Abstract
We have developed a defined system to characterize the role of SR proteins and exonic enhancers in directly promoting splice-site interactions across an intron. Using RNA affinity chromatography, we find that SR proteins alone are sufficient to promote the specific association of the enhancer-containing exon 5 with the adjoining exon 6 from avian cardiac troponin-T. Direct visualization of this exon/exon association by electron spectroscopic imaging shows it to be highly specific. Furthermore, using in vivo characterized mutants of exon 5, we also show that this exon/exon association depends on the splicing enhancer within exon 5. These results suggest a model by which SR proteins may function through exonic enhancers to directly promote exon bridging.
During splicing of precursor messenger RNA (pre-mRNA), splice sites that are short and poorly conserved must be properly chosen and brought together across introns, some as large as 100 kb (reviewed in ref. 1). This process is modulated during alternative splicing in which as many as hundreds of different mRNAs are formed from single genes by alternative selection of different splice sites. Research to date has focused on identifying cis sequences in pre-mRNAs and trans-acting factors involved in alternative splicing; it follows that characterizing their mechanism of action will enhance our understanding of how splice sites are chosen.
Two factors that appear to play a role in alternative pre-mRNA splicing are exonic splicing enhancers and SR proteins. Exonic enhancers have been found to promote splicing in a variety of pre-mRNAs from a number of species; they are characteristically composed of purine-rich sequences that reside within alternatively spliced exons (reviewed in refs. 2 and 3). Exonic enhancers have also been shown to bind to members of the SR protein family of essential splicing factors that promote a wide array of splicing events (reviewed in ref. 4). Individual SR proteins, for instance, have been shown to have distinct functions in promoting alternative 5′ splice site selection (5). As well, SR proteins are sufficient to protect pre-mRNAs from splicing inhibition by certain competitor RNAs (6). These results indicate that both SR proteins and exonic enhancers may have an early role in the process of splice-site selection.
One mechanism by which SR proteins and exonic enhancers may influence splice-site selection is to recruit the splicing machinery to particular splice sites. Critical to the assembly of the splicing machinery is a series of base-pairing interactions between five small nuclear RNAs [U1, U2, U4, U5, and U6 small nuclear RNA (snRNAs)] and the pre-mRNA (reviewed in ref. 7). SR proteins have been shown to be important for the earliest snRNA/pre-mRNA associations that can be detected in splicing. For example, SR proteins directly interact with the U1 small nuclear ribonucleoprotein (snRNP) and promote its association to the 5′ splice site (8, 9). In addition, SR proteins, bound to exonic enhancers, appear to promote an interaction of the U2 snRNA with the 3′ splice site (10–13). These studies indicate that SR proteins and exonic enhancers facilitate the interaction of snRNPs with the pre-mRNA during splicing.
Though much is known about the mechanisms by which individual splice sites interact with the splicing machinery, it is still unclear how the 5′ and 3′ splice sites are correctly brought together before catalysis. Studies showing that SR proteins and exonic enhancers can activate trans splicing indicated that these molecules may function to promote 5′/3′ splice-site associations (14, 15). However, these experiments, performed in complex nuclear extracts, did not distinguish whether these molecules can be functioning directly or indirectly by promoting snRNP/pre-mRNA interactions. In this study, we hypothesized that both SR proteins and exonic enhancers may have a direct role in this process. To test this, we used in vivo characterized mutants of an exonic enhancer to develop two assays to study exon/exon associations. First, using RNA affinity chromotography, we have found that SR proteins, in a defined system, are sufficient to promote the specific association of two exons that are spliced together in vivo. Second, using electron spectroscopic imaging, we have directly visualized this SR protein-mediated exon/exon association and find that it is highly specific. These results indicate that SR proteins and exonic enhancers function not only to mediate snRNP/pre-mRNA interactions, but also to directly promote exon/exon associations.
MATERIALS AND METHODS
Assembly of the RNA Affinity Columns.
Two oligonucleotides were annealed together to create a double-stranded exon 5-UP DNA (16) sequence, which was ligated into the EcoRV site of Bluescript (KS+). A XhoI/EcoRI fragment containing two copies of the R17-binding site was ligated between the SmaI/EcoRI sites 5′ to the exon 5-UP sequence (17). Site-directed mutagenesis was used to create the R17-DOWN (referred to as INAC; ref. 16) and R17-exon 6 (18) fusion DNAs from the R17-UP plasmid. The sequence of the R17 fusion RNA is shown below with the R17 binding sequence underlined. The sequences of the exons are shown in the next section.
R17-exon fusion RNAs: GGGCGAAUUGGAGCUCGACCGCGGUGGCGGCCGCUCUAGAACUAGUGGAUCCCCCUCGAGCAGCUGAAGCUUGCAGCUGCAGGUCGACCUAGAAAACAUGAGGAUCACCCAUGUCUGCAGGUCGACUCUAGAAAACAUGAGGAUCACCCAUGUCUGCAGGUGACCUAGAGGAUCCCCGGGUACCGAGCUCGAAUCGAUAUUCAGEXON.
R17-exon fusion RNAs were transcribed with T7 polymerase (19) from R17-UP and R17-DOWN DNAs linearized with Bbs1 and R17-exon 6 DNA linearized with ApaI. For each column, 5 μg of RNA was bound to 10 μg of R17-GST protein affixed to 70 μl of glutathione agarose similar to previous methods (20). Before complex assembly, columns were washed once in 400 μl of buffer S (20 mM Hepes, pH 7.65/250 mM KCl/4 mM MgCl2/0.01% Triton X-100/1 mM DTT).
Complex Assembly, RNA, and Protein Extraction.
The RNA-affinity columns were incubated for 1 hr at 30°C in a volume of 220 μl in 20 mM Hepes, pH 7.65/220 mM KCl/3.6 mM MgCl2/3.6 mM ATP/4.5 mM creatine phosphate with 100 μl of various extracts. For the immunoblot analysis, 100 μl of HeLa nuclear extract was added (21) along with 4.9 μg of calf thymus SR proteins (22). For the northern blot analysis, 100 μl of HeLa S100 extract was added (21) with or without 10.4 μg of HeLa SR proteins (22). For the defined system exon/exon binding studies, 100 μl of a solution of 200 ng/ml BSA, 50 ng/ml E. coli tRNA was added with or without 2.6 μg of calf thymus or HeLa SR proteins. The final concentration of SR proteins in the defined system was 12 μg/ml, whereas the SR protein concentration in standard splicing reactions with HeLa nuclear extract is approximately 30 μg/ml (23). After incubation, columns were washed four times at 4°C in buffer S with 0.1% Triton X-100. All washes were 400 μl.
To some columns, various radiolabeled RNAs were added at the same time as SR proteins. UP exon 5, DOWN exon 5, and HET RNAs were transcribed as previously described (16). Control RNA was transcribed from Bluescript (KS+) linearized with XbaI (Figs. 2 A–C, and 3) or XhoI (Fig. 2D). The Exon 6, Int1, Int2, Int3, and Int4 RNAs were transcribed from synthetic oligonucleotide templates. The exon 6 RNA consists of the entire sequence of wild-type avian cardiac Troponin-T (cTnT) exon 6 (18). The sequences of intron fragments 1–4 are in the intron between exons 5 and 6 of avian cTnT at positions 240–296 (Int1), 132–188 (Int2), 624–679 (Int3), and 323–377 (Int4) (18). The sequences of these RNAs are shown below; underlined are heterologous sequences that are derived from the transcription template:
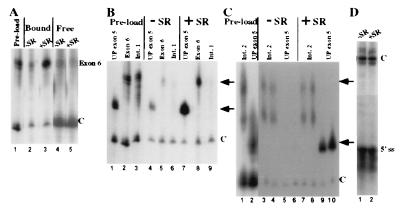
Figure 2.
SR proteins, in a defined system, promote an association of exons that are spliced together in vivo that is specific with respect to intronic RNAs. (A) Association of exon 6 with R17-UP columns in the presence and absence of SR proteins. The relative amount of exon 6 and control RNA added to R17-UP columns is shown (lane 1). The signals from exon 6 and the control RNA are shown for complexes assembled in the absence (lane 2) or presence (lane 3) of SR proteins at a concentration similar to that in standard in vitro splicing reactions. Shown are the signals for supernatant fractions from the complexes shown in lanes 2 and 3 (lanes 4 and 5, respectively). (B and C) Association of intron fragments Int1 and Int2 with R17-UP columns in the presence and absence of SR proteins. (B) The relative amount of UP exon 5, exon 6, and Int1 RNA added to R17-UP affinity columns is shown (lanes 1, 2, and 3, respectively). The signals from the UP exon 5, exon 6, and Int1 RNAs are shown from complexes assembled in either the absence (B, lanes 4–6, respectively), or presence (lanes 7–9, respectively) of SR proteins. (C) The relative amount of UP exon 5 and Int2 RNA added to R17-UP affinity columns are shown (lanes 1 and 2, respectively). The signals from the UP exon 5 and Int2 RNAs are shown from complexes assembled in duplicate in the absence (lanes 3 + 4 and 5 + 6, respectively) and presence (lanes 7 + 8 and 9 + 10, respectively) of SR proteins. (D) The signal from the 5′ splice site RNA is shown from R17-UP affinity complexes assembled in the absence (lane 1) and presence (lane 2) of SR proteins. Because of the small size of the 5′ splice site, a larger control RNA was used in this experiment. In all experiments, one-third of each complex was loaded on each lane. The relative migration of the exon RNAs and the internal control RNAs is indicated to the right of each figure by arrows and Cs, respectively.
Exon 6: GGUCAGGAGGAUCAGGUAGACGAGGAGGAAGAGGAGACAGAGGAAACCACGGCAGAAG; UP exon 5: GGGCGAAUUGGUCAGAAGAGGAAGAAGAAGAAGAGGAAGACGACGGU; DOWN exon 5: GGGCGAAUUGGUCAGAAGGGAGGGAAUGGCUUGAGGAAGACGACGGU; HET exon: GGGCGAAUUGGUCGUUCACAACCAUCUAAAGCAAGAUGUCUGAGU; intron fragment 1 (Int1): GGCUGUGGGUCCAAACGUCUCCUGCAGGACCUGCGGGCUCUGACAGAGGACUCUCGUG; intron fragment 2 (Int2): GGCGCUCUCCCCAUCCUGCUGUGCCAACCUGCUCUCAGUUCUGUGCUUUCUGUCUUCC; intron fragment 3 (Int3): GGGCUGUAGGGAGCCAGCAGGAGCUGCGGCCGUCCUACUGACCCUGUCCUUAUUGCAG; intron fragment 4 (Int4): GGGAUGCUCCAGCAGUGUCAUGCAGGAGAUUUAUGCCAUACAGUUUUGCUCUCUGCUG; control polylinker (C): GGGCGAAUUGGAGCUCCACCGCGGUGGCGGCCGCUCUAG.
All of these RNAs were transcribed with T7 polymerase and labeled by using [α-32P]GTP. The RNAs for a given experiment were transcribed in parallel with the ratios of labeled GTP/cold GTP set such that all the RNAs would have the same molar-specific activity. RNAs were purified from 5% urea/PAGE gels and quantified by using a scintillation counter. Equal amounts of the exon/intron RNAs were mixed with a set amount of control RNA to make normalized RNA samples. A part of these samples were saved as preload, and the rest was added in equal amounts to RNA affinity columns. Additionally, to some columns, 2 ng of end-labeled 5′ splice-site oligo wt41: 5′-AAG/GUAAGUAdT-3′ was added (24).
Each resin was transferred to a clean tube before RNA or protein extraction. RNA was extracted as previously described (21). For snRNA analysis, one-sixth of each sample was loaded onto a 6 M urea/10% acrylamide gel. snRNA Northern blotting was performed similar to previous methods (25, 26) except that the probes were the following end-labeled oligonucleotides: U2, 5′-GGAGGTACTGCAATACC; U1, 5′-TCCCCTGCCAGGTAAGTAT; U4, 5′-ACTGCCACTGCGCAAAGCT; U5, 5′-TGGGTTAAGACTCAGAGTT; U6, 5′-ATGGAACGCTTCACGAATTT.
For radiolabeled RNA-binding studies analysis, one-third of each sample was loaded onto a 6 M urea/5% acrylamide gel. In some experiments, gels were exposed to a PhosphorImager and the relative signals were quantified with imagequant (Molecular Dynamics). Protein was extracted from the columns by incubating each resin with 40 μl protein sample buffer (27). The protein was resolved on SDS/polyacrylamide gels and was transferred to nitrocellulose and probed by immunoblotting as previously described (27).
Electron Spectroscopic Imaging.
To create the UP5/WT6 plasmid, a PCR fragment of the cTnT genomic construct ΔPB.SA (28), which begins 5′ of exon 5 by 306 nt and ends 3′ of exon 6 by 48 nt was generated and ligated into the EcoRI/XbaI sites of Bluescript (KS+). Site-directed mutagenesis was used to create the HET5/WT6 and UP5/HET6 DNAs from the UP5/WT6 plasmid. cTnT RNAs were transcribed with T7 polymerase as described previously (19).
RNA at a concentration of 50 nM and SR protein at a concentration of 250 nM were incubated in a 40 μl volume for 1 hr at 30°C in a reaction buffer containing 20 mM Hepes (pH 7.65), 200 mM KCl, 4 mM MgCl2, 2.5 mM insulin, 4 mM ATP, and 5 mM creatine phosphate. Five microliters of the reaction mix was placed directly onto electron microscope (EM) grids or diluted up to 5-fold in reaction buffer. The 1,000-mesh EM grids were previously coated with a 3-nm-thick carbon film to support the molecules and glow-discharged immediately before exposure to the reaction mix (29). After 30 sec, excess material was washed from the grids with water (GIBCO). Most of the water was removed with filter paper before staining with 50 μM uranyl acetate for 30 sec. Excess stain was then removed with a water wash, and the grids were allowed to air dry.
The grids were examined in a Zeiss EM 902 transmission electron microscope equipped with an imaging spectrometer (29). A 400-μm condenser aperture, 90-μm objective aperture, and 20-eV energy-selecting slit aperture were used. Images were recorded at ×13,000 magnification at 120 eV energy loss to take advantage of the contrast contributed by uranium because of its O4,5 ionization edge. The images were recorded on electron image film (Kodak, SO-163) and were developed at full-strength developer (Kodak, D-19) for 15 min.
RESULTS
We have examined the specific associations of exon 5 from avian cTnT with multiple components involved in pre-mRNA splicing. The alternatively spliced exon 5 has been shown to have a purine-rich exonic splicing enhancer that has dramatic effects on exon 5 inclusion (28). In avian muscle cells transfected with a minigene of cTnT, exon 5 is included at ≈75%, whereas two mutants of exon 5, UP and DOWN (previously referred to as INAC), are included at >98% and <2%, respectively. These mutations have similar effects on exon 5 inclusion in vitro (16, 30). These experiments indicated that splicing components may associate with the active enhancer-containing exon. To test this hypothesis, we assembled complexes on these two exon 5 RNAs by using affinity chromatography. The affinity columns were generated by using fusion RNAs that contain a 32-nt exon 5 sequence placed downstream of two copies of the binding site for R17 coat protein from R17 phage RNA (17). The binding sites were used to tether the exon 5 RNA to glutathione agarose via an R17 coat protein/glutathione S-transferase fusion protein (17, 20).
A Subset of SR Proteins Associates with UP Exon 5 and Promotes Its Association with the U1 snRNP.
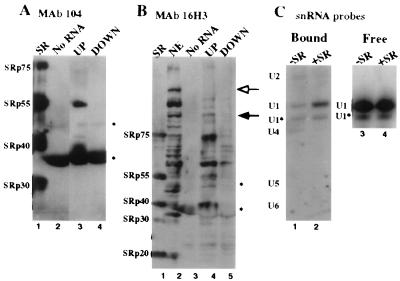
To identify subsets of splicing factors that associate with R17-UP affinity columns, we incubated them with splicing-competent nuclear extracts under standard splicing conditions (31). The columns were then washed to isolate purified complexes. We first asked whether distinct SR proteins bind to the R17-UP affinity columns. Immunoblotting analysis with the anti-SR protein monoclonal antibody mAb 104 (Fig. 1A) shows that R17-UP complexes contain significant amounts of SRp40 and SRp55 and some SRp75; but neither SRp30a nor SRp30b were detectable (lane 3). In contrast, all of these SR proteins are detected in a preparation of SR proteins (lane 1), and no SR proteins are detected in complexes assembled either with R17-DOWN RNA or without column RNA (lanes 4 and 2, respectively). SRp20 was not detectable in any lanes of this blot, but by 16H3 immunoblot, SRp20 does not appear to associate with R17-UP columns (see below). These results are in agreement with previous results that show specific UV cross-links of a subset of SR proteins with a functional exon 5 RNA (16). Additionally, immunoblot analysis with mAb 16H3, which binds a larger family of alternating arginine proteins, shows that R17-UP complexes have a higher affinity than R17-DOWN columns for a subset of non-SR protein-alternating arginine proteins that are present in the starting extract (Fig. 1B, compare lanes 2 and 4). These results indicate that a subset of alternating arginine proteins, including a subset of SR proteins, specifically associates with the R17-UP RNA.
Figure 1.
A subset of SR proteins associate with UP exon 5 and promote its association with the U1 snRNP. Immunoblotting signals from mAb 104 and mAb 16H3 are shown for RNA-affinity complexes isolated from HeLa nuclear extract without RNA lane 2 (A) and lane 3 (B), with the R17-UP RNA lane 3 (A) and lane 4 (B), and with R17-DOWN RNA lane 4 (A) and lane 5 (B). Fifty percent of a complex assembled with 5 μg of column RNA was loaded in each lane. The two bands marked with asterisks on the right of each blot, the lower of which is the R17-GST protein, are due to signal that is independent of primary antibody. Lane 1 of each blot contains 20 μg SR proteins. (B) Lane 2 shows immunoblotting signal from 3 μl HeLa nuclear extract. The outlined arrow to the right indicates a band that is abundant only in starting extract and not the complexes. The solid arrow to the right indicates a band that is present in both starting extract and purified complexes. (C) Northern blot hybridization signals from anti-U snRNA oligonucleotides are shown for R17-UP RNA-affinity complexes isolated from HeLa S100 with (lane 2) or without (lane 1) SR proteins. RNA was also extracted from supernatants of the incubation reactions and probed by Northern blotting with a DNA oligonucleotide antisense to the U1 snRNA. Shown are the resulting hybridization signals for supernatant fractions from the complexes shown in lanes 1 and 2 (lanes 3 and 4, respectively).
Previous studies have demonstrated a role for SR proteins in the association of the U1 snRNA with 5′ splice sites (8, 9). Given that the U1 snRNP has been shown to bind to exonic enhancers (32, 33), we were interested in whether this U1 snRNP/pre-mRNA association was also dependent on SR proteins. The splicing extract S100 contains all of the splicing snRNPs; however, S100 lacks SR proteins and is not competent for splicing unless supplemented with SR proteins. Northern blot analysis of complexes formed in S100 (Fig. 1C) shows that the addition of SR proteins resulted in an elevated amount of U1 snRNA (8-fold, blot performed in triplicate) and a slight increase in the amount of U1* (<2 fold), a U1 snRNA breakdown product (34) present in the complexes (compare lanes 2 and 1). In contrast, the addition of SR proteins resulted in a slight decrease in the amounts of U2 and U4 snRNAs present in the complexes (compare lanes 2 and 1). The U5 and U6 snRNAs were not detectable in any of the complexes. Northern blot analysis of supernatant fractions from these incubation reactions shows that the amount of U1 and U1* snRNAs present at the end of the incubation was the same whether SR proteins were present or not (lanes 3 and 4). This result indicates that SR proteins influenced the affinity of the U1 or U1* snRNPs for the affinity columns rather than their stability in the incubation reaction. Therefore, SR proteins can promote the specific association of the U1 snRNP to an enhancer-containing exon.
SR Proteins Alone Are Sufficient to Promote the Specific Association of the Enhancer-Containing Exon 5 with the Adjoining Exon 6 from Avian cTnT.
We were next interested in whether SR proteins may function at exonic enhancers to promote other RNA/RNA interactions required for splicing. While surveying for potential interactions by sequence examination, we found that purine-rich SR protein binding sites tend to be present not only in the enhancer-containing exon of a pre-mRNA, but also in flanking exons (Table 1). We hypothesized that SR proteins could bind these sequences in exons and promote not only snRNP/pre-mRNA associations, but also exon/exon associations.
Table 1.
Purine-rich sequences tend to be present not only in the alternative exon of a pre-mRNA but also in flanking exons
| Enhancer-containing exon | Purine-rich sequences in flanking exon |
|---|---|
| IgM exon M2 | Upstream exon M1: (17/116) gctGAGGAGGAAGGctt |
| BGH exon 5 | Upstream exon 4: (20/162) tatGAGAAGctGAAGGAcct |
| ASLV env | Distal 5′ exon (23/379) gttGGAAGAcGGGAAGGAAGccc |
| dsx female | Upstream exon 3: (15/139) ggcGAAtcGAAGAGg/gu |
| Fibronectin EDIIIA | Upstream exon −1: (25/114) ctcAGAAtccAAGcGGAGAGAGtca |
| Downstream exon +1: (16/118) cccAAGGAGAAGAccg | |
| Caldesmon exon 5 | Downstream exon 6: (20/78) ag/gGGAGAAGAGAAGGGAActa |
The cDNA and genomic sequences of representative mRNAs containing well characterized exonic enhancers were surveyed for additional purine-rich exonic sequences (38–44). Shown are purine-rich sequences found in exons that flank enhancer-containing exons. The numbers in parentheses are the number of exonic nucleotides shown per the number of nucleotides in the exon. Purine residues are in capital letters. Points where sequences extend into introns are marked by a slash.
To address this possibility, we asked whether SR proteins alone, without splicing extracts, could promote the association of exon 5 with the adjoining exon 6 from cTnT. Radiolabeled exon 6 RNA was incubated with R17-UP affinity columns along with a fixed amount of radiolabeled control RNA both in the presence or absence of SR proteins. To assay for relevant interactions, the concentration of SR proteins in these experiments is 12 μg/ml, which is on the same order as that present in standard splicing reactions with HeLa nuclear extract (≈30 μg/ml; ref. 23). After several washes, RNA was extracted from the columns and resolved on a 5% urea/PAGE gel. In these experiments (Fig. 2A), the addition of SR proteins resulted in a large increase in the amount of exon 6 that bound to R17-UP columns (lanes 2 and 3). In contrast, four fragments of the intron between exons 5 and 6, covering 34% of the intron, and a 5′ splice site RNA (24), were unaffected by SR proteins for their association with R17-UP columns (Fig. 2 B–D, and data not shown). Analysis of supernatant fractions from these incubation reactions shows that the amounts of exonic, intronic, and control RNAs present at the end of the incubations were the same with or without SR proteins (Fig. 2A, lanes 4 and 5, and data not shown). This result indicates that SR proteins do not influence the stability of the these RNAs in the incubation reaction. Together, these results indicate that SR proteins alone are sufficient to promote an interaction between exon 5 and the adjoining exon 6 in a manner that is specific with respect to intronic RNAs.
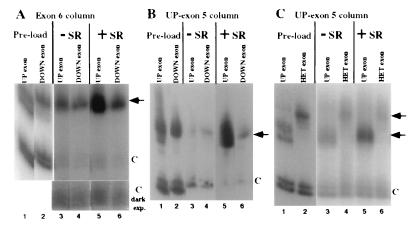
We hypothesized that SR proteins may activate exon 5 inclusion by promoting exon bridging between exons 5 and 6. One prediction of this model is that the efficiency of the SR protein-mediated exon 5/exon 6 interaction should correlate with the splicing enhancer strength of exon 5. To test this prediction, we compared two in vivo-characterized mutants of exon 5, UP and DOWN, for their association with exon 6 columns. In these experiments (Fig. 3A), the addition of SR proteins reproducibly resulted in a 5-fold increase in the amount of UP exon 5 (lanes 3 and 5) and a 2-fold increase in the amount of DOWN exon 5, which bound to R17-exon 6 columns (lanes 4 and 6). Similar results were observed when R17-UP exon 5 was used as the column RNA (Fig. 3B). Assays of RNA levels in supernatant fractions as in Fig. 2 indicate that SR proteins did not influence the stability of the RNAs in these experiments (data not shown). These results indicate that the efficiency of the SR protein-mediated exon 5/exon 6 interaction correlates with the enhancer strength of exon 5.
Figure 3.
SR protein-mediated exon/exon associations are enhancer-dependent. Shown is relative association of UP exon 5, DOWN exon 5, and HET exon with R17-exon columns in the presence and absence of SR proteins. The relative amount of UP exon 5 and DOWN exon 5 RNA added to R17-exon 6 (A) or R17-UP affinity columns (B) is shown (lanes 1 and 2, respectively). The signals from the UP exon 5 and DOWN exon 5 RNAs are shown from complexes assembled either in the absence (lanes 3 and 4, respectively) or presence (lanes 5 and 6, respectively) of SR proteins. A darker exposure of the control RNA is shown for the complexes assembled with R17-exon 6 (lanes 3–6). (C) The relative amount of UP exon 5 and HET exon RNA added to the R17-UP affinity columns is shown (lanes 1 and 2, respectively). The signals from the UP exon 5 and HET exon RNAs are shown from complexes assembled either in the absence (lanes 3 and 4, respectively) or presence (lanes 5 and 6, respectively) of SR proteins. In all experiments, one-third of each RNA-affinity complex was loaded on each lane. The relative migration of the exon RNAs and the internal control RNAs is indicated to the right of each figure by arrows and Cs, respectively.
The SR Protein-Mediated Exon 5/Exon 6 Association Can Be Visualized by Electron Spectroscopic Imaging.
The above results show that SR proteins can promote the association of the 30-nt alternative exon 5 with the 58-nt constitutive exon 6. The finding that intron fragments do not show SR protein-dependent binding in the affinity experiments leads to the idea that SR proteins bring the two short exons together and, as a result, loop out the 683-nt intron. To test this hypothesis, we used electron spectroscopic imaging (ESI) to analyze the structure of exon 5/exon 6 cTnT pre-mRNAs incubated with SR proteins. The advantage of ESI is that very high image contrast of small molecules can be achieved by imaging at an energy loss specific to phosphorus for unstained nucleic acids, or at an energy loss specific for uranium when the specimen is stained with uranyl acetate (35, 36). Thus, RNA and protein can be distinguished with ESI by relative electron energy losses (36).
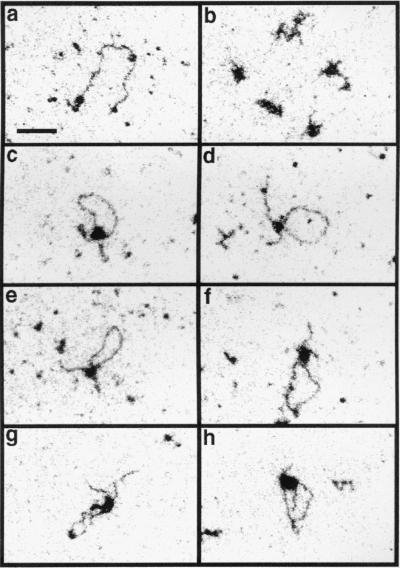
ESI imaging was performed on cTnT pre-mRNAs incubated with SR proteins at a molar ratio of 5 (protein:RNA) under similar conditions as in Fig. 2. In these experiments three kinds of structures were observed. The most prevalent RNA molecules are those not complexed with protein but highly compact because of secondary structure (Fig. 4b). This morphology is the only type seen when no protein was added in a mock reaction (data not shown). The second class is composed of linear molecules, some appearing naked and some with protein molecules attached (Fig. 4a). Contour lengths of the apparently naked molecules were measured. Ninety percent of the molecules measured (n = 58) were within 12% of the mean length. From these measurements, the distance between bases was calculated to be 0.23 nm. This value depends on a number of variables, including the affinity of the RNA for the carbon film. A third class of complexes involves those in which the associated protein appears to cause a loop in the RNA (Fig. 4 c–h).
Figure 4.
Electron spectroscopic images of SR protein/cTnT pre-mRNA incubation reactions show that SR proteins are able to create a loop in the RNA between the two functional exons. SR proteins and UP5/WT6 RNA were incubated under similar conditions as in Fig. 4, and fractions of these reactions were placed directly on EM grids and analyzed with a Zeiss EM 902 transmission electron microscope. The images were recorded at 120 eV. Linear molecules are shown in a, and structures compacted because of RNA secondary structure are shown in b. Loop structures involving the RNA with two functional exons are shown in c–f, one loop involving the RNA with the mutation in exon 5 is shown in g, and RNA with a mutation in exon 6 is shown in h. (Bar = 55 nm.)
To characterize the role of exon sequences in the formation of structures visualized by ESI, we compared the frequency of each of these structures for an RNA containing two functional exons with RNAs containing a mutation in either exon 5 or exon 6. These mutant RNAs were created by replacing either the 30-nt exon 5 or the 58-nt exon 6 with the 30-nt HET exon. The HET sequence both lacks enhancer function in vivo for exon 5 inclusion, exon 4/5, and exon 5/6 splicing (28), and also is unresponsive to SR proteins for associating with R17-UP exon 5 columns (Fig. 3C). The percentage of RNA molecules that were not complexed with protein but were compacted by secondary structure were 82% for the RNA with two functional exons (898 molecules were counted in one experiment), 93% for the RNA with a mutant exon 5 (1,410 molecules), and 95.6% for the RNA with a mutant exon 6 (1,848 molecules). Of the remaining structures of the other two morphological classes, 9.8% of the molecules containing two functional exons had a single loop and both ends of the molecule could be discerned (Fig. 4 c–f). For the molecules with an exon 5 mutant and those with an exon 6 mutant, the fraction that had a single loop, respectively, was 38 and 25% of that seen with the RNA with two functional exons.
We then compared the contour length measurements of the looped structures formed between the different RNA molecules. The predicted exon/exon association structure of the cTnT pre-mRNA would be a loop 683 nt long with two tails, one 306 nt long, and one 48 nt long. For the RNA molecule with two functional exons, 81% of the looped molecules that were randomly sampled by photography had a loop that was within 15% of the expected length (examples shown in Fig. 4 c–f). As well, the lengths of the two ends generally agreed with the expected values, although occasionally one of the ends was not visible. In contrast, the frequency of molecules with loop sizes within 15% of the expected value was only 5.9% for the RNA containing an exon 5 mutant and 5.3% for the RNA molecule containing an exon 6 mutant. Examples are shown in Fig. 4 g (exon 5 mutant) and h (exon 6 mutant). From this analysis, we conclude that SR proteins are able to create a loop in the RNA between the two functional exons, and this activity is at least 50-fold lower with either mutant form of the RNA.
DISCUSSION
The proper association of 5′ and 3′ splice sites during pre-mRNA splicing is a critical aspect of gene expression. Previous imaging of this association has been restricted to electron microscopic observations of nascent transcripts on in vivo chromatin spreads (37). These studies showed that stable RNP particles can form at two neighboring splice sites that subsequently coalesce to loop out the intron. We have developed a defined system to characterize the role of individual splicing factors in this process. Using two methods, RNA affinity chromatography and electron spectroscopic imaging, we have shown that SR proteins are sufficient to promote the specific association of two exons, cTnT exons 5 and 6, which are spliced together in vivo. This interaction is likely to be physiologically relevant because mutations of exon 5 that are known to affect alternative inclusion of this exon have similar effects on the exon 5/exon 6 association. The results presented here indicate that SR proteins influence the formation of a functional splicing complex by promoting the bridging of exons.
Splice sites, in addition to associating with one another, must also interact with the splicing machinery to allow for catalysis. It appears that SR proteins also function during this process by promoting a number of pre-mRNA/snRNA interactions. For example, SR proteins bound to exonic enhancers can promote an interaction of the U2 snRNA with the 3′ splice site (10–13). In this study, we show that SR proteins also can promote an association of the U1 snRNP with an exonic enhancer. It is interesting to note that SR proteins had a lesser effect on U1*, a U1 snRNA degradation product that lacks the 5′ cap and the first 5′ 6 nt (34). This result indicates that the 5′ end of U1, which can base pair with 5′ splice-site sequences, is not absolutely essential for the association of the U1 snRNP with exonic enhancers, but is, however, required for an efficient interaction. Given that the column RNA does not have a recognizable 5′ splice site, these results indicate that the 5′ end of the U1 snRNA may either function to interact with some other sequence within the exonic RNA or, alternatively, may be required for the efficient interaction of the U1 snRNP with SR proteins. These results are consistent with other reports that U1* is deficient in assembling into RNP complexes (25). Regardless of the details of the U1 snRNP/exon association, it is clear that part of the mechanism of SR protein function is to promote snRNP/pre-mRNA interactions.
The findings presented here, as well as the studies of others, suggest that SR proteins mediate RNA/RNA interactions critical for splicing, including both exon/exon and snRNA/pre-mRNA interactions. Determination of the nature of these interactions will enable a more complete description of SR protein function. One possible model is that the association of two RNA sequences occurs when SR proteins bound to different RNA sequences associate with one another. Consistent with this hypothesis, phosphatase treatment of SR proteins, which is likely to debilitate these interactions, inhibits SR protein-mediated exon/exon associations (data not shown). Similarly, one SR protein and another RNA-binding protein could each interact with different RNAs and with each other. This model is supported by studies that show that SR proteins can bind to a number of RNA-binding proteins, including themselves (reviewed in ref. 4).
The identification of an exon bridging complex also provides a model for studying the regulation of cTnT alternative splicing. Regulation could occur at the level of either restricting the SR protein/exon 5 interaction or the subsequent interaction of an SR protein-bound exon 5 with exon 6. It is also possible that regulation could occur later in the assembly of a functional splicing complex. Future studies will focus on whether SR protein-mediated exon bridging plays an important role in cTnT alternative splicing.
Acknowledgments
We thank members of the Roth lab and L. B. Roth for comments on the manuscript and Drs. A. Beyer, T. Cooper, and K. Neugebauer for their insightful comments on this work. This work was supported by National Institutes of Health Grant GM48435-01A2 to M.B.R. and a grant from the Natural Sciences Engineering and Research Council (Canada) to D.P.B.-J. J.M.S. is supported by National Research Service Award T32 GMO7270 from the National Institute of General Medical Sciences.
ABBREVIATIONS
- snRNA
small nuclear RNA
- cTnT
avian cardiac Troponin-T
- ESI
electron spectroscopic imaging
- EM
electron microscope
- snRNP
small nuclear ribonucleoprotein
References
- 1.Moore J M, Query C C, Sharp P A. In: The RNA World. Gestleland R F, Atkins J F, editors. Cold Spring Harbor, NY: Cold Spring Harbor Lab. Press; 1993. pp. 303–358. [Google Scholar]
- 2.Berget S M. J Biol Chem. 1995;270:2411–2414. doi: 10.1074/jbc.270.6.2411. [DOI] [PubMed] [Google Scholar]
- 3.Black D L. RNA. 1995;1:763–771. [PMC free article] [PubMed] [Google Scholar]
- 4.Valcarcel J, Green M R. Trends Biochem Sci. 1996;21:296–301. [PubMed] [Google Scholar]
- 5.Zahler A M, Neugebauer K M, Lane W S, Roth M B. Science. 1993;260:219–222. doi: 10.1126/science.8385799. [DOI] [PubMed] [Google Scholar]
- 6.Fu X-D. Nature (London) 1993;365:82–85. doi: 10.1038/365082a0. [DOI] [PubMed] [Google Scholar]
- 7.Madhani H D, Guthrie C. Annu Rev Genet. 1994;28:1–26. doi: 10.1146/annurev.ge.28.120194.000245. [DOI] [PubMed] [Google Scholar]
- 8.Kohtz J D, Jamison S F, Will C L, Zuo P, Luhrmann R, Garcia-Blanco M A, Manley J L. Nature (London) 1994;368:119–124. doi: 10.1038/368119a0. [DOI] [PubMed] [Google Scholar]
- 9.Zahler A M, Roth M B. Proc Natl Acad Sci USA. 1995;92:2642–2646. doi: 10.1073/pnas.92.7.2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laviguer A, La Branche H, Kornblihtt A R, Chabot B. Genes Dev. 1993;7:2405–2417. doi: 10.1101/gad.7.12a.2405. [DOI] [PubMed] [Google Scholar]
- 11.Valcarcel J, Gaur R K, Singh R, Green M R. Science. 1996;273:1706–1709. doi: 10.1126/science.273.5282.1706. [DOI] [PubMed] [Google Scholar]
- 12.Wang Z, Hoffmann H M, Grabowski P J. RNA. 1995;1:21–35. [PMC free article] [PubMed] [Google Scholar]
- 13.Zuo P, Maniatis T. Genes Dev. 1996;10:1356–1368. doi: 10.1101/gad.10.11.1356. [DOI] [PubMed] [Google Scholar]
- 14.Chiara M D, Reed R. Nature (London) 1995;375:510–513. doi: 10.1038/375510a0. [DOI] [PubMed] [Google Scholar]
- 15.Bruzik J P, Maniatis T. Proc Natl Acad Sci USA. 1995;92:7056–7059. doi: 10.1073/pnas.92.15.7056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ramchatesingh J, Zahler A M, Neugebauer K M, Roth M B, Cooper T A. Mol Cell Biol. 1995;15:4898–4907. doi: 10.1128/mcb.15.9.4898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bardwell V J, Wickens M. Nucleic Acids Res. 1990;18:6587–6594. doi: 10.1093/nar/18.22.6587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cooper T A, Ordahl C P. J Biol Chem. 1985;260:11140–11148. [PubMed] [Google Scholar]
- 19.Grodberg J, Dunn J J. J Bacteriol. 1988;170:1245–1253. doi: 10.1128/jb.170.3.1245-1253.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tian M, Maniatis T. Cell. 1993;74:105–114. doi: 10.1016/0092-8674(93)90298-5. [DOI] [PubMed] [Google Scholar]
- 21.Dignam J D, Lebovitz R M, Roeder R J. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zahler A M, Lane W S, Stolk J A, Roth M B. Genes Dev. 1992;6:837–847. doi: 10.1101/gad.6.5.837. [DOI] [PubMed] [Google Scholar]
- 23.Zahler A M, Neugebauer K M, Stolk J A, Roth M B. Mol Cell Biol. 1993;13:4023–4028. doi: 10.1128/mcb.13.7.4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Konforti B B, Konarska M M. RNA. 1995;1:815–827. [PMC free article] [PubMed] [Google Scholar]
- 25.Wassarman D A, Steitz J A. Proc Natl Acad Sci USA. 1993;90:7139–7143. doi: 10.1073/pnas.90.15.7139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peculis B A, Steitz J A. Cell. 1993;73:1233–1245. doi: 10.1016/0092-8674(93)90651-6. [DOI] [PubMed] [Google Scholar]
- 27.Neugebauer K M, Stolk J A, Roth M B. J Cell Biol. 1995;129:899–908. doi: 10.1083/jcb.129.4.899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu R, Teng J, Cooper T A. Mol Cell Biol. 1993;13:3660–3674. doi: 10.1128/mcb.13.6.3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bazett-Jones D P. Microbeam Anal. 1993;2:69–79. [Google Scholar]
- 30.Cooper T A. J Biol Chem. 1992;267:5330–5338. [PubMed] [Google Scholar]
- 31.Krainer A R, Maniatis T, Ruskin B, Green M R. Cell. 1984;36:993–1005. doi: 10.1016/0092-8674(84)90049-7. [DOI] [PubMed] [Google Scholar]
- 32.Staknis D, Reed R. Mol Cell Biol. 1994;14:7670–7682. doi: 10.1128/mcb.14.11.7670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Watakabe A, Tanaka K, Shimura Y. Genes Dev. 1993;7:407–418. doi: 10.1101/gad.7.3.407. [DOI] [PubMed] [Google Scholar]
- 34.Lerner M R, Boyle J A, Mount S M, Wolin S L, Steitz J A. Nature (London) 1980;283:220–224. doi: 10.1038/283220a0. [DOI] [PubMed] [Google Scholar]
- 35.Bazett-Jones D P. Electron Microsc Rev. 1991;5:37–58. doi: 10.1016/0892-0354(92)90004-a. [DOI] [PubMed] [Google Scholar]
- 36.Bazett-Jones D P, Leblanc B, Herfort M, Moss T. Science. 1994;264:1134–1137. doi: 10.1126/science.8178172. [DOI] [PubMed] [Google Scholar]
- 37.Beyer A L, Osheim Y N. Genes Dev. 1988;2:754–756. doi: 10.1101/gad.2.6.754. [DOI] [PubMed] [Google Scholar]
- 38.Early P, Rogers J, Davis M, Calame K, Bond M, Wall R, Hood L. Cell. 1980;20:313–319. doi: 10.1016/0092-8674(80)90617-0. [DOI] [PubMed] [Google Scholar]
- 39.Woychik R P, Camper S A, Lyons R H, Horowitz S, Goodwin E C, Rottman F M. Nucleic Acids Res. 1982;10:7197–7210. doi: 10.1093/nar/10.22.7197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schwartz D E, Tizard R, Gilbert W. Cell. 1983;32:853–869. doi: 10.1016/0092-8674(83)90071-5. [DOI] [PubMed] [Google Scholar]
- 41.Burtis K C, Baker B S. Cell. 1989;56:997–1010. doi: 10.1016/0092-8674(89)90633-8. [DOI] [PubMed] [Google Scholar]
- 42.Kornblihtt A R, Vibe-Pederson K, Baralle F E. Nucleic Acids Res. 1984;12:5853–5868. doi: 10.1093/nar/12.14.5853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hayashi K, Yano H, Hashida T, Takeuchi R, Takeda O, Asada K, Takahashi E, Kato I, Sobue K. Proc Natl Acad Sci USA. 1992;89:12122–12126. doi: 10.1073/pnas.89.24.12122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Humphrey M B, Bryan J, Cooper T A, Berget S M. Mol Cell Biol. 1995;15:3979–3988. doi: 10.1128/mcb.15.8.3979. [DOI] [PMC free article] [PubMed] [Google Scholar]