Abstract
The in vitro selection for an intramolecular AUG-cleaving hammerhead-like ribozyme is described. One of the ribozymes selected was found to cleave after this triplet, both intramolecularly and intermolecularly, with rates comparable to the rate of the native GUC-cleaving hammerhead ribozyme. Although the selection was designed for cleavage 3′ of the AUG triplet, the ribozyme also cleaves 3′ of the AUA triplet. AUU and AUC triplets are, however, not cleaved, and thus the selected ribozyme is purine-specific for the third position in the triplet. In addition, cleavage 3′ of the AAG triplet has been observed, thus the central U is not essential. Nuclease digestion indicates that the selected ribozyme has a secondary structure similar to that of the native hammerhead ribozyme, although with an altered core and stem–loop II sequence. All nucleotides in the core, except one, are essential for activity. The nucleotides in loop II are sensitive to changes and cannot, as in the hammerhead ribozyme, be replaced by other sequences or a nonnucleotide linker. Thus there are differences between these two ribozymes even though they have similar two-dimensional structures. The new ribozyme enlarges the application of hammerhead ribozymes for the inhibition of gene expression by extending the range of cleavable triplets.
The hammerhead ribozyme is one of the smallest ribozymes known and has thus attracted much attention for the study of structure–function relationships in catalytic RNAs as well as for its potential for the sequence-specific inhibition of gene expression (1–3). The hammerhead cleaves RNA sequence-specifically adjacent to the general triplet sequence NUH, where N is any nucleotide and H can be A, U, or C. Cleavage 3′ to a guanosine such as in GUG is very slow (4.3 × 10−5 min−1) compared with the best triplet substrate GUC (1 min−1) (4). Although the x-ray structure of this ribozyme has been solved and a mechanism proposed (5, 6), the question of what determines its specificity for the NUH sequence is still largely unresolved. It is presumed that the inability to cleave 3′ of a G is due to an unfavorable interaction with the cytosine at position 3 in the core, which prevents the reaching of or stabilizing of the transition state (4, 7).
One way of obtaining an insight into this problem might be to compare sequences of hammerhead ribozymes with different triplet cleaving specificities. In previous publications it was demonstrated, by in vitro selection, that the native hammerhead sequence has been optimized by natural evolution for GUC cleavage (8–12). It was of interest to see what changes have to be imposed on the native hammerhead sequence for it to cleave 3′ of AUG, which usually resists cleavage, and thus arrive at a hammerhead with a new specificity. To achieve this, an in vitro selection was undertaken, where the number of randomized positions in the starting pool corresponded to the number of nucleotides typically found in the core and stem–loop II region of a hammerhead ribozyme. This would allow all possible sequence permutations to be explored in the search for a hammerhead which is able to cleave 3′ of AUG. Recent examples and reviews of the in vitro selection method demonstrate the power of such an approach (13–23).
MATERIALS AND METHODS
Templates and Primers.
Oligodeoxyribonucleotides and oligoribonucleotides were chemically synthesized and purified as previously described (11, 24). The ribozyme containing the hexaethyleneglycol linker was prepared with the 18-O-dimethoxytritylhexaethyleneglycol, 1-[(2-cyanoethyl)-(N,N-diisopropyl)]-phosphoramidite (Glen Research, Sterling, VA). The following primers were used in the selection: primer 1, 5′-TGGTGCAAGCTTAATACGACTCACTATAGGGAGACTGTCTAGATCATGAGGATGCTA-3′; and primer 2, 5′-TCTCGGATCCTGCAGATCATNNNNNNNNNNNNNNNNNNNNNNAGGATTAGCATCCTCAT-3′. These two primers were used for the initial Sequenase reaction, and primer 1 also functioned as a restoration primer for the re-introduction of the lost sequence after cleavage. RT-primer, 5′-TCTCGGATCCTGCAGATCAT-3′; and PCR-primer, 5′-TGGTGCAAGCTTAATACGACTCA-3′, where the HindIII (5′-end) and BamHI and PstI (3′-end) restriction sites are indicated in boldface; the T7 promoter is underlined; the ribozyme cleavage triplet is in italics; and N indicates the randomized nucleotides.
RNA Selection.
RNA selection was performed as previously described (11) with modifications to the initial two cycles of selection. Transcriptions from pool 0 and pool 1 were performed on a larger scale (10 times in 500 μl and 1 time in 250 μl, respectively) for 12 h, and PCR amplification (step 5) was omitted in each case. Successive transcriptions were performed at 100-μl volume. Transcription reactions were for decreasing periods of time from 12 h (first cycle) to 1 min (13th cycle), where reactions for short periods were quenched by addition of EDTA (75 mM final concentration). All transcriptions were performed with 1 μM DNA. DNA template in the PCR mixture was at least 1 × 10−15 M and the minimum concentration of RNA template was 1 × 10−8 M for reverse transcription. The concentrations were determined assuming a molar extinction coefficient at 260 nm of 6,600 per nucleotide.
Cloning and sequencing were performed as described (11) except that the double-stranded DNA (dsDNA) was digested with HindIII and BamHI. Clones from pools 7 and 10 were tested for transcript self-cleavage from linearized plasmids. For clones from pool 13 plasmid DNA was amplified by PCR, using the RT- and PCR-primers, and then transcribed. Rates of intramolecular cleavage of transcripts were determined as described but with 10 units of enzyme per μl (25). Active clones were sequenced.
Nuclease Digestion.
A full-length transcript for the intramolecularly cleaving ribozyme was generated by transcription at 12°C, to minimize cleavage, for 8 h (26) and was 3′-end labeled with 32pCp. Limited nuclease digestions with RNase A, RNase T1, and nuclease S1 were performed as described (27, 28). Alkaline hydrolysis of labeled RNA was performed in 50 mM NaHCO3 at 100°C for 7.5 min.
Cleavage Kinetics.
Cleavage kinetics of intermolecularly cleaving ribozymes were performed with chemically synthesized ribozyme and substrate in 50 mM Tris⋅HCl, pH 8.0/10 mM MgCl2 with 25–500 nM ribozyme and 12.5 nM substrate for single turnover, and 25–500 nM substrate with 1.25–25 nM ribozyme for multiple turnover kinetics (29). Data were fitted to the Michaelis–Menten and Eadie–Hofstee equations by KaleidaGraph (Synergy Software, Reading, PA). Rate constants are the average of several measurements with less than 20% variation. Km values varied by 2-fold at most.
RESULTS
The procedure reported here to select for intramolecular AUG cleaving sequences is based on a previously reported selection of alternative GUC cleaving hammerhead sequences (11). An initial pool of dsDNAs containing 22 randomized positions flanked by two constant regions was synthesized. These were transcribed, with transcription being initiated with guanosine 5′-[γ-thio]triphosphate, to produce a pool of potential ribozymes. Self-cleavage of active ribozymes occurred during transcription, and the cleavage products were then separated from intact transcripts by means of a mercury gel (30). The recovered cleavage products were then reverse transcribed and amplified by PCR to give a pool of cDNAs of the selected ribozymes. This selection procedure was repeated for 13 cycles, during which a selection pressure was exerted by progressively reducing the time of transcription from 12 h for the first six cycles, to 6 h for the seventh, 1 h for the eighth, 30 min for the ninth and tenth, 5 min for the eleventh, and 1 min for the twelfth and thirteenth cycles.
The dsDNA from pools 7, 10, and 13 was cloned and the plasmids were isolated from a random selection of white colonies. Only plasmids that contained active ribozymes were sequenced. These were identified by transcribing the dsDNA produced on PCR amplification of the plasmid insert. Ribozymes sequenced from pools 7 and 10 showed no homology. The rate of intramolecular cleavage of the most active sequence from each of these pools was 0.03 and 0.06 min−1, respectively. Of 70 clones picked from pool 13, only 20 contained plasmids with inserts of the correct length, which were identified by agarose gel electrophoresis, and from these 16 yielded active ribozymes (Fig. 1).
Figure 1.
Sequences and rates of intramolecular cleavage of ribozymes derived from different clones. Underlined sequences are complementary and form stem II.
These 16 active ribozymes could be divided into two groups: one group contained transcripts with 22 nucleotides in the randomized region and the second group contained a deletion in this region, with only 21 nucleotides being present. Several sequences were represented twice, such as those from clones 13/29, 13/36, and 13/40. One of the most active sequences, clone 13/40, with kobs(cis) of 0.52 min−1, was chosen for further study. In comparison, under these conditions the intramolecular cleavage of the native hammerhead with a GUC triplet had kobs(cis) of 0.66 min−1, indicating that the cleavage activity of the selected ribozyme, Rz13/40, with an AUG triplet is comparable. A transcript of only 50 nucleotides, without the extra sequences in stem III used for cloning, showed a kobs(cis) of 0.32 min−1.
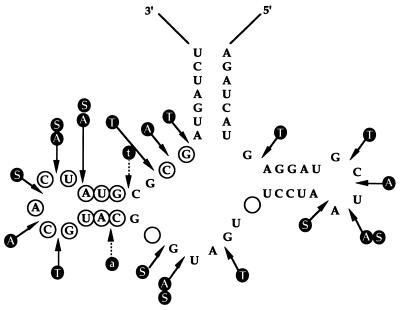
To acquire some information on the secondary structure of Rz13/40, a limited nuclease digestion of the 3′-end-labeled uncleaved ribozyme was performed. Rz13/40 was prepared by transcription in such a way that cleavage was minimized and intact ribozyme could be isolated (26). Limited digestion of the transcript with RNase A and T1 and nuclease S1, which indicates single-stranded regions, gave a digestion pattern consistent with Rz13/40 adopting a hammerhead-like secondary structure (Fig. 2). Because the structure of Rz13/40 resembles the hammerhead so closely, the same numbering system has been adopted, with positions 3 and 9 in the core being considered vacant. The data from the nuclease digestion corresponded fairly well with the mfold structure (data not shown) except for a few discrepancies. mfold indicated base pairing in the core region between U7 and G14, G8 and C13, GL2.1 and UL2.5, and U4 and G17, although there is cleavage at these positions by the nucleases.
Figure 2.
Limited nuclease digestion of full-length transcript of Rz13/40. Letters in filled circles indicate the following: A, cleavage with RNase A; S, with nuclease S1; T, with RNase T1; t, weak cleavage with RNase T1; and a, weak cleavage with RNase A. Nucleotide symbols in open circles, positions different from conventional hammerhead ribozyme; empty circles, position not occupied in Rz13/40.
To further characterize the selected ribozyme for intermolecular cleavage, Rz13/40 was divided into a catalytic portion and the corresponding substrate strand (Fig. 3). The initial ribozyme construct was designed with 5 and 6 base pairs each in stems I and III, respectively, for annealing to the substrate. Stems of this length were chosen because they had been previously shown to give reliable kinetic data with the hammerhead (31). This intermolecular version of Rz13/40 gave a very high K′m (1.1 mM) and low k′cat (0.1 min−1) under single-turnover conditions and was inactive under multiple-turnover conditions. As there was no doubt about the intramolecular cleavage of Rz13/40, the lack of catalytic activity must be associated with the design of the intermolecular construct.
Figure 3.
Ribozyme 13/40 for intermolecular cleavage. Broken line differentiates the two constructs that differ in length of helix III.
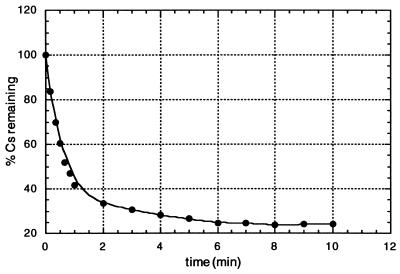
After analysis of the ribozyme–substrate complex by native gel electrophoresis (7) it was concluded that formation of the complex was inefficient under the conditions used. A second ribozyme construct was synthesized, where stem III was extended to 10 base pairs. Native gel analysis confirmed the formation of the ribozyme–substrate complex, and the ribozyme cleaved its corresponding substrate efficiently under both multiple- and single-turnover conditions (Fig. 4 and Table 1). The discrepancy between kcat and k′cat (0.14 and 0.88 min−1, respectively) is presumably the result of product inhibition, which is a common effect observed with the native hammerhead. The time course of cleavage was followed for about 12 half-lives under single-turnover conditions (Fig. 5). Reactions were first-order up to 60% cleavage within the first minute and reached a total of approximately 80%. First-order end points with the conventional hammerhead are commonly 70–75% (32). There was no activity in the absence of Mg2+. A small plateau of activity was reached between 5 and 10 mM Mg2+, but activity rose from there approximately 2.5-fold up to 100 mM. Such an increase of activity is also observed for the native hammerhead ribozyme, with the magnitude depending on the pH of the reaction mixture (33).
Figure 4.
Single-turnover cleavage of Rz13/40 as a function of ribozyme concentration. Conditions were as described in Materials and Methods.
Table 1.
Rate constants for ribozyme 13/40 cleavage at various triplets
| Cleavable triplet | Single-turnover
|
||
|---|---|---|---|
| Multiple-turnover
|
k′cat, min−1 | ||
| kcat, min−1 | Km, nM | ||
| A16.2U16.1G17↓A1.1 | 0.14 | 30.33 | 0.88 |
| A16.2U16.1A17↓A1.1 | 0.10 | 34.09 | 0.38 |
| A16.2U16.1U17↓A1.1 | NA | NA | 0.007 |
| A16.2U16.1C17↓A1.1 | NA | NA | NA |
| A16.2A16.1G17↓A1.1 | 0.19 | 108.7 | 0.54 |
| A16.2U16.1G17↓U1.1 | 0.08 | 80.06 | 0.40 |
| A16.2U16.1G17↓G1.1 | 0.46 | 172.05 | 0.68 |
| U16.2U16.1G17↓A1.1 | ND | ND | 0.83 |
| G16.2U16.1G17↓A1.1 | ND | ND | 1.19 |
The cleavage position is indicated by an arrow. Reaction conditions were as described in Materials and Methods. NA, no activity detectable; ND, not determined; deviations from the AUG-A sequence are underlined.
Figure 5.
Time course of single turnover of Rz13/40 reaction.
The site of substrate cleavage was confirmed as being 3′ to G of the AUG triplet by comparison of the ribozyme cleavage product, obtained under single-turnover conditions after 30 min, with the products of a limited alkaline hydrolysis of the 5′-end-labeled substrate, and with a partial RNase T1 digestion. The cleavage product was also shown to be the 2′,3′-cyclic phosphate by hydrolysis with cyclic nucleotide phosphodiesterase (CNPase) (34).
The effects of mutations around the cleavage site have also been investigated under single- and multiple-turnover conditions (Table 1). Surprisingly, the AUA triplet was also cleaved quite efficiently even though it was not selected for, again yielding a 2′,3′-cyclic phosphate (see above). Triplets terminating in a uridine or cytidine were cleaved either extremely slowly or not at all. Thus this ribozyme is purine nucleoside-specific. It was of interest to test whether Rz13/40 required a central U in the triplet for cleavage. Interestingly the triplet AAG was cleaved by a ribozyme with U at position 15.1 with a k′cat of 0.54 min−1 under single-turnover conditions and under multiple-turnover conditions showed a kcat of 0.19 min−1. In addition, the importance of the nucleotide directly adjacent to the cleavage site was examined. Replacing the nucleotide at position 1.1 with either U or G had only a small effect on the rate of cleavage by ribozymes with the complementary nucleotide at position 2.1 (Table 1).
To investigate the properties and sequence requirements of the selected ribozyme Rz13/40, mutations were made in stem–loop II and the core region of the ribozyme. Removal of a single base pair (10.3⋅11.3) in stem II reduced activity to 0.001 min−1, and changing loop II to GAAA, UUUUU, or a 17-atom polyethyleneglycol linker also drastically reduced activity (Fig. 6). Successively replacing each of the eight nucleotides in the core region by A, or G8 by C, or A6 by G, led to loss of activity with the exception of position 5, where activity was reduced only 5-fold. However, replacement of G5 by pyrimidine nucleotides again led to inactivity.
Figure 6.
Single-turnover rates of intermolecular cleavage for mutated Rz13/40. Boxed region, loop II.
DISCUSSION
The aim of this work was to select, in vitro, an AUG-cleaving ribozyme, starting with a randomized motif based on the hammerhead ribozyme. This project was undertaken to extend the cleavage specificity of the hammerhead ribozyme to G-terminating triplets and to gain insight into the cleavage specificities by comparison of the sequences of the new and the native hammerhead. After 13 rounds of selection, two families of closely related active ribozymes were identified. Of these two classes of ribozymes, the ribozymes containing a one-nucleotide deletion in the randomized region were the better intramolecular cleavers.
A ribozyme, Rz13/40, was selected from this class for further study because it was one of the more active ribozymes, with a rate similar to the native hammerhead cleaving 3′ of the GUC triplet. Initially, the secondary structure of Rz13/40 was probed by limited nuclease digestion, which indicated that the selected ribozymes adopted a secondary structure similar to the hammerhead. Though there are secondary structure similarities with the native hammerhead, there are also differences. The sequence 5′-GCGCG at positions 11.2 to 14 is present in all ribozymes from pool 10 onwards, indicating the importance of this sequence to ribozyme activity, just as the GAAA sequence is in the native hammerhead (10). Other differences include the “vacancies” at positions 3 and 9 in the core. On the basis of the secondary structure, in-trans-cleaving ribozymes were designed (Fig. 3) so that the properties of the ribozyme could be investigated. Surprisingly, the specificity of this ribozyme was broader than envisaged, as cleavage was observed 3′ to A as well as to G. No cleavage 3′ to U and C could be detected. Whether this indicates a necessity for base pairing between U4 and the nucleotide at the cleavage site, as possible with AUG and AUA, is uncertain at present. The central U of triplets, which is invariably required for hammerhead cleavage, could be changed to an A without loss of activity.
Next, the sequence specificity of the core region was investigated. It revealed that replacing any of the nucleotides in the core with A, or A6 by G, led to loss of activity, except for position 5 where, besides the original G, an A was also tolerated. Thus, there is a strong similarity with the hammerhead in that the core sequence at most positions is indeed invariant. It is interesting to note that from the selected ribozymes position 8 in the core would appear to tolerate some variation (G to C). When the in-trans-cleaving ribozyme was synthesized, changing the sequence at this position inactivated the ribozyme. Thus it appears that variation at position 8 is tolerated only when there is a concomitant change in the sequence at position 10.2 in stem II. There is actually less freedom in Rz13/40 than in the hammerhead, where position 7 is tolerant of any nucleotide.
Stem–loop II in Rz13/40 has more stringent sequence requirements than the hammerhead ribozyme. In the hammerhead, a variety of structures are tolerated: the stem of active ribozymes can be reduced to two base pairs (24), and the loop can be of virtually any size and even contain nonnucleotide linkers (24, 35). In the selected ribozyme Rz13/40, less variation in the stem–loop structure is observed. Here, stem–loop II consists of two G⋅C base pairs adjacent to the core, followed by two A⋅U base pairs, and finally terminating in a five-nucleotide loop. The length of the stem appears to have a large effect on catalytic efficiency, because the removal of a single base pair (10.3⋅11.3) abolishes activity.
There are some variations in loop II of Rz13/40 at positions L2.4 and L2.5 that do not interfere with activity. However, replacing the entire loop by GAAA, UUUUU, or a 17-atom polyethyleneglycol linker reduces activity drastically (Fig. 6). This is contrary to what is observed with the hammerhead ribozyme, where these changes are tolerated very well (24, 29, 35). These results taken together indicate a somewhat different role of the stem–loop II structural element for the two ribozymes. A possible interaction between stem I and stem–loop II has been considered for some hammerhead variants but has not been unequivocally established (36). A detailed discussion of structural similarities between the two ribozymes will have to await further structural investigations. These might also then provide further insight into the determinants of cleavage specificity.
The new triplet specificity should be useful for the application of the hammerhead ribozyme family for the inhibition of gene expression, as it expands the targets on a mRNA for cleavage beyond that of the native hammerhead ribozyme (3). Although the conventional NUH triplets might be considered sufficient for a wide application, the accessibility of hammerhead ribozymes to mRNAs is very restricted, as it is for oligonucleotides in general (37–39). For example, screening of a human acetylcholinesterase transcript for GUC and CUC triplets revealed that only 5 of the potential 55 sites were accessible for ribozyme cleavage (40). Thus the specificity of the new ribozyme for cleavage at purines, apparently independent of the nature of the neighboring nucleotides, expands greatly the number of potential cleavage sites and thus has a considerable advantage over the native NUH-cleaving ribozyme for this application.
Acknowledgments
This work was aided by fellowships from the Alexander von Humboldt-Stiftung to N.K.V. and O.F. and by financial support of the Deutsche Forschungsgemeinschaft.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviation: dsDNA, double-stranded DNA.
References
- 1.Symons R H. Annu Rev Biochem. 1992;61:641–671. doi: 10.1146/annurev.bi.61.070192.003233. [DOI] [PubMed] [Google Scholar]
- 2.Thomson J B, Tuschl T, Eckstein F. In: RNA Catalysis, Nucleic Acids and Molecular Biology. Eckstein F, Lilley D M J, editors. Vol. 10. Berlin: Springer; 1996. pp. 173–196. [Google Scholar]
- 3.Birikh K R, Heaton P A, Eckstein F. Eur J Biochem. 1997;245:1–16. doi: 10.1111/j.1432-1033.1997.t01-3-00001.x. [DOI] [PubMed] [Google Scholar]
- 4.Baidya N, Uhlenbeck O C. Biochemistry. 1997;36:1108–1114. doi: 10.1021/bi962165j. [DOI] [PubMed] [Google Scholar]
- 5.Pley H W, Flaherty K M, McKay D B. Nature (London) 1994;372:68–74. doi: 10.1038/372068a0. [DOI] [PubMed] [Google Scholar]
- 6.Scott W G, Murray J B, Arnold J R P, Stoddard B L, Klug A. Science. 1996;274:2065–2069. doi: 10.1126/science.274.5295.2065. [DOI] [PubMed] [Google Scholar]
- 7.Ruffner D E, Stormo G D, Uhlenbeck O C. Biochemistry. 1990;29:10695–10702. doi: 10.1021/bi00499a018. [DOI] [PubMed] [Google Scholar]
- 8.Nakamaye K L, Eckstein F. Biochemistry. 1994;33:1271–1277. doi: 10.1021/bi00171a030. [DOI] [PubMed] [Google Scholar]
- 9.Ishizaka M, Ohshima Y, Tani T. Biochem Biophys Res Commun. 1995;214:403–409. doi: 10.1006/bbrc.1995.2301. [DOI] [PubMed] [Google Scholar]
- 10.Thomson J B, Sigurdsson S T, Zeuch A, Eckstein F. Nucleic Acids Res. 1996;24:4401–4406. doi: 10.1093/nar/24.22.4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vaish N K, Heaton P A, Eckstein F. Biochemistry. 1997;36:6495–6501. doi: 10.1021/bi963134r. [DOI] [PubMed] [Google Scholar]
- 12.Tang J, Breaker R R. RNA. 1997;3:914–925. [PMC free article] [PubMed] [Google Scholar]
- 13.Breaker R R. Chem Rev. 1997;97:371–390. doi: 10.1021/cr960008k. [DOI] [PubMed] [Google Scholar]
- 14.Osborne S E, Ellington A D. Chem Rev. 1997;97:349–370. doi: 10.1021/cr960009c. [DOI] [PubMed] [Google Scholar]
- 15.Abelson, J. N., ed. (1996) Methods Enzymol. 267.
- 16.Wright M C, Joyce G F. Science. 1997;276:614–617. doi: 10.1126/science.276.5312.614. [DOI] [PubMed] [Google Scholar]
- 17.Santoro S W, Joyce G F. Proc Natl Acad Sci USA. 1997;94:4262–4266. doi: 10.1073/pnas.94.9.4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang F, Yarus M. Biochemistry. 1997;36:6557–6563. doi: 10.1021/bi970475b. [DOI] [PubMed] [Google Scholar]
- 19.Faulhammer D, Famulok M. J Mol Biol. 1997;269:188–202. doi: 10.1006/jmbi.1997.1036. [DOI] [PubMed] [Google Scholar]
- 20.Ekland E H, Bartel D P. Nature (London) 1996;382:373–376. doi: 10.1038/382373a0. [DOI] [PubMed] [Google Scholar]
- 21.Frank D N, Ellington A E, Pace N R. RNA. 1996;2:1179–1188. [PMC free article] [PubMed] [Google Scholar]
- 22.Lorsch J R, Szostak J W. Acc Chem Res. 1996;29:103–110. doi: 10.1021/ar9501378. [DOI] [PubMed] [Google Scholar]
- 23.Hager A J, Pollard J D, Szostak J W. Chem Biol. 1996;3:717–725. doi: 10.1016/s1074-5521(96)90246-x. [DOI] [PubMed] [Google Scholar]
- 24.Tuschl T, Eckstein F. Proc Natl Acad Sci USA. 1993;90:6991–6994. doi: 10.1073/pnas.90.15.6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Long D M, Uhlenbeck O C. Proc Natl Acad Sci USA. 1994;91:6977–6981. doi: 10.1073/pnas.91.15.6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frank D N, Ellington A E, Pace N R. RNA. 1996;2:1179–1188. [PMC free article] [PubMed] [Google Scholar]
- 27.Hodgson R A, Shirley J N J, Symons R H. Nucleic Acids Res. 1994;22:1620–1625. doi: 10.1093/nar/22.9.1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Loria A, Pan T. RNA. 1996;2:551–563. [PMC free article] [PubMed] [Google Scholar]
- 29.Thomson J B, Tuschl T, Eckstein F. Nucleic Acids Res. 1993;21:5600–5603. doi: 10.1093/nar/21.24.5600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Igloi G. Biochemistry. 1988;27:3842–3849. doi: 10.1021/bi00410a048. [DOI] [PubMed] [Google Scholar]
- 31.Fedor M J, Uhlenbeck O C. Biochemistry. 1992;31:12042–12054. doi: 10.1021/bi00163a012. [DOI] [PubMed] [Google Scholar]
- 32.Hertel K J, Herschlag D, Uhlenbeck O C. Biochemistry. 1994;33:3374–3385. doi: 10.1021/bi00177a031. [DOI] [PubMed] [Google Scholar]
- 33.Dahm S, Derrick W B, Uhlenbeck O C. Biochemistry. 1993;32:13040–13045. doi: 10.1021/bi00211a013. [DOI] [PubMed] [Google Scholar]
- 34.Heaton P A, Eckstein F. Nucleic Acids Res. 1996;24:850–853. doi: 10.1093/nar/24.5.850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Benseler F, Fu D-J, Ludwig J, McLaughlin L. J Am Chem Soc. 1993;115:8483–8484. [Google Scholar]
- 36.Clouet-d’Orval B, Uhlenbeck O C. Biochemistry. 1997;36:9087–9092. doi: 10.1021/bi9710941. [DOI] [PubMed] [Google Scholar]
- 37.Millner N, Mir K U, Southern E M. Nat Biotechnol. 1997;15:537–541. doi: 10.1038/nbt0697-537. [DOI] [PubMed] [Google Scholar]
- 38.Lima W F, Brown-Drivers V, Fox M, Hanecak R, Bruice T W. J Biol Chem. 1997;272:626–638. [PubMed] [Google Scholar]
- 39.Ho S P, Britton D H O, Behrens D L, Leffet L M, Hobbs F W, Miller J A, Trainor G L. Nucleic Acids Res. 1996;24:1901–1907. doi: 10.1093/nar/24.10.1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Birikh K R, Berlin Y A, Soreq H, Eckstein F. RNA. 1997;3:429–437. [PMC free article] [PubMed] [Google Scholar]