (19) in the region external to the loxR site. Expression of the Cre recombinase induces recombination between the lox sites resulting in excision of the central region of the transposon. The final product contains a 5-bp duplication caused by transposon insertion in addition to a 274-bp (mTn-3xHA) or 262-bp (mTn-4xHA) element. This element consists of a single loxR site and sequences encoding three or four copies of the HA epitope, flanked by the Tn3 terminal repeats (Fig. 1). The mTn-3xHA-derived element also contains a sequence encoding the factor Xa cleavage site. When the transposon has inserted into a gene to generate an in-frame fusion of lacZ or GFP coding sequences, the excision event results in insertion of 93 amino acids (mTn-3xHA) or 89 amino acids (mTn-4xHA) into the protein. We designate these insertions HAT tags.
Mutagenesis of Yeast Genes.
Transposons mTn-3xHA/lacZ and mTn-4xHA/lacZ were tested by mutagenesis of the yeast SPA2 gene. SPA2 encodes a nonessential protein that localizes to sites of polarized growth; spa2 mutants exhibit defects in polarized growth processes, including formation of the mating projection (20, 26). mTn-3xHA/lacZ has also been used to mutagenize the ARP100 and SER1 genes (Fig. 2). ARP100 is an essential gene whose encoded protein localizes to the spindle pole body region (N. Burns and M.S., unpublished data), and SER1 encodes 3-phosphoserine aminotransferase, which is required for serine biosynthesis (21). mTn-3xHA/GFP was tested by mutagenesis of the BDF1 gene, which encodes a chromatin-associated bromodomain protein (6, 22). In each case, a plasmid carrying the gene (or a region thereof) was mutagenized in E. coli by shuttle mutagenesis. DNA containing the transposon was then excised from the plasmid and transformed into yeast, where it replaced the chromosomal locus by homologous recombination.
With both mTn-3xHA/lacZ and mTn-4xHA/lacZ, about 10% of transformants were identified as producing β-gal fusion protein (44/500 mTn-4xHA/lacZ transformants for SPA2 and 92/900, 25/200, and 62/600 mTn-3xHA/lacZ transformants for SPA2, ARP100, and SER1, respectively). Strains expressing the reporter genes were used for further analyses. The approximate position of the transposon insertion in these strains was determined by size analysis of PCR products obtained from their genomic DNA (Materials and Methods). In some instances PCR products were sequenced, enabling exact identification of insertion points (Fig. 2).
Efficiency of Cre-Mediated loxR–loxP Recombination.
Although efficient Cre-mediated recombination between loxP elements has been demonstrated in S. cerevisiae (17), recombination between a modified loxR element and loxP has not previously been analyzed in yeast. Studies in bacteriophage λ indicated that loxR–loxP recombination may occur at a much lower rate (27). Therefore, the efficiency of Cre-mediated excision was determined for 11 strains carrying mTn-3xHA/lacZ and 9 strains carrying mTn-4xHA/lacZ. Eight strains tested were transformants of diploid strain Y800, the remainder were transformants of the haploid derivative Y800-1Ta. For all 20 strains, >90% of cells showed loss of the mTn URA3 marker when cre expression was induced by growth in galactose (see Materials and Methods). In contrast, <1% of cells exhibited loss of the marker when cre transcription was repressed by glucose. These frequencies are similar to those reported in previous studies of loxP elements (16).
The sequence spanning the recombinant loxR junction was analyzed in PCR products obtained from a total of 12 SPA2 and ARP100 mutant strains. In all cases, the novel junction had been formed correctly.
Generation of Epitope-Tagged Proteins.
The ability of the mTns to generate functional, epitope-tagged versions of proteins was tested by mutagenesis of the SPA2 and ARP100 genes. Mutagenized SPA2 DNA was transformed into haploid strain Y800-1Ta. Strains containing an HAT-tagged SPA2 gene were derived from nine transformants carrying mTn-3xHA/lacZ and eight transformants carrying mTn-4xHA/lacZ. One strain proved to contain a fusion that is out-of-frame with respect to SPA2, but in-frame with an internal methionine initiator codon, and was excluded from further analysis. The remaining 16 strains represent at least 6 independent insertion events for each transposon (Fig. 2). Analysis of mating projection formation upon exposure to pheromone revealed that the Spa2::β-gal protein is nonfunctional in all 16 strains that carry spa2::lacZ fusions. However, the HAT-tagged Spa2 protein (Spa2–HAT) appears to be fully functional in 13 of the 16 derivative strains (data not shown). One strain shows a defect identical to that seen in a spa2Δ mutant, and was found to contain an HAT insertion in the coiled-coil region of the Spa2 protein (codon 423 of Spa2p; Fig. 2). Two strains show a partial defect in mating projection formation; one contains an insertion in the region of codon 840, the other in the region of codon 960 (Fig. 2).
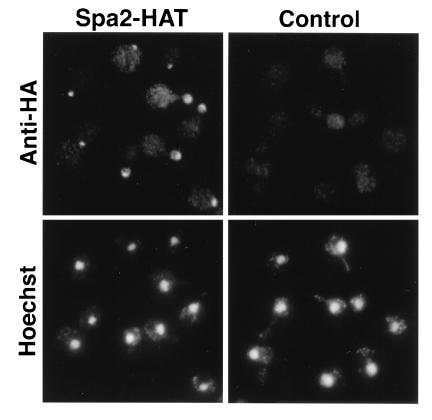
Localization of the Spa2–HAT protein was analyzed by indirect immunofluorescence of whole cells using antibody directed against the HA epitope. Native Spa2p localizes to presumptive bud sites, bud tips, and to the cell neck in dividing cells (20, 26). Fifteen of the 16 HAT-tagged versions of the protein showed correct localization; one did not localize. An example of the staining pattern observed in 15 strains is shown in Fig. 3. No difference in quality or intensity of immunofluorescence was observed depending on whether three or four copies of the HA epitope were present, and the intensity of the signals was similar to that obtained using antibodies prepared to authentic Spa2p (20, 26). The nonlocalizing Spa2–HAT represents an insertion at codon 423, in the coiled-coil region of Spa2p (Fig. 2); this insertion also caused a spa2 phenotype (see above). The requirement for an intact coiled-coil region is in agreement with studies indicating that this segment of the Spa2 protein is necessary for its localization (B. Santos and M. S., unpublished data).
Figure 3.
Immunolocalization of an HAT-tagged Spa2 protein. (Left) A strain containing Spa2–HAT. (Right) An isogenic wild-type strain. (Upper) Localization of a monoclonal antibody directed against the HA epitope. Cells containing Spa2–HAT show staining of presumptive bud sites, bud tips, and the cell neck in dividing cells. (Lower) Fluorescence of the same cells stained with the DNA-binding dye, Hoechst 33258.
The Arp100 protein is essential for cell viability (N. Burns and M.S., unpublished data). Therefore, DNA from ARP100 mutagenized with mTn-3xHA/lacZ was transformed into diploid yeast strain Y800. Twenty β-gal-producing strains were analyzed. Two contained insertions in a region of a neighboring gene present on the mutagenized plasmid, and one represents a fusion that is out-of-frame with respect to ARP100 but in-frame with an internal methionine initiator codon. Four strains were derived from a single unusual transposition event, indicating that a “bottleneck” occurred during the mutagenesis procedure.
For the remaining 13 strains, HAT-tagged derivatives were sporulated and tetrads were dissected. In five strains, representing two independent insertions (Fig. 2), the HAT tag causes an inviability phenotype that can be complemented by introduction of the wild-type gene on a plasmid. The recombinant loxR junction was sequenced and found to be correct in all five. In the remaining eight strains, representing six independent insertions, each tetrad contained four viable spores, indicating that a functional Arp100 protein is being produced. Mendelian segregation of the HAT-tagged copy of ARP100 in these strains was confirmed by PCR analysis of DNA from viable spores. The Arp100–HAT protein could be detected by both immunoblot analysis and immunoprecipitations from yeast cell extracts using an antibody directed against the HA epitope (data not shown). Correct subcellular localization of Arp100–HAT to the spindle pole body region in all eight viable haploid strains was confirmed by immunofluorescence analysis (N. Burns and M.S., unpublished data).
Use of the Novel Transposon mTn-3xHA/GFP to Create a GFP Fusion Protein.
The utility of mTn-3xHA/GFP was examined by mutagenesis of the BDF1 gene. Localization of Bdf1p to the nucleus has been shown previously using antibodies either to the native protein or to the β-gal portion of a Bdf1–β-gal fusion protein (6, 7).
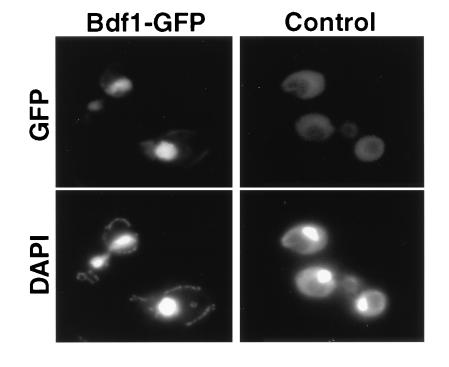
The 2058-bp BDF1 coding region was mutagenized with mTn-3xHA/GFP and transformed into diploid yeast strain Y1279. Individual yeast transformants were grown in liquid culture and then examined directly by fluorescence microscopy for localization of GFP. Of 38 transformants examined, 4 showed fluorescence of the nucleus, and therefore presumably contain in-frame fusions. An example of Bdf1–GFP localization is shown in Fig. 4. The remainder of transformants exhibited a low level of background fluorescence, similar to the wild-type strain.
Figure 4.
Localization of a Bdf1–GFP fusion protein. (Left) A strain containing bdf1::mTn3xHA/GFP. (Right) An isogenic wild-type strain. (Upper) Fixed, spheroplasted cells viewed for GFP fluorescence. (Lower) Fluorescence of the same cells stained with the DNA-binding dye, 4′,6-diamidino-2-phenylindole (DAPI).
Use of the HAT Tag to Create a Conditional Phenotype.
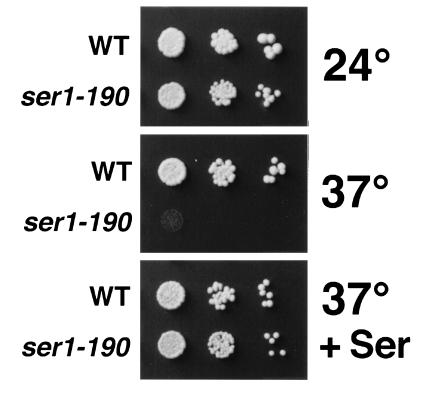
Introduction of the small HAT insertion might be a useful method for generating conditional mutations. This possibility was investigated by mutagenesis of the SER1 gene. ser1 strains are auxotrophic for serine (Ser−; ref. 21), providing a convenient assay for function. A region comprising 322 codons of the 395-amino acid Ser1 protein was mutagenized with mTn-3xHA/lacZ and transformed into haploid yeast strain W303-1B. HAT-tagged derivatives were obtained from 38 strains that produced a β-gal fusion protein and were Ser−. These strains were assayed for growth on SC medium lacking serine (SC-Ser). At 24°C, nine strains were Ser+, 10 were Ser−, and 19 showed impaired growth (Serw). At 37°C, three Ser− strains became Serw, and eight Serw strains became Ser+. Additionally, one Ser+ strain and five Serw strains became Ser−. Conditional auxotrophy for serine was most marked in a strain containing an HAT tag insertion approximately at codon 190 (ser1-190). Growth of the ser1-190 strain under different conditions is shown in Fig. 5.
Figure 5.
Growth of a Ser1–HAT tagged strain. Serial dilutions (left to right) of a cell suspension from strain W303-1B (WT) or a transformant containing an HAT tag at approximately codon 190 of Ser1p (ser1-190) were placed on solid medium and grown under various conditions. (Top) SC-Ser medium, 24°C. (Middle) SC-Ser medium, 37°C. (Bottom) SC medium; 37°C.
The site of HAT tag insertion was determined for four strains that were unconditionally Ser− and four that were Ser+. The Ser− strains carried insertions in the N-terminal third of Ser1p, whereas the Ser+ strains had insertions in the central region of the protein. The site of insertion was also determined for the six strains that became Ser− at 37°C. These represent at least five independent insertions, and all of the insertions fall in the first half of Ser1p (Fig. 2).
Mutagenesis of a Yeast Genomic Library.
We have previously described (7) a yeast genomic library mutagenized by insertion of an mTn3-lacZ/LEU2 transposon (3). In addition to its use for analyses of gene expression, protein localization, and function (7), this insertion library has been widely used as a mutagenic agent because it generates a tagged locus (16). The genomic library has been mutagenized with the three new transposons described in this report, and a “rescue” plasmid (pRSQ2–LEU2) has been constructed that enables identification of the site of mTn insertion in a given yeast transformant [see Burns et al. (7) for experimental approach]. In addition to providing an alternative selectable marker for yeast, the additional features present in the new transposons enhance the utility of the insertion library.
DISCUSSION
A set of multifunctional transposons has been constructed that allows many different types of information about a gene to be obtained from a single, simple mutagenesis procedure. We have demonstrated the capabilities of these mTns by mutagenesis of four yeast genes. Insertion of either mTn-3xHA/lacZ or mTn-4xHA/lacZ created reporter fusions in the SPA2 gene; in addition, mTn-3xHA/lacZ was tested with success on the ARP100 and SER1 genes. In the case of SPA2 and SER1, strains carrying these insertions displayed a mutant phenotype, as expected from a disruption allele. Using Cre-mediated excision of the transposon to create the smaller HAT tag insertion, accurate immunolocalization was achieved for both the Spa2 and Arp100 proteins. Correct subcellular localization was also observed for several Bdf1–GFP fusion proteins generated by mutagenesis of the BDF1 coding region with mTn-3xHA/GFP. Finally, several novel conditional alleles of the SER1 gene were created by insertion of sequences encoding the HAT tag.
Several techniques for mutagenesis of yeast genes have been described previously. Hoekstra et al. (18) originally developed an mTn shuttle mutagenesis system that utilizes cre–lox recombination to produce small insertion mutations, while PCR and other methods are now available for directly tagging genes (28). More cumbersome methods for generating conditional mutations also exist (e.g., ref. 9).
The transposon-based method described here has several advantages over the system developed by Hoekstra et al. (18). The latter relied on lox sites for resolving cointegrates in E. coli. In our laboratories, this led to a high level of transposon rearrangement, presumably due to the presence of four lox sites in the cointegrates. Our transposons use the Tn3 res system for cointegrate resolution and do not experience this problem. In addition, the incorporation of sequences encoding the HA epitope, for which many antibody reagents are commercially available, allows the rapid immunodetection of the tagged protein. Finally, the presence of a factor Xa recognition site in the HAT tag should allow protein products to be cleaved with the factor Xa protease.
The advantage of transposon mutagenesis over PCR and other methods for epitope tagging of proteins is the ease and low cost with which a large number of independent tags can be created and screened. Analysis of three genes mutagenized with the new mTns indicates that the transposons show minimal target specificity (Fig. 2), in agreement with previous observations (2). The ability to screen a range of epitope-tag insertion sites can be crucial in the case of proteins that lose function when foreign epitopes are placed at conventional sites (usually the amino or carboxyl terminus). For example, loss of function due to a carboxyl-terminal tag will occur in many isoprenylated proteins (e.g., ref. 29). The Arp100 protein is nonfunctional when tagged at the carboxyl terminus (N. Burns and M. S., unpublished data); however, five functional epitope-tagged versions of Arp100p were successfully generated by the transposon system. In addition, the PCR-based method of epitope tagging (28) depends on infrequent spontaneous excision events to remove the URA3 marker and return the mutated gene to the desired chromosomal context. By contrast, the Cre-mediated excision event that generates the HAT tag is extremely efficient (ref. 16 and this work), occurring 100- to 1000-fold more frequently than mitotic crossing over or gene conversion (which can also generate a Ura− phenotype). Spontaneous excision of the new transposons is limited by the lack of directly repeated sequences: loxR and loxP share only 16 bases of identity, and sequences encoding the repeated HA epitope were designed to avoid direct reiteration.
The ease with which a large number of independent, marked insertions can be created and screened is also advantageous for mutagenesis. Perhaps surprisingly, given the relatively large size of the HAT tag (about 90 amino acids), many HAT insertions allow the target protein to retain function. Six out of 8 strains with independent insertions in the essential protein Arp100 are viable, while 13 of 16 strains with insertions in the Spa2 protein display normal morphology on exposure to mating pheromone, and 28 out of 38 insertions in Ser1p allow at least some degree of function. In the cases where the exact position of the HAT insertion was known (eight insertions in Arp100p and three in Spa2p; Fig. 2) the region was analyzed using the program geneworks (IntelliGenetics) to predict hydrophobicity and probability of surface exposure. As might be expected, all insertions that do not disrupt function of the protein (six in Arp100p, two in Spa2p) lie in regions predicted to be hydrophilic and either on the surface of the protein or at the transition from surface to interior. Conversely, insertions that disrupt function (two in Arp100p, one in Spa2p) occur in predicted hydrophobic, internal regions.
In the case of Ser1p, the correlation between the position of the HAT tag and the growth phenotype indicates that the tag may be a useful tool for analysis of functional domains in proteins, a feature noted previously for the transposon described by Hoekstra et al. (18). While the Ser1 protein shows identity to other phosphoserine aminotransferases along its entire length (21), HAT insertions causing a complete loss of function were in the amino-terminal region, and insertions causing no phenotype fell in the central region.
The successful generation of conditional alleles of the SER1 gene demonstrated an additional utility for the HAT tag. From only 38 in-frame insertions tested, several strains with a temperature-sensitive Ser− phenotype were identified. The temperature-sensitive growth defect of one HAT insertion strain, ser1-190, was very pronounced (Fig. 4), making it an excellent candidate for genetic analyses or biochemical studies of Ser1p function.
One difference between this epitope-tagging system and many conventional methods is that the HAT tag is formed directly in the chromosome rather than by first constructing a plasmid. To shuttle the desired mutations into other strains, the allele must be recovered by cloning or plasmid-rescue techniques (16). Alternatively, incorporation of a yeast origin of replication, a centromere and a yeast selectable marker into the pHSS6 vector used for transposon mutagenesis allows insertions to be screened and recovered as plasmid-borne elements (Jun-Yi Leu and G.S.R., unpublished data).
In summary, the primary advantage of these new transposons is that many types of information and a variety of constructs can be obtained from a single procedure. Null mutations, conditional alleles, mutations in different domains of a protein, epitope-tagged alleles, and reporter fusions can all be generated from a single mutagenesis, allowing rapid acquisition of a great deal of information about a gene and its encoded protein. The transposons are expected to be useful both for analyses of individual genes and for systematic characterization of the yeast genome.
Acknowledgments
We thank B. Manning and L. Vallier for comments on the manuscript, and P. Chua, B. Futcher, F. Heffron, B. Sauer, and R. Tsien for providing strains and plasmids. E.-Y. Choi participated in the early stages of this work. Sequencing was performed by the Keck Sequencing Facility at Yale University. This work was supported by National Institutes of Health Grant HD32637.
Footnotes
Abbreviations: β-gal, β-galactosidase; GFP, green fluorescent protein; HA, influenza virus hemagglutinin epitope; HAT tag, HA/transposon tag; mTn, mini-transposon.
Data deposition: The sequences reported in this paper have been deposited in the GenBank data base [accession nos. U54828U54828 (mTn-3xHA/lacZ), U54829U54829 (mTn-4xHA/lacZ), U54830U54830 (mTn-3xHA/GFP), and U64693U64693 (pRSQ2-LEU2)].
References
- 1.Bassett D E, Boguski M S, Spencer F, Reeves R, Goebl M, Heiter P. Trends Genet. 1995;11:372–373. doi: 10.1016/s0168-9525(00)89109-x. [DOI] [PubMed] [Google Scholar]
- 2.Seifert H S, Chen E Y, So M, Heffron F. Proc Natl Acad Sci USA. 1986;83:735–739. doi: 10.1073/pnas.83.3.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hoekstra M F, Seifert H S, Nickoloff J, Heffron F. Methods Enzymol. 1991;194:329–342. doi: 10.1016/0076-6879(91)94025-8. [DOI] [PubMed] [Google Scholar]
- 4.Rockmill B, Roeder G S. Genes Dev. 1991;5:2392–2404. doi: 10.1101/gad.5.12b.2392. [DOI] [PubMed] [Google Scholar]
- 5.Chun K T, Goebl M G. Genetics. 1996;142:39–50. doi: 10.1093/genetics/142.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chua P, Roeder G S. Mol Cell Biol. 1995;15:3685–3696. doi: 10.1128/mcb.15.7.3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burns N, Grimwade B, Ross-Macdonald P B, Choi E-Y, Finberg K, Roeder G S, Snyder M. Genes Dev. 1994;8:1087–1105. doi: 10.1101/gad.8.9.1087. [DOI] [PubMed] [Google Scholar]
- 8.Field J, Nikawa J, Broek D, Macdonald B, Rodgers L, Wilson I A, Lerner R A, Wigler M. Mol Cell Biol. 1988;8:2159–2165. doi: 10.1128/mcb.8.5.2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sikorski R S, Boeke J D. Methods Enzymol. 1991;194:302–329. doi: 10.1016/0076-6879(91)94023-6. [DOI] [PubMed] [Google Scholar]
- 10.Prasher D C, Eckenrode V K, Ward W W, Prendergast F G, Cormier M J. Gene. 1992;111:229–233. doi: 10.1016/0378-1119(92)90691-h. [DOI] [PubMed] [Google Scholar]
- 11.Heim R, Prasher D C, Tsien R Y. Proc Natl Acad Sci USA. 1994;91:12501–12504. doi: 10.1073/pnas.91.26.12501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tyers M, Tokiwa G, Nash R, Futcher B. EMBO J. 1992;11:1773–1784. doi: 10.1002/j.1460-2075.1992.tb05229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Newman B J, Grindley N D. Cell. 1984;38:463–469. doi: 10.1016/0092-8674(84)90501-4. [DOI] [PubMed] [Google Scholar]
- 14.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 15.Guthrie C, Fink G R. Methods Enzymol. 1991;194:933. doi: 10.1016/0076-6879(91)94058-k. [DOI] [PubMed] [Google Scholar]
- 16.Burns N, Ross-Macdonald P, Roeder G S, Snyder M. In: Microbial Genome Methods. Adolph K W, editor. Boca Raton, FL: CRC; 1996. pp. 298–308. [Google Scholar]
- 17.Sauer B. Mol Cell Biol. 1987;7:2087–2096. doi: 10.1128/mcb.7.6.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoekstra M F, Burbee D, Singer J, Mull E, Chiao E, Heffron F. Proc Natl Acad Sci USA. 1991;88:5457–5461. doi: 10.1073/pnas.88.12.5457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Magnusson S, Peterson T E, Sottrup-Jensen L, Claeys H. In: Proteases and Biological Control. Reich E, Rifkin D B, Shaw E, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1975. pp. 123–149. [Google Scholar]
- 20.Snyder M. J Cell Biol. 1989;108:1419–1429. doi: 10.1083/jcb.108.4.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Belhumeur P, Fortin N, Clark M W. Yeast. 1994;10:385–389. doi: 10.1002/yea.320100311. [DOI] [PubMed] [Google Scholar]
- 22.Lygerou Z, Conesa C, Lesage P, Swanson R N, Ruet A, Carlson M, Sentenac A, Seraphin B. Nucleic Acids Res. 1994;22:5332–5340. doi: 10.1093/nar/22.24.5332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Casadaban M J, Martinez-Arias A, Shapira S K, Chou J. Methods Enzymol. 1983;100:293–308. doi: 10.1016/0076-6879(83)00063-4. [DOI] [PubMed] [Google Scholar]
- 24.Niedenthal R K, Riles L, Johnston M, Hegemann J H. Yeast. 1996;12:773–786. doi: 10.1002/(SICI)1097-0061(19960630)12:8%3C773::AID-YEA972%3E3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 25.Wilson I A, Niman H L, Houghten R A, Cherenson A R, Connolly M L, Lerner R A. Cell. 1984;37:767–778. doi: 10.1016/0092-8674(84)90412-4. [DOI] [PubMed] [Google Scholar]
- 26.Snyder M, Gehrung S, Page B D. J Cell Biol. 1991;114:515–532. doi: 10.1083/jcb.114.3.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sternberg N, Hamilton D, Hoess R. J Mol Biol. 1981;150:487–507. doi: 10.1016/0022-2836(81)90376-4. [DOI] [PubMed] [Google Scholar]
- 28.Schneider B L, Seufert W, Steiner B, Yang Q H, Futcher A B. Nucleic Acids Res. 1995;11:1265–1274. doi: 10.1002/yea.320111306. [DOI] [PubMed] [Google Scholar]
- 29.Ziman M, O’Brien J M, Ouellette L A, Church W R, Johnson D I. Mol Cell Biol. 1991;11:3537–3544. doi: 10.1128/mcb.11.7.3537. [DOI] [PMC free article] [PubMed] [Google Scholar]