Abstract
A hybrid dysgenesis syndrome occurs in Drosophila virilis when males from an established laboratory strain are crossed to females obtained from the wild, causing the simultaneous mobilization of several different transposable elements. The insertion sequence responsible for the mutant phenotype of a dysgenic yellow allele has been characterized and named Penelope. In situ hybridization and Southern analyses reveal the presence of more than 30 copies of this element in the P-like parental strain, whereas Penelope is absent in all M-like strains tested. Penelope contains one 2.5-kb-long ORF that could encode products with homology to integrase and reverse transcriptase. Northern analysis and whole-mount in situ hybridization show strong induction of a 2.6-kb RNA in the ovaries of dysgenic females that is expressed at very low levels in the parental strains or in the progeny from the reciprocal cross. Injection of Penelope-containing plasmids into preblastoderm embryos of an M-like strain results in mutant progeny caused by insertion of Ulysses and perhaps other transposons, suggesting that Penelope expression might be responsible for the observed dysgenesis syndrome and the simultaneous mobilization of other transposable elements.
Keywords: Drosophila, hybrid dysgenesis, transposable element, genomic instability
Hybrid dysgenesis in Drosophila melanogaster results in high sterility and mutation rates, male recombination, segregation distortion, and chromosome nondisjunction (1–3). The transposase-encoding P element is responsible for the P–M hybrid dysgenesis syndrome in this species (4, 5). A second hybrid dysgenesis system, designated I–R, also leads to similar abnormalities. Although the dysgenic traits that arise in P–M and I–R crosses are similar, the nature of the transposable elements involved is very different. The I transposable element differs from the P element in that it encodes a protein with sequence similarities to reverse transcriptase (RT) (6). Some dysgenic traits have also been observed in systems involving the hobo family of transposable elements, which can promote high rates of chromosomal rearrangements and other dysgenic traits (7).
A similar dysgenic syndrome takes place in Drosophila virilis in unidirectional crosses between males of a strain named 160 and females of strain 9 (8). These two strains are respectively designated P-like and M-like based on the parallels in their behavior with P and M strains in D. melanogaster. The above cross results in characteristic traits in the progeny such as a high level of gonadal sterility in F1 males and females, chromosomal nondisjunction and rearrangements, male recombination, and the occurrence of multiple visible mutations, although it was shown that neither P nor I elements are present in this species (8). A white mutation (wd9) isolated from the progeny of a dysgenic cross has been characterized, and the insertion sequence responsible for the mutant phenotype has been isolated (9, 10). This sequence is a 10.6-kb long terminal repeat-containing retrotransposon named Ulysses. The transcription pattern of Ulysses is the same in the progeny of dysgenic and reciprocal crosses (M.B.E., unpublished data), suggesting that induction of the phenomenon does not correlate with Ulysses expression. In addition, some of the mutations obtained in the progeny of dysgenic crosses contain a copy of and element other than Ulysses at the mutant locus, indicating that different transposable elements are responsible for the mutant phenotypes. Two of these mutations have been characterized at the molecular level and found to be caused by the insertion of novel transposable elements. The sn10 dysgenic allele is caused by insertion of a mariner/Tc1-like element named Paris, whereas Helena, a LINE-like element, is responsible for the sn25 dysgenic mutation (11). These two elements are present in both the P-like strain 160 and the M-like strain 9 and are therefore not good candidates as causative agents of the dysgenesis syndrome.
Here we describe the characterization of a fourth transposable element named Penelope, which is found in a yellow mutation induced by hybrid dysgenesis in D. virilis. In contrast to the distribution of Ulysses, Paris, and Helena, multiple copies of Penelope are present in the P-like strain 160 but not in the M-like strain 9, suggesting that Penelope might be responsible for the observed dysgenesis syndrome. Consistent with this hypothesis, microinjection of plasmids containing the Penelope element into embryos of an M-like strain results in high incidence of mutations, some of which appear to be the result of Ulysses insertion.
MATERIALS AND METHODS
Flies were maintained on standard medium at 25°C. Wild-type strain 9 was collected in Batumi (Georgia, former Soviet Republic), wild-type strain 2 was collected in Kutaisi (Georgia, former Soviet Republic) in 1970, wild-type strain Krasnodar was collected in Krasnodar City (Russia) in 1982, and wildtype strain Pasadena was obtained from Bowling Green, OH. These strains give high percentage of F1 sterility when crossed with males of strain 160, but no effects are observed when crossed to strain 9. By analogy with the P–M hybrid dysgenesis syndrome, we have designated these strains M-like. Strain 160 is an established laboratory strain from Japan that carries the following recessive markers: b (broken cross-veins) in chromosome 2, tb (tiny bristles) and gp-l2 (gap in longitudinal wing vein 2) in chromosome 3, cd (cardinal eye color) in chromosome 4, pe (peach eye color) in chromosome 5, and gl (glossy eye) in chromosome 6. This strain gives rise to a dysgenic syndrome when crossed to females of the M-like strains described above; by analogy with P–M hybrid dysgenesis we will refer to it as a P-like strain.
DNA from wild-type and mutant strains of D. virilis was prepared as described (12). Southern analyses were carried out by standard procedures (13). Genomic libraries from parental and mutant strains were prepared by partial Sau3A digestion with subsequent ligation into the BamHI site of λDash (Stratagene). Libraries were screened with random primer-labeled probes. Clones of interest were sequenced with Sequenase (Amersham). Biotinylated probes containing Penelope sequences were used for in situ hybridization as described by Lim (14). RNA was isolated by homogenizing 1 g of frozen flies in 4 ml of RNAzol-B solution (Tel-Test, Friendswood, TX). After a heat treatment at 65°C for 10 min, the extract was subjected to chromatography on two sequential oligo(dT)-cellulose columns. Poly(A)+ RNAs were subjected to Northern analysis using standard procedures (13). In situ hybridization to whole-mount ovaries was carried out as described (15). Injections were carried out by standard procedures (4). DNA (100 μg/ml) in 5 mM KCl and 0.1 mM potassium phosphate (pH 6.8) was injected into preblastoderm stage embryos (0–2 h) of the M-like strain 9 using an Eppendorf model 5242 microinjector.
RESULTS
A Dysgenic Mutation Is Caused by a New Transposable Element Named Penelope.
Several mutations were recovered among the F2 and F3 generations resulting from dysgenic crosses between D. virilis females of strain 9 and males of strain 160, including alleles of singed (sn), yellow (y), white (w), and Delta (Dl) (8). In particular, a yellow allele designated yd arose as a single male from a cross between the strains described above. As a first step in determining the molecular basis of this yellow mutation, genomic DNA was prepared from yd flies and both parental strains, digested with HindIII, and subjected to Southern analysis using a fragment from the D. melanogaster yellow locus as a probe. The probe hybridized to an 8.5-kb fragment in both parental strains (Fig. 1A). This band is absent in the mutant and is replaced by an 11-kb fragment, suggesting that the mutant gene has been rearranged or contains an insertion of DNA sequences. To determine the nature of these sequences, the yellow gene was isolated from yd mutant flies and sequences responsible for the yd phenotype were identified by comparing wild-type and mutant clones. The yellow-containing clone was labeled with biotin-UTP and hybridized to polytene chromosomes from the two parental strains. These experiments reveal one hybridization site, corresponding to the yellow locus in D. virilis (17), in the chromosomes of the M-like strain 9, and more than 30 hybridization sites in the P-like strain 160 (Fig. 2 A and B; not all sites are visible). Hybridization sites other than that corresponding to the yellow gene must be due to the presence of insertion sequences similar to that within the yd locus used as a hybridization probe. Characterization of these sequences showed they correspond to a new transposable element we have named Penelope.
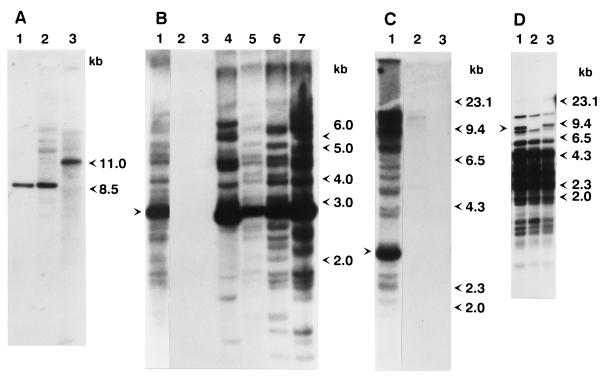
Figure 1.
Southern blot analysis of genomic DNA from wild-type and mutant strains. (A) Ten micrograms of total DNA from strain 9 (lane 1), strain 160 (lane 2), and the yd mutant strain (lane 3) was digested with HindIII and subjected to Southern blot analysis using the SalI–EcoRI fragment of a D. melanogaster yellow-containing clone (16) as a probe. (B) Genomic DNA from various D. virilis strains and mutants isolated from the progeny of dysgenic crosses was digested with XhoI and probed with the XhoI fragment of Penelope. Lanes: 1, strain 160; 2, strain 2; 3, strain 9; 4, white mutant; 5, sn25 mutation; 6, revertant of sn25 mutation; 7, yd mutation. The arrow indicates the position of the 2.8-kb band. (C) Genomic DNA of various D. virilis strains was digested with BamHI and subjected to Southern blot analysis. Lanes: 1, strain 160; 2, Pasadena strain; 3, Krasnodar strain. The arrow indicates the position of a prominent 2.7-kb band present in all strains containing Penelope. (D) DNA from strain 9 (lane 2) and two independent droop mutants (lanes 1 and 3) obtained from the progeny of embryos injected with Penelope-containing clones were digested with EcoRI and HindIII and hybridized with a probe containing an internal SalI–BamHI fragment of the Ulysses element. The arrow indicates an additional restriction fragment seen in both droop mutant strains.
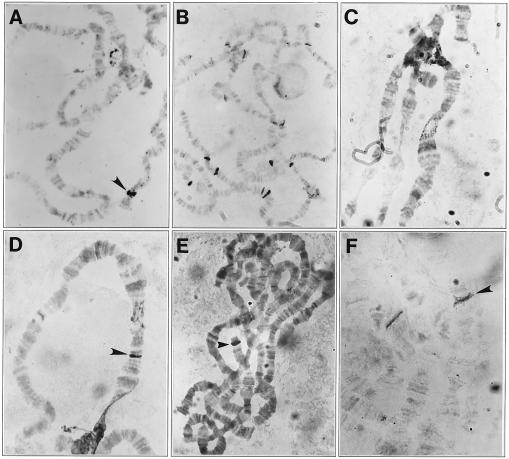
Figure 2.
In situ hybridization of Penelope and Ulysses elements to D. virilis polytene chromosomes. (A) Hybridization of a yellow-containing clone to chromosomes of the M-like strain 9. The arrowhead indicates the only labeled site corresponding to the yellow locus in the D. virilis X-chromosome. (B) Hybridization of the yellow-containing clone to chromosomes of the P-like strain 160; multiple sites are seen due to hybridization of Penelope. (C) Hybridization of the SalI–BamHI fragment of Ulysses with the proximal end of chromosome X of D. virilis strain 9. (D) Hybridization of Ulysses with polytene chromosomes from a droop mutant obtained in the progeny of injected embryos of strain 9; the arrowhead indicates the appearance of an additional site of hybridization. (E) Hybridization of Penelope (clone p1) with strain 9 chromosomes; the only site of hybridization in the 49B section of chromosome 4 is indicated by the arrowhead and is due to the presence of flanking sequences in the clone. (F) Hybridization of the same clone with the chromosomes of strain D8, an unstable strain displaying a Delta phenotype obtained in the progeny of embryos injected with the Penelope element (see Table 1). An additional site of hybridization in chromosome 5 indicated by an arrow resulted from the insertion of Penelope sequences.
Penelope Elements Are Structurally Highly Polymorphic in D. virilis.
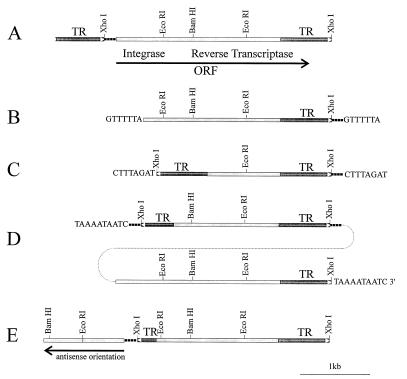
We have sequenced several copies of Penelope isolated from the P-like 160 parental strain or from singed or yellow alleles that arose in the progeny of dysgenic crosses. All copies of the Penelope family studied so far are different in their structure. Fig. 3 summarizes the diversity of structures as determined by DNA sequencing. It is evident that all the elements depicted in this figure have a variable 5′ region and a constant central core region. The Penelope-containing clone py2 isolated from the yellow locus of the yd mutation does not have a 5′ repeat (see below), is truncated in the 5′ region, and is flanked by 7-bp target site duplications (Fig. 3B). The copy of Penelope present in clone p6 is flanked by two long terminal direct repeats of 690 bp and has a complete core region between the two flanking repeats (Fig. 3A). Clone p17 has two terminal repeats in inverse orientation, and the 3′ repeat has an additional 34 bp at its right end and resembles the DIRS elements isolated from Dictyostelium (18) (Fig. 3C). This copy is truncated in the central core region and is flanked by 8-bp target site duplications. Clone p1 contains the largest Penelope copy isolated so far, and it has an organization suggestive of a tandem array of two copies of Penelope (Fig. 3D). The core region of the rightmost copy is identical to that of clone p6, whereas the leftmost copy is truncated in the 5′ region and the deletion includes part of the 5′ terminal repeat. Clone p1, isolated from a genomic library obtained from a strain carrying the dysgenic sn25 mutation, was localized by in situ hybridization to the 49B region of chromosome 4 that does not contain Penelope sequences in the parental P-like strain 160. The fact that p1 is flanked by a 10-bp target site duplication suggests that the array of two copies of Penelope either moved as a unit or that a second element inserted into a pre-existing one; this second element might have inserted by a different mechanism, as no base pair duplications are observed flanking the inserted sequences. Another interesting clone, designated psn25-4, was also isolated from the sn25 strain (Fig. 3E). This copy is very similar in organization to that of clone p17. It also contains two terminal repeats of unequal length and is truncated in the 5′ part of the core region. Unlike all other clones mentioned above, the left end of this copy is composed of 1200 bp from the 5′ core region of Penelope in inverted orientation. We failed to detect any target site duplications at the ends of this Penelope copy. The repeats flanking Penelope elements sometimes carry an additional 34 bp. This tail may be located at the very right end of the 3′ repeat, as is the case for the Penelope copy in py2 and p17. In contrast, this sequence is found at the 5′ repeat in clones psn25-4 and p6, while clone p1 has two such sequences at the ends of two terminal repeats in opposite orientation.
Figure 3.
Schematic representation of the structure of different Penelope copies isolated from various genomic libraries. (A) Penelope from strain 160 containing terminal repeats in direct orientation and a complete ORF (p6). (B) Penelope element inserted in the yellow locus in the dysgenic yd allele (clone py2). (C) Penelope from strain 160 with two repeats in inverse orientation (p17). (D) Copy of Penelope isolated from strain sn25 obtained from a dysgenic cross; this copy contains two tandemly arranged central cores carrying two terminal repeats in direct orientation and one in inverted orientation (clone p1). The dotted line represents the genomic tandem organization. (E) Penelope from a dysgenic mutant strain with part of the core region in antisense orientation (clone psn25-4).
Genomic Organization of the Penelope Element.
To determine the number of copies of Penelope present in the genome with the different structures depicted in Fig. 3, genomic DNA isolated from D. virilis wild-type and mutant strains was examined by Southern blot analysis using Penelope as a probe (Fig. 1 B and C). No hybridization was detected in four different M-like strains, strains 2 and 9, Krasnodar and Pasadena, used in this experiment (Fig. 1 B and C, lanes 2 and 3). Significant fragment length polymorphism was detected in strain 160 and several mutant strains obtained in the progeny of dysgenic crosses (Fig. 1B). In addition to bands of single-copy intensity, a strong hybridization signal corresponding to a band of 2.8 kb is particularly apparent. The intensity of this band gives an indication of the number of Penelope copies with a complete core region, and hence an ORF flanked by two terminal repeats. This is the structure of the Penelope element present in clones p1 and p6. As described above, clone p1 has a complex organization that might be the result of an insertion of one Penelope element into a second copy of Penelope, a characteristic behavior of DIRS elements (18). To estimate the frequency of such tandemly arranged structures in the chromosomes of different D. virilis strains, DNA was digested with BamHI at a site located in the middle of the Penelope-conserved core region. In addition to multiple signals corresponding to single copy equivalents, a prominent band of 2.7 kb is evident in strain 160 (Fig. 1C). The size of the prominent 2.7-kb BamHI fragment closely corresponds to the distance between the two BamHI sites of the Penelope copy in clone p1, which includes two tandemly repeated core regions. The intensity of this band suggests that approximately half of the Penelope copies present in the P-like strain 160 contain this element with the tandem arrangement (Fig. 3D), supporting the suggestion that Penelope tends to insert into or adjacent to pre-existing copies. This characteristic of Penelope might help explain the diversity of structures found for independent isolates of this element. The structure of an intact Penelope element might then be composed of the core region flanked by two terminal repeats arranged in inverted orientations (Fig. 3C). Other observed structures could then be explained by insertion of new copies of Penelope into pre-existing ones in a direct or reversed orientation, followed by homologous recombination at different locations within the composite elements. This type of mechanism could give rise to the different structures shown in Fig. 3. Southern blot analyses shown in Fig. 1 also support a direct correlation between the M-like or P-like behavior of the strains analyzed and the presence of Penelope elements in their genomes. Strain 160, as well as the four dysgenic mutant strains examined, behave as P-like strains; all these five strains contain over 30 copies of Penelope. On the other hand, strains 2, 9, Krasnodar, and Pasadena behave as M-like strains, and none of these strains contain copies of Penelope. The correlation between the presence of Penelope and P-like behavior suggests that Penelope might be responsible for the induction of the dysgenic syndrome.
Penelope Encodes Proteins Homologous to Retroviral Integrase and RT.
The most complete core region of a Penelope element was recovered from clone p6 (Fig. 3A). The DNA sequence of this copy of Penelope was determined and deposited in the GenBank data base under accession number U49102U49102. This element contains a single ORF of 2.5 kb that starts at the 3′ end of the 5′ repeat and extends 480 bp into the 3′ terminal repeat (Fig. 3). The predicted protein sequence of 220 amino acids shows sequence similarity to the integrase and RT proteins of eukaryotic retroviruses and retrotransposons (19, 20). Fig. 4 shows an alignment of the putative integrase of Penelope with other representative retrotransposons. Even though the Penelope sequence differs considerably from all known integrases, the conservation of structural sequence features allows the identification of the His-His-Cys-Cys Zn finger motif and the DD35E motif characteristic of the active site of integrases (19) (Fig. 4). The Zn finger of the putative integrase protein contains a Ser instead of the canonical Cys residue; in addition, the distance between the second Asp and the Glu residues in the DD35E motif is 44-amino acid residues instead of 35. These alterations observed in the Penelope integrase might alter its binding specificity or its ability to cut DNA.
Figure 4.
Sequence alignments of retroviral and Penelope-encoded proteins. (A) Sequence alignment of representative integrases of various retroelements with the putative integrase of Penelope. Amino acids identical or chemically similar to those of the putative Penelope protein are shown as gray boxes. Residues that form part of the Zn finger motif have been indicated by a + symbol, whereas amino acids in the DD35E motif are indicated by ∗. (B) Sequence alignment of RTs of various retroelements and the RT of Penelope. Dotted lines separate conserved blocks of RTs described by Xiong and Eickbush (21). Identical or chemically conserved residues are indicated as shaded boxes. Chemically similar amino acids are grouped as follows: A, S, T, P, and G; N, D, E, and Q; H, R, and K; M, L, I, and V; F, Y, and W (22).
We have also located the N-terminal end of a putative RT-like protein at position 1759 of the Penelope p6 nucleotide sequence. This RT extends to nucleotide position 2700, close to the 5′ end of the 3′ repeat. Fig. 4 shows an alignment of the Penelope RT-like protein with five selected RTs from different retroelements. Because the amino acid sequence similarity of the putative Penelope RT to all other known categories of RT sequences is, on average, only about 14%, we based the alignment on groups of conserved amino acid residues that can be identified in all RT-like sequences available (21, 23). The sequence motive YVDDL identifies the catalytic center of RT (24). RTs are connected via a tether peptide to an RNase H unit, but Penelope seems to lack a tether and the RNase H domain.
Transcription of Penelope Elements.
To investigate the expression of Penelope, Northern blots of poly(A)+ RNA isolated from whole flies, ovaries, or carcasses were hybridized with a Penelope probe. Neither of the parental strains 160 or 9 show appreciable accumulation of Penelope-encoded transcripts, whereas both females and males obtained from a dysgenic cross show high levels of several Penelope-encoded RNAs (Fig. 5A); females of the reciprocal cross show very low levels of Penelope RNAs. One prominent band of 2.6 kb corresponds well to the predicted size for an RNA that starts at the beginning of the core region and terminates at the end of the 3′ terminal repeat, in agreement with the canonical structure we have proposed for Penelope (see above). The rest of the larger bands observed in the Northern blot analysis (Fig. 5A) might be due to transcription of Penelope elements that contain two or more copies in various orientations or transcription from external promoters located in adjacent sequences.
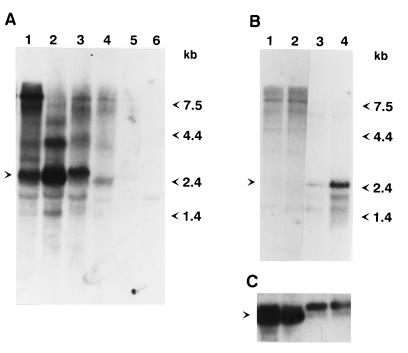
Figure 5.
Analysis of Penelope expression in parental strains and reciprocal hybrids. (A) Transfer analysis of RNA isolated from parental strains and dysgenic hybrids using the 32P-labeled XhoI fragment of Penelope as a probe. Lanes: 1, dysgenic females; 2, dysgenic males; 3, females from the reciprocal cross; 4, males from the reciprocal cross; 5, strain 160 flies; 6, strain 9 flies. The position of the 2.6-kb transcript induced in dysgenic hybrids is indicated by the arrow. (B) Five micrograms of poly(A)+-containing RNA isolated either from ovaries or from fly carcasses lacking ovaries was subjected to Northern blot analysis as described above. Lanes: 1, carcasses of females from the reciprocal cross without ovaries; 2, carcasses of dysgenic females without ovaries; 3, ovaries isolated from females of the reciprocal cross; 4, ovaries isolated from dysgenic females. The position corresponding to the 2.6-kb transcript induced in the ovaries is indicated by the arrow. (C) The same blot was rehybridized with a Drosophila actin gene and is shown as a marker for the amount of RNA. The position of the actin RNA is indicated by the arrow.
To study the significance of the differential expression of Penelope in the dysgenic progeny versus the parental strains, RNA samples were obtained from carcasses and ovaries of dysgenic and reciprocal cross females and subjected to Northern blot analysis (Fig. 5B). Both samples of ovarian RNA show the presence of a 2.6-kb band that is considerably more abundant in dysgenic than nondysgenic females. This RNA is not present in carcasses from individuals resulting from either cross (Fig. 5B). These results indicate a correlation between Penelope RNA expression in the ovaries and the induction of the dysgenic syndrome, suggesting that both phenomena might be causally related, and that Penelope transcription might be a prerequisite for its mobilization and subsequent dysgenesis. The slight accumulation of the 2.6-kb transcript in the ovaries of reciprocal cross females correlates with a low level of hybrid dysgenesis, such as sterility and spontaneous mutations, in the progeny of this cross (8). The high molecular weight transcripts observed in RNA samples from whole individuals (Fig. 5A) do not accumulate in ovaries, and their expression seems to be limited to somatic tissue. This suggests that Penelope expression is under the control of a germ line-specific promoter, and that other transcripts observed in carcasses might originate from promoters located in sequences adjacent to the insertion sites of this element. To confirm the results of Northern blot analysis, we carried out whole-mount in situ hybridization using a labeled Penelope RNA probe that detects sense RNA. Strong labeling of oocytes and nurse cells at all stages of oogenesis was observed in egg chambers of dysgenic females, whereas no significant hybridization was present in the oocytes of control females from the M-like strain 9 (data not shown). These results support the idea of a primary role for Penelope in the induction of hybrid dysgenesis in D. virilis.
Injection of Penelope-Containing Clones into D. virilis Embryos Causes Germ-Line Mutations.
To further investigate the role of Penelope in the induction of the dysgenic syndrome, three different clones containing Penelope were injected into early embryos of strain 9. The p1 and p6 clones contain a putative intact copy of Penelope plus flanking sequences corresponding to the chromosomal location where the particular copy of Penelope had inserted. If Penelope plays a role in inducing the dysgenic syndrome, injection of these plasmids should give rise to new mutations caused by Penelope or one of the other elements previously found in dysgenic alleles (Ulysses, Paris, or Helena). The p17 clone lacks half of the core region of Penelope, including the putative integrase, and it probably represents a defective copy; injection of this plasmid into M-like embryos should have no mutagenic effect, and it was used in these experiments as a control. After injection, F0 females were crossed individually to males of strain 9 and the F1 generation was examined for any changes in phenotype. F1 flies were crossed inter se to establish putative mutant strains. The results are summarized in Table 1. Mutant phenotypes were detected after inter se crosses of the F1 progeny to make homozygous any recessive mutations. A total of 19 mutations were obtained in the progeny of 175 embryos injected with the p1 and p6 clones, whereas no mutants were observed in the progeny of 23 embryos injected with the p17 plasmid. Six of the mutations display phenotypes not previously described in D. virilis and characterized by spread wings (three independent alleles), pointed wings (two independent mutations), and deformed legs with extra hairs (two alleles). Thirteen additional mutants had phenotypes similar to previously described mutations. These mutations were assigned to specific genes based on the characteristics of the mutant phenotypes and allelism established by crosses with known mutations (Table 1). The most striking feature was the frequent occurrence of Delta mutations; interestingly, the Dl locus is a hot spot for mutations occurring as a result of dysgenic crosses (8). In situ hybridization studies showed that 11 of the mutations obtained correlate with the presence of Ulysses at the corresponding cytogenetic locus. This conclusion is based on the close correlation between the known genetic location of the mutant locus and the cytogenetic location of Ulysses insertion. For example, the droop mutation has been mapped to the 18C region of chromosome 1 (17), and the droop alleles obtained in the injection experiments contain a copy of Ulysses at this location (Fig. 2 C and D). The observation of new sites of Ulysses hybridization is not due to polymorphisms in Ulysses insertion sites in strain 9, as no hybridization sites were observed for this element in over 100 individuals of strain 9 examined by in situ hybridization (data not shown). Southern blot analysis showed that a new restriction fragment hybridizes with Ulysses in both independent droop mutants (Fig. 1D), confirming the results of in situ studies. Therefore, the injection of Penelope clones into embryos of strain 9 mimics the dysgenic crosses leading to Ulysses mobilization. None of the mutations tested was associated with the presence of Penelope, and the possible mobilization of Paris and Helena was not determined. Because not all insertions will result in a visible mutation, the number of established mutant strains listed in Table 1 is clearly an underestimate of the actual frequency of transposition events. For example, new sites of insertion of Penelope can be observed among the progeny of injected individuals (Fig. 2 E and F), suggesting that injection of this element can both promote its own transposition as well as that of Ulysses, although no mutations caused by Penelope insertion were identified. In fact, ≈100 additional mutant phenotypes were identified among the progeny of injected individuals, but these flies were sterile and stocks could not be established for further study.
Table 1.
Summary of results from injection experiments
| Penelope clone | Fertile F0 females | F1 flies examined | Mutant strains established | Phenotype of mutation | Mutations with Ulysses insertion |
|---|---|---|---|---|---|
| p6 | 57 | 20,000 | 8 | droop (3) | 3 (18C) |
| triangle (1) | 1 (13D) | ||||
| ebony (1) | |||||
| Spread wings (2) | 2 (22C) | ||||
| Pointed wings (1) | |||||
| p1 | 118 | 55,800 | 11 | Delta (4) | |
| clipped (1) | |||||
| apricot (1) | |||||
| small bristles (2) | 2 (15CD) | ||||
| Deformed legs (2) | 2 (45E) | ||||
| Spread wings (1) | 1 (22C) | ||||
| Pointed wings (1) | |||||
| p17 | 23 | 9,000 | 0 | 0 | 0 |
Preblastoderm embryos of D. virilis strain 9 were injected with three different Penelope-containing plasmids, and the progeny were analyzed for visible phenotypes. Phenotypes of mutations are indicated in italics when they correspond to a previously characterized mutation; numbers of independent alleles isolated are indicated in parentheses. The numbers of these alleles containing a copy of Ulysses at the insertion site are also indicated, with the chromosomal locations in parentheses.
DISCUSSION
The Penelope transposable element described here is a good candidate for the causative agent of hybrid dysgenesis in D. virilis. Most copies of Penelope isolated so far are flanked by repeats; in some cases these repeats are arranged in the same orientation whereas other Penelope copies contain inverted repeats. The functional Penelope unit probably contains the core region flanked by direct terminal repeats. This peculiar organization, coupled with extremely high structural polymorphism and instability of isolated clones, resembles the DIRS elements described in Dictyostelium, which also tend to jump into copies of themselves (18). The comparison of Penelope RT and integrase with corresponding proteins of retroviruses and retrotransposons supports the idea of a unusual organization for this element. No category of RT-like proteins was found to exceed 21% amino acid identity with the putative Penelope RT (data not shown), suggesting that Penelope does not fit into any of the previously defined families of retrotransposons (25). This assumption is also supported by the unusual organization of Penelope-encoded genes, in which the integrase is closely associated with the 5′ long terminal repeat followed by an unidentified protein and then an RT-like protein.
Several lines of evidence implicate Penelope in the hybrid dysgenesis syndrome observed in D. virilis. First, the absence of this element in the four M-like strains analyzed. Second, the pattern of tissue-specific expression of Penelope in parental and hybrid flies demonstrate strong induction of transcription of this element in the ovaries of dysgenic females as has been observed in the case of I–R dysgenesis (26). Finally, injection experiments using different Penelope clones provide support for a causative role for Penelope in the syndrome. Injection of Penelope-containing plasmids into preblastoderm embryos of an M-like strain results in the induction of visible mutations, some of which are due to Ulysses insertion; the appearance of transposable element-induced mutations is part of the hybrid dysgenesis syndrome. In addition, new Penelope insertion sites were also observed in the progeny of injected individuals, indicating that Penelope can promote both its own mobilization and that of Ulysses. The mechanism by which Penelope induces the mobilization of other transposable elements to bring about the dysgenic syndrome might be related to the synthesis of Penelope-encoded proteins in the germ line of the parents of mutant individuals. Penelope RNA is present in the nurse cells and oocyte of dysgenic females, and injection of Penelope-containing plasmids in the posterior end of preblastoderm embryos should ensure the presence of Penelope products in the pole cells and germ line of the injected flies. Subsequent mobilization of Penelope might be simply due to the availability of Penelope RNA in the germ line of dysgenic and injected individuals as substrate for reverse transcription and integration. Other transposable elements mobilized during dysgenesis should be expressed in the germ line independent of Penelope, and the rate limiting step for their integration might be the lack of protein products that can be supplied by Penelope—i.e., RT or integrase. Because the Paris element that comobilizes with Penelope during dysgenesis is Tc1-like in structure, it is difficult to explain a role for RT in the mobilization of this element. This leaves Penelope integrase as the most likely candidate for causing mobilization of other elements.
Acknowledgments
The work reported here was supported by Public Health Service Award GM35463 from the National Institutes of Health and American Cancer Society Grant DB-7F to V.G.C., and by a Fogarty International Research Center Award from the Fogarty International Center to M.B.E.
Footnotes
References
- 1.Hiraizumi Y. Proc Natl Acad Sci USA. 1971;68:7369–7373. [Google Scholar]
- 2.Sved J A. Aust J Biol Sci. 1976;29:375–388. doi: 10.1071/bi9760375. [DOI] [PubMed] [Google Scholar]
- 3.Kidwell M G, Kidwell J F, Sved J A. Genetics. 1977;86:333–351. [Google Scholar]
- 4.Rubin G M, Kidwell M G, Bingham P M. Cell. 1982;29:987–994. doi: 10.1016/0092-8674(82)90462-7. [DOI] [PubMed] [Google Scholar]
- 5.O’Hare K, Rubin G M. Cell. 1983;34:25–35. doi: 10.1016/0092-8674(83)90133-2. [DOI] [PubMed] [Google Scholar]
- 6.Finnegan D J. In: Mobile DNA. Berg D E, Howe M M, editors. Washington, DC: Am. Soc. for Microbiol.; 1989. pp. 503–521. [Google Scholar]
- 7.Blackman R E, Gelbart W M. In: Mobile DNA. Berg D E, Howe M M, editors. Washington, DC: Am. Soc. for Microbiol.; 1989. pp. 523–529. [Google Scholar]
- 8.Lozovskaya E R, Scheinker V S, Evgen’ev M B. Genetics. 1990;126:619–623. doi: 10.1093/genetics/126.3.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scheinker V S, Lozovskaya E R, Bishop J, Corces V G, Evgen’ev M B. Proc Natl Acad Sci USA. 1990;87:9615–9619. doi: 10.1073/pnas.87.24.9615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Evgen’ev M B, Corces V G, Lankenau D-H. J Mol Biol. 1992;225:917–924. doi: 10.1016/0022-2836(92)90412-d. [DOI] [PubMed] [Google Scholar]
- 11.Petrov D A, Schutzman J L, Hartl D L, Lozovskaya E R. Proc Natl Acad Sci USA. 1995;92:8050–8054. doi: 10.1073/pnas.92.17.8050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zelentsova E S, Kraev A S, Evgen’ev M B. Chromosoma. 1986;93:469–476. [Google Scholar]
- 13.Sambrook J, Fritch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 14.Lim J K. Drosophila Inf Serv. 1993;72:73–77. [Google Scholar]
- 15.Lankenau S, Corces V G, Lankenau D-H. Mol Cell Biol. 1994;14:1764–1775. doi: 10.1128/mcb.14.3.1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Geyer P K, Corces V G. Genes Dev. 1987;1:996–1004. doi: 10.1101/gad.1.9.996. [DOI] [PubMed] [Google Scholar]
- 17.Gubenko I S, Evgen’ev M B. Genetica. 1982;65:127–139. [Google Scholar]
- 18.Capello J, Cohen S M, Lodish H F. Mol Cell Biol. 1984;4:2207–2213. doi: 10.1128/mcb.4.10.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kulkosky J, Jones K S, Katz R A, Mack J P G, Skalka A M. Mol Cell Biol. 1992;12:2331–2338. doi: 10.1128/mcb.12.5.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Skalka A M. In: The Dynamic Genome. Fedoroff N, Botstein D, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1992. pp. 173–183. [Google Scholar]
- 21.Xiong Y, Eickbush T H. EMBO J. 1990;9:3353–3362. doi: 10.1002/j.1460-2075.1990.tb07536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schwartz R M, Dayhoff M O. In: Atlas of Protein Structure. Dayhoff M O, editor. Vol. 5. Washington, DC: Natl. Biomed. Res. Found.; 1978. pp. 353–358. [Google Scholar]
- 23.Xiong Y, Eickbush T H. Mol Biol Evol. 1988;5:675–690. doi: 10.1093/oxfordjournals.molbev.a040521. [DOI] [PubMed] [Google Scholar]
- 24.Kohlstaedt L A, Wang J, Friedman J M, Rice P A, Steitz T A. Science. 1992;256:1783–1790. doi: 10.1126/science.1377403. [DOI] [PubMed] [Google Scholar]
- 25.Boeke J D, Corces V G. Annu Rev Microbiol. 1989;43:403–433. doi: 10.1146/annurev.mi.43.100189.002155. [DOI] [PubMed] [Google Scholar]
- 26.Chaboissier M C, Busseau I, Prosser J, Finnegan D J, Bucheton A. EMBO J. 1990;9:3557–3563. doi: 10.1002/j.1460-2075.1990.tb07566.x. [DOI] [PMC free article] [PubMed] [Google Scholar]