Abstract
Escherichia coli-derived recombinant human glycosylation inhibiting factor (rhGIF) contains three cysteine residues (Cys-57, -60, and -81). All SH groups in the cysteine residues are free, and the GIF molecule had no biologic activity. Carboxymethylation of the SH group of Cys-60 in the molecule resulted in the generation of bioactivity, although the activity of the carboxymethylated GIF was 10- to 20-fold less than that of suppressor T cell (Ts)-derived GIF. However, treatment of the inactive rhGIF with ethylmercurithiosalicylate or 5,5′-dithiobis(2-nitrobenzoic acid) (DTNB) resulted in the generation of derivatives whose bioactivity was comparable to that of the Ts-derived bioactive GIF. The activity of these derivatives was lost by treatment with DTT. Isolation and chemical analysis of the DTNB-treated GIF derivative revealed that binding the 5-thio-2-nitrobenzoic acid group with Cys-60 was responsible for the generation of the highly bioactive derivative. Inactive cytosolic GIF from mammalian cells could also be converted to bioactive derivative by treatment with the SH reagent, while Ts-derived bioactive GIF was inactivated by DTT. These results, together with an x-ray crystal structure of GIF molecules, strongly suggest that the generation of bioactivity of GIF in Ts cells is due to posttranslational modifications that result in conformational changes in the molecule.
Keywords: SH reagent, conformational structure, suppressor T cells
We have previously described glycosylation inhibiting factor (GIF), a 13-kDa cytokine, as a product of suppressor T (Ts) cells (1, 2) and a subunit of antigen-specific Ts cell factors (3, 4). After molecular cloning of this cytokine, however, we realized that various cell line cells produced the 13-kDa peptide, which reacted with polyclonal antibodies against recombinant GIF (rGIF). Nevertheless, only the peptide secreted by Ts cells demonstrated GIF bioactivity (5). Escherichia coli-derived rGIF was also inactive. It was found that Ts cells contained a substantial quantity of inactive GIF peptide in cytosol (5) and that the amino acid sequence of the inactive peptide was identical to that of the bioactive homologue (6). No difference was detected by SDS/PAGE analysis among the bioactive GIF, inactive GIF in cytosol, and E. coli-derived rGIF. These findings suggested to us the possibility that generation of bioactive GIF by Ts cells is due to posttranslational modifications of the peptide and that heterogeneity of GIF in bioactivity is due to a conformational transition of the same peptide (5).
Deduced amino acid sequence of recombinant GIF indicated that the GIF peptide contains three cysteine residues (Cys-57, -60, and -81) (7). One might expect the formation of an intrachain disulfide bond to change the conformation of GIF molecules. We were also interested in the sequence of Cys-57-Xaa-Xaa-Cys-60, because such a structure may be involved in interprotein association (8). Cysteine residues in the Cys-Xaa-Xaa-Cys sequence may also be involved in the coordination of a metal ion that mediates protein–protein interaction (9). Green and Chambon (10) provided evidence that such a sequence is essential in the DNA-binding domain of steroid hormone receptors. It was also predicted that cysteine residues may be required for particular conformation of some proteins (8). We anticipate that the interaction of a cysteine residue with a heavy metal or the association of protein–peptide with the sequence may affect the conformation of GIF. Because of such possibilities, the present experiments were undertaken to determine the effect of SH reagents on the bioactivity of GIF. The results show that modification of Cys-60 in an inactive GIF peptide results in the generation of bioactivity.
MATERIALS AND METHODS
Purified Human GIF Preparations.
Recombinant human GIF (rhGIF) expressed in E. coli was purified from a soluble fraction of the cells by HPLC using a CM-Sepharose column (6), and purified GIF was further fractionated on a CM-5PW column (Tosoh, Tokyo) with NaCl gradient to recover the major protein peak. The final GIF preparation gave a single 13-kDa band in SDS/PAGE analysis. Amino acid sequence analysis showed that the GIF peptide lacked the first methionine in the deduced sequence (7). Inactive hGIF peptide in culture supernatant of the 2FH2 cell line (i.e., a stable transfectant of hGIF cDNA in murine Ts hybridoma 231F1 cells) (5) was purified by absorption of the supernatant with rabbit IgG (RGG)-coupled Affi-Gel 10 (Bio-Rad), followed by elution of the column with 0.1 M glycine HCl buffer (pH 3.0) (6). Bioactive hGIF in the flow-through fraction from RGG-coupled Affi-Gel 10 was recovered using the monoclonal anti-hGIF antibody, 388F1, coupled to Affi-Gel 10 by the procedure described in ref. 6. Cytosolic, inactive GIF in the lysates of the 2FH2 cells was isolated by affinity purification using 388F1-coupled Affi-Gel 10 (5). All of the inactive GIF from culture supernatants, cytosolic GIF preparations, and a bioactive GIF preparation gave a single band of 13 kDa in SDS/PAGE analysis and silver staining. Immunoblotting with polyclonal anti-GIF was carried out by the procedures described previously (6).
Treatment of Purified GIF Preparations with SH Reagents.
Purified rhGIF (1.0 mg/ml) was incubated overnight at room temperature with 18 mM iodoacetate (Sigma) at pH 8.2 for carboxymethylation of free SH groups. The sample was applied to a Sephadex G-25 column to remove excess iodoacetate, and the GIF protein was recovered by elution with 20 mM sodium acetate buffer (pH 5.5). The same purified rhGIF was treated with ethylmercurithiosalicylate (EMTS) (Sigma) or 5,5′-dithiobis(2-nitrobenzoic acid) (DTNB) (Sigma). EMTS was added to 100 to ≈130 μg/ml of rhGIF in PBS (pH 7.0), and DTNB was added to rhGIF in 0.1 M Tris·HCl buffer (pH 7.2) at the final concentration of 100 to ≈250 μM. The reaction mixtures were kept at 4°C for 24 hr and then extensively dialyzed against PBS. In some experiments, 1 mg/ml rhGIF in 50 mM Tris·HCl buffer (pH 8.5) was incubated overnight at room temperature with 3.3 mM DTNB in the presence of 4% acetonitrile.
Fractionation of GIF Derivatives and Determination of SH Groups.
Recombinant GIF treated with a SH reagent was fractionated by using a HPLC system (System Gold, Beckman). A 0.2–0.5 ml sample in 20 mM acetate buffer (pH 5.5), 10 mM phosphate buffer (pH 6.5), or 10 mM phosphate buffer (pH 6.5) containing 0.08 M NaCl was applied to a CM-5PW column, and proteins were eluted by a linear gradient of NaCl up to 0.5 M at a flow rate of 0.5 ml/min.
The number of free SH groups per molecule of GIF and its derivatives were determined by the method of Glazer et al. (11). Samples were concentrated to ≈10 mg/ml and suspended in 800 μl of 0.1 M Tris·HCl buffer (pH 8.0) containing 8 M urea and 1 mM EDTA. After absorption was measured at 280 nm, 50 μl of 0.1 M DTNB in 50 mM phosphate buffer (pH 7.0) was added to the solution. The reaction mixture was kept at room temperature for 10 min, and absorption at 412 nm was determined using a DTNB solution in 50 mM phosphate buffer as a control. Molar concentrations of GIF and SH groups were calculated using molar extinction coefficients of GIF at 280 nm and DTNB at 412 nm of 14,355 and 14,290 M−1·cm−1, respectively.
Mass Spectrometric Analysis.
The HPLC system for liquid chromatography-electrospray ionization/MS was a HP 1090 liquid chromatograph (Hewlett–Packard) with a C8 column (Inertsyl 0.5 × 50 mm; GL Sciences, Tokyo). After application of a sample, the column was washed for 5 min with 18% acetonitrile containing 0.1% formic acid, and proteins were eluted with a linear gradient from 18% to 45% acetonitrile containing 0.1% formic acid at a flow rate of 150 μl/min. One-tenth of the effluent was split by a flow-frit for the flow rate of 15 μl/min, and the eluate was directed to the electrospray probe through a fused-silica capillary. Mass spectra were obtained on a triple-quadrapole mass spectrometer (VG Quattro II, VG Organic, Manchester, U.K.) equipped with an electrospray ionization source. The potential of spray needle was held at 3.15 kV. Positive ion masses were recorded at a cone voltage setting of 25 V. Multiple-charged species were measured by scanning over a mass range of m/z 800-1700 in 3.5 sec, and the molecular mass was calculated using the maxent program (VG Organic).
Partial Hydrolysis and Peptide Map.
Approximately 5 μg of rGIF or its derivative in 0.5 ml of 0.2 M sodium acetate buffer (pH 4.0) was hydrolyzed with 40 ng of pepsin (Sigma) for 7 hr at room temperature. Peptides were recovered by reverse phase chromatography on a super octadecylsilane column (0.2 × 5 cm; Tosoh) equilibrated with a mixture of 95% (vol/vol) solution A (0.05% trifluoroacetic acid) (Sigma), and 5% (vol/vol) solution B (0.02% trifluoroacetic acid/70% isopropanol/30% acetonitrile). The column was washed with the mobile phase solution for 5 min at a flow rate of 0.2 ml/min, and peptides were eluted from the column with a linear gradient of 5–25% solution B over a period of 45 min, and then with 100% solution B for an additional 5 min. Amino acid sequence of each peptide recovered from the column was determined using a gas-phase amino acid sequencer (PPSQ 10; Shimadzu).
ELISA Assays and Detection of GIF Bioactivity.
Detailed procedures of ELISA assays have been described (6). Briefly, microplates (Maxi Sorp; Nunc) were coated with serial 2-fold dilutions of GIF or its derivatives. After blocking, plates were incubated for 2 hr with polyclonal anti-GIF at 2 μg/ml, followed by incubation for 2 hr with horseradish peroxidase-coupled F(ab′)2 of donkey anti-rabbit IgG (Amersham). ELISA signals were developed by adding substrate and determined by absorption at 405 nm.
Bioactivity of GIF peptides was detected by their ability to cause mouse T-cell hybridoma 12H5 cells to switch from forming glycosylated IgE-binding factor (IgE-BF) to forming unglycosylated IgE-BF. Detailed procedures for the assay have been described (12). IgE-BF in culture filtrates was fractionated on lentil lectin-Sepharose. When the 12H5 cells were cultured in the absence of GIF, essentially all IgE-BF formed by the cells bound to the lectin and was recovered by elution with 0.2 M α-methyl d-mannoside. In the presence of a sufficient concentration of GIF, most of the IgE-BF formed by the cells was not retained on the column. To titrate bioactivity of a sample, aliquots of 12H5 cells were cultured with serial 2-fold dilutions of the sample, and the bioactivity of a GIF derivative was expressed by the minimum concentration of the 13-kDa peptide required for switching the 12H5 cells for the formation of unglycosylated IgE-BF. A dilution of the sample was taken as GIF-positive if IgE-BF in the flow-through fraction was more than that present in the eluate.
RESULTS
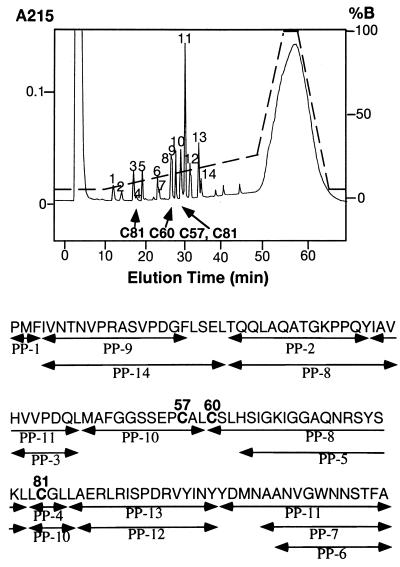
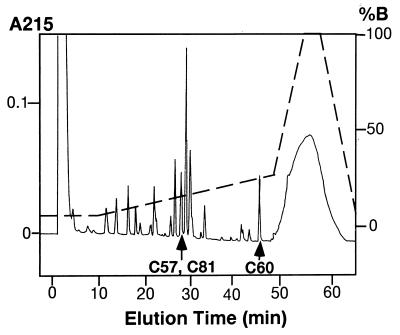
Experiments were carried out to determine whether the rhGIF peptide may or may not possess an intrachain disulfide bond. Peptic digests of rhGIF were fractionated by reverse-phase chromatography. The profile of the peptides and amino acid sequence of each peptide are shown in Fig. 1. We detected four peptides, each of which contained a single cysteine residue. A Cys-60-containing peptide was present in peak 8. Peak 10 consisted of a peptide containing Cys-57 as the major component and another peptide containing Cys-81 as a minor component. However, the majority of Cys-81 was recovered in a peptide present in peak 4, suggesting that a disulfide bond does not exist between Cys-57 and Cys-81. Indeed, the elution profile of the peptides from the super ODS column was not affected by pretreatment of the peptic digests with 10 mM DTT for 30 min before the reverse-phase chromatography. Absence of an intrachain disulfide bond in rhGIF was confirmed by the presence of three titratable SH groups per 13 kDa GIF molecule, as measured by the DTNB procedure (data not shown).
Figure 1.
HPLC chromatogram of pepsin digests of E. coli-derived rhGIF. The column was eluted with a linear gradient of solution B from 5% to 25%, indicated by the broken line. Peptide peaks, determined by absorption at 215 nm, are numbered. cysteine-containing peptides are indicated by arrows. Some of the peptide peaks, such as PP-8, PP-10, and PP-11, contained two different peptides. Amino acid sequences of all peptides recovered from the column are shown below the chromatogram.
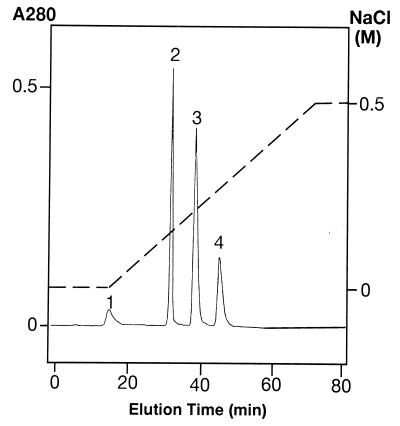
Attempts were made to carboxymethylate SH groups in rGIF. A rhGIF preparation was treated with iodoacetate and fractionated on a CM-5PW column at pH 5.5 (see Materials and Methods). As shown in Fig. 2, four protein peaks were obtained. The elution time of peak 4 was identical to that of untreated rhGIF applied to the same column. The number of free SH groups per 13-kDa GIF molecule recovered in peaks 1, 2, 3, and 4 were 2.08, 2.41, 2.69, and 2.95, respectively. The results indicate that the peak 4 protein represents unmodified GIF, and the peak 1 protein is a derivative in which one SH group per GIF molecule is carboxymethylated.
Figure 2.
Fractionation of carboxymethylated rhGIF on a CM-5PW column. Proteins were eluted from the column with a linear gradient of NaCl, indicated by the dotted line. Elution time of peak 4 corresponded to that of untreated GIF applied to the same column.
Previous studies of the x-ray crystal structure of GIF indicated that rhGIF crystalizes as a trimer (13). Indeed, the average molecular weight of rhGIF in solution, determined by analytical ultracentrifugation and light-scattering photometry, was 32,013 and 36,490, respectively, indicating that the majority of rGIF in physiological solution is in the form of trimer (T. Tomura, H.W., T.N., T.M., and K.I., unpublished results). If the proteins in the first three peaks in Fig. 2 represent carboxymethylated derivatives of GIF trimer, the numbers of carboxymethylated groups per trimer in peaks 1, 2, and 3 would be 2.8, 1.8, and 0.9, respectively. Thus, it appears that peak 2 and peak 3 may represent the GIF trimer, in which one SH group in two of the three subunits or one SH group in one of the three subunits was carboxymethylated.
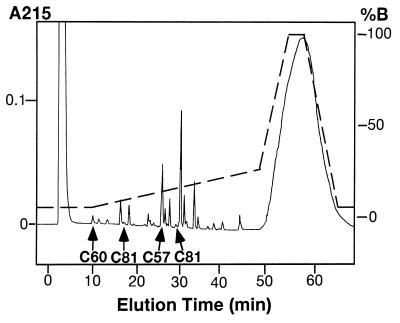
To determine which cysteine residue in rGIF molecules was carboxymethylated by the treatment with iodoacetic acid, the peak 1 protein from the CM-5PW column (Fig. 2) was digested with pepsin and subjected to peptide mapping. The profile of the peptides, shown in Fig. 3, was essentially the same as that of untreated GIF (see Fig. 1), except that the peptide containing Cys-60 (PP-8 in Fig. 1) was not detectable in the peptic digests of peak 1 protein. Instead, a peptide-containing carboxymethylated Cys-60 was recovered in an earlier fraction eluted at 10 min (Fig. 3). The yield of this peptide indicated that essentially all Cys-60 in the peak 1 protein was carboxymethylated. In contrast, the yields of the other three cysteine-containing peptides from the carboxymethylated protein were comparable to those recovered from untreated GIF. Thus, the results indicated that only Cys-60 was carboxymethylated under the experimental conditions employed.
Figure 3.
Peptide map of pepsin digests of carboxymethylated rhGIF. Peak 1 protein shown in Fig. 2 was digested with pepsin, and the peptide map was obtained by the same method described in Fig. 1 under identical conditions. Absorbance at 215 nm is shown by the solid line, and the percentage of solution B for elution is shown by the broken line. Elution time of cysteine-containing peptides are indicated by arrows. The amino acid sequence of the Cys-60-containing peptide, recovered in the first peak, was LeuCysSerLeu, and this peptide contained one carboxymethyl group. Amino acid sequences of two Cys-81-containing peptides and that of a Cys-57-containing peptide were identical to those shown in Fig. 1.
The GIF bioactivity of the carboxymethylated derivatives was assessed. As shown in Table 1, carboxymethylation of Cys-60 in each monomeric subunit resulted in a >16-fold increase in its bioactivity. The significantly less bioactivity observed in peak 2 and peak 3 proteins compared with that observed in peak 1 protein corresponded to the fact that only two or one subunit in the GIF trimer was carboxymethylated in the peak 2 and peak 3 derivatives, respectively.
Table 1.
Conversion of inactive rhGIF to bioactive derivatives by treatment with a SH reagent
In view of the findings that carboxymethylation of the SH group in Cys-60 resulted in the generation of GIF bioactivity, we determined possible effects of the other SH reagents on the activity of rGIF. We treated aliquots of 100 μg/ml rGIF with 0.05–0.25 mM EMTS. As shown in Table 1, the EMTS treatment markedly increased the bioactivity of rGIF in a dose-dependent manner. Repeated experiments confirmed that treatment of 130 μg/ml (10 μM) of inactive rGIF with 0.2–0.25 mM EMTS reproducibly gave GIF preparations that showed the bioactivity at a concentration of 5 ng/ml. Generation of a highly bioactive GIF derivative was also achieved by the treatment of inactive rhGIF with DTNB. GIF preparations treated with 0.1 to ≈0.25 mM DTNB showed the bioactivity at a concentration of 8–10 ng/ml. To confirm that the generation of bioactive GIF was due to modification of a SH group(s), a portion of the bioactive GIF derivatives, which were prepared by the treatment with either 0.2 mM EMTS or 0.25 mM DTNB, was incubated overnight with 5 mM DTT at 4°C. Measurement of the GIF bioactivity showed that most of the bioactivity was lost because 1 μg/ml of the 13-kDa peptide in the reduced samples was required for the detection of GIF bioactivity.
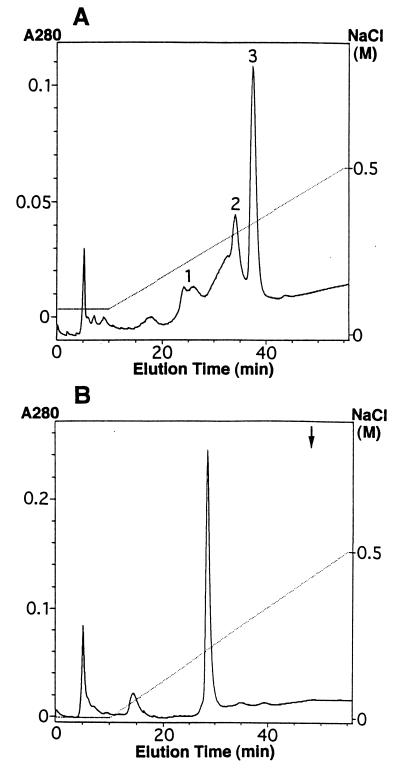
Attempts were made to isolate the bioactive GIF derivatives. Thus, 10 μM E. coli-derived rhGIF was treated with 0.25 mM EMTS, and the preparation was fractionated on a CM-5PW column at pH 6.5. Comparisons of the elution profile, shown in Fig. 4A, with that of untreated GIF from the same column indicated that the last protein peak (peak 3) recovered at 36–38 min represented unmodified GIF, while peak 2, recovered at 31–34 min, did not exist in the original GIF preparation. Measurement of the concentration of GIF in each fraction by ELISA, followed by determination of bioactivity of the fractions, indicated that the GIF derivative in peak 2 showed bioactivity at 2 ng/ml (Table 1). The minimum concentration of GIF in the last peak fraction for the detection of bioactivity was 500 ng/ml. The results clearly showed that the protein in peak 2 represented the bioactive derivative of GIF. Similar experiments were carried out with DTNB-treated GIF. Recombinant GIF (10 μM) was treated with 0.25 mM DTNB and fractionated on a CM-5PW column. The chromatogram showed one major protein peak obtained between 26 and 29 min (Fig. 4B). Under the experimental conditions used, unmodified GIF was undetectable. Determination of bioactivity of the GIF derivative recovered from the CM-5PW column revealed that 5 ng/ml of the derivative was sufficient for the detection of bioactivity (Table 1).
Figure 4.
Fractionation of EMTS-treated GIF and DTNB-treated GIF on a CM-5PW column. The starting buffer for the fractionation of EMTS-treated GIF (A) was 10 mM phosphate buffer (pH 6.5) containing 0.08 M NaCl, while that for DTNB-treated GIF (B) was the same buffer without NaCl. Proteins were eluted by a linear gradient of NaCl, indicated by a broken line. The arrow represents the elution time of untreated GIF applied to the same column.
Determination of molecular weight species in a EMTS-treated GIF preparation by mass spectrometry showed that the EMTS derivatives consisted of four species with molecular weights of 12,342, 12,570, 12,799, and 13,027. Considering that the molecular weight of the ethylmercuryl group is 229 and that the molecular weight of rhGIF calculated from the amino acid sequence is 12,344, the EMTS-treated GIF appears to consist of GIF molecules that contain 0–3 ethylmercuryl groups per 12.3-kDa peptide. In contrast, the DTNB-treated GIF preparation was quite homogenous. Treatment of rhGIF with 3.3 mM DTNB followed by chromatography on a CM-5PW column yielded a single protein peak similar to that shown in Fig. 4B. Mass spectrometric analysis of this protein fraction yielded a single peak of molecular weight 12,541, which is consistent with a protein that contains one group of thionitrobenzoic acid (molecular weight 197) per 12.3-kDa GIF molecule. It was confirmed that the purified DTNB derivative could switch the 12H5 cells to form unglycosylated IgE-BF at a concentration of 3–5 ng/ml. Next, this GIF derivative was digested with pepsin to obtain a peptide map. As shown in Fig. 5, the elution time of the peptide Cys-60–Leu-79 (PP-8 in Fig. 1) was markedly retarded, and amino acid sequence of the peptide demonstrated the association of thionitrobenzoic acid with the peptide. Elution times of the peptide Met-48–Leu-59 (PP-10 in Fig. 1), which contained Cys-57, and of Leu-80–Leu-84 (PP-10), which contained Cys-81, were identical to those obtained from untreated rGIF, and neither of these peptides contained thionitrobenzoic acid. In the peptide map of the DTNB derivative, neither Leu-80–Leu-83 (PP-4 in Fig. 1) nor Leu-84–Tyr-99 (PP-13) was detected. Instead, the yield of Leu-80–Leu-84 (PP-10) and Ala-85–Tyr-99 (PP-12) markedly increased as compared with the yield of the same peptides from unmodified GIF, indicating that the cleavage between Leu-83 and Leu-84 did not occur in the DTNB-treated GIF. The results indicated that under the experimental conditions employed, DTNB reacted exclusively with Cys-60 and failed to react with either Cys-57 or Cys-81.
Figure 5.
Peptide map of a pepsin digest of a DTNB-derivative of rhGIF. The derivative isolated by chromatography on a CM-5PW column (see Fig. 4B) was digested with pepsin, and a peptide map was obtained under the same conditions as that in Fig. 1. Absorbance at 215 nm is shown by the solid line, and percentage of solution B for elution is given by the broken line. A Cys-60-containing peptide was recovered in the peak, which is indicated by an arrow. Amino acid sequence of the peptide was identical to that shown in Fig. 1 (PP-8). A Cys-57-containing peptide and a Cys-81-containing peptide were recovered in the same peak, indicated by an arrow. Amino acid sequences of these peptides were identical to those shown in Fig. 1 (PP-10).
We have previously shown that rhGIF as well as inactive GIF protein released from Ts hybridomas and cytosolic GIF have affinity for Affi-Gel 10 and RGG-coupled Affi-Gel 10, while bioactive GIF secreted from the Ts hybridomas fails to be retained on RGG-coupled Affi-Gel 10 (6). Thus, we determined whether the bioactive derivative prepared by the EMTS treatment of E. coli-derived rhGIF may have affinity for RGG-coupled Affi-Gel 10. An aliquot of the rGIF was treated with 0.25 mM EMTS, and 5 μg of the preparation was mixed overnight with 1 ml RGG-coupled Affi-Gel 10 in the presence of 0.5 mg/ml BSA (6). Proteins retained on RGG-coupled Affi-Gel 10 were recovered by acid elution, and both the flow-through fraction and acid eluate fraction were assessed for the presence of GIF by SDS/PAGE and immunoblotting with anti-GIF. Approximately two-thirds of the EMTS-treated GIF was recovered in the acid eluate. The GIF peptide in the flow-through fraction showed bioactivity at a concentration of 2–3 ng/ml, while 400 ng/ml of the peptide in the acid eluate fraction was required for detection of GIF bioactivity. These results indicate that the bioactive derivative of rGIF shares the same properties with native bioactive GIF from Ts hybridomas with respect to its lack of affinity for Affi-Gel 10.
We wondered whether a cysteine residue(s) in mammalian cell-derived GIF may also be involved in converting inactive GIF peptide to bioactive GIF. To test this possibility, we determined the effect of SH reagents on the inactive GIF from the 2FH2 cells. Both the inactive GIF peptide in the culture supernatant and cytosolic GIF failed to show bioactivity at a concentration of 0.5–1.0 μg/ml. An aliquot of these preparations was incubated for 24 hr with 0.25 mM EMTS or DTNB, and bioactivity of the treated preparations was determined. The results showed that at a concentration of 10 ng/ml of the 13-kDa protein of the EMTS-treated and DTNB-treated preparations, bioactivity was demonstrable, indicating that not only E. coli-derived rGIF but also mammalian cell-derived inactive GIF could be converted to bioactive derivatives by treatment with either EMTS or DTNB.
Finally, we determined the effect of reducing reagent on the bioactive GIF in the culture supernatant of the 2FH2 cells. The preparation was obtained by preabsorption of the culture supernatant with RGG-coupled Affi-Gel 10, followed by affinity purification of the bioactive species in the flow-through fraction using anti-GIF 388F1-Affi-Gel. As expected, 5 ng/ml of this GIF in the preparation was sufficient for the detection of bioactivity. The preparation was then treated overnight with 5 mM DTT at 4°C. Estimation of the concentration of GIF peptide by ELISA and determination of bioactivity showed that even at the concentration of 300 ng/ml, the l3-kDa peptide in the reduced material failed to show bioactivity.
DISCUSSION
The present experiments clearly showed that E. coli-derived rGIF does not contain an intrachain disulfide bond. This finding is supported by tertiary structure of GIF revealed by x-ray crystallography (13), which indicated that distances between two ionized S atoms in Cys-57 and Cys-60, Cys-57 and Cys-81, and Cys-60 and Cys-81 are ≈10 Å, 13 Å, and 9 Å, respectively. Because the interatomic distance has to be 2 Å or less for the formation of a disulfide bond (14), an intrachain disulfide bond cannot be formed in the GIF peptide.
The crystal structure of GIF protein may also explain the preferential reaction of a SH reagent with Cys-60. E. coli-derived rGIF molecules form a barrel-shaped trimer of the 13-kDa GIF protein, and the barrel consists of three β-sheets. Six α-helices, two of which belong to each monomer, surround the outside of the barrel (13). In this structure, the SH group of Cys-57 is directed toward the inside of the barrel, and the SH group of Cys-81, which is in one of the α-helices, is directed toward the β-sheet. In contrast, the SH group of Cys-60, which is in one of the β-strands, is sticking out between the two α-helices. Reaction of a SH reagent with this SH group indicates that the reagents could go between the two α-helices. The present experiments showed that both iodoacetic acid and DTNB reacted preferentially with Cys-60. One may expect that EMTS also reacts with Cys-60. However, mass analysis of the EMTS-treated GIF demonstrated the presence of three molecular species that contained one to three ethylmercuryl groups per GIF monomer. These results are not surprising because EMTS could react not only with SH groups but also with amino groups at high pH levels. When the rGIF crystal was incubated with EMTS, cysteine as well as Asp-17 and His-63 were modified (unpublished results). Because of heterogeneity of the EMTS derivatives, we did not pursue further studies on the structural basis of the generation of bioactivity by the EMTS treatment. Nevertheless, reversal of highly bioactive EMTS derivatives to inactive GIF by treatment with DTT indicates that the binding of ethylmercuryl group to SH group(s) is responsible for the generation of bioactivity.
An important principle obtained in the present experiments was that modification of the SH group of Cys-60 in the inactive rGIF resulted in the generation of bioactivity. An identical amino acid sequence shared by the bioactive derivatives and inactive GIF, and lack of an intrachain disulfide bond in these molecules, collectively suggest that bioactivity depends on their tertiary structure. It should be noted that specific bioactivity of the derivatives is different depending on the chemical group bound to the same sulfur atom in Cys-60. Carboxymethylation of the SH group resulted in a 16- to 20-fold increase of bioactivity, while formation of a disulfide bond with 5-thio-2-nitrobenzoic acid increased the activity by 200-fold. Indeed, the minimum concentration of the DTNB derivative for the detection of GIF bioactivity was comparable to that of bioactive GIF secreted from Ts hybridomas (6). The quantitative difference in the bioactivity between the carboxymethylated GIF and the DTNB derivative suggests that the generation of bioactivity is not due to the loss of the free SH group or to the formation of a particular chemical bond with the sulfur atom. Previous studies by Kato et al. (13) indicated high conformational flexibility in the α-helices and adjacent loop regions in the rhGIF molecules. One may speculate that binding of a chemical group to the sulfur atom in Cys-60, which is sticking out between the two α-helices, would increase the distance between the α-helices and may change their conformation, and that such conformational changes may be responsible for the generation of bioactivity. This speculation is supported by the fact that the binding of a large group, such as 5-thio-2-nitrobenzoic acid, to Cys-60 was more effective in increasing the bioactivity than the binding of a smaller carboxymethyl group to the same sulfur atom.
Bioactive derivatives of rGIF are artificial products. However, the bioactive derivatives of rGIF and Ts-derived bioactive GIF may share common conformational structures. Previous experiments, together with the present findings, have shown that both the inactive cytosolic GIF and E. coli-derived rGIF had affinity for Affi-Gel 10 (6), while the bioactive GIF from Ts cells and bioactive derivatives of rhGIF did not. The present experiments also showed that treatment of inactive, cytosolic GIF with EMTS or DTNB resulted in the generation of bioactivity, while bioactive GIF in the culture supernatant of the same cells was inactivated by the treatment with DTT, suggesting that SH groups in the cytosolic GIF molecules are involved in conversion of the inactive GIF to bioactive GIF in Ts cells. Previous experiments provided evidence that the bioactive GIF from Ts cells possesses the I-J antigenic determinant, and that the I-J phenotype of GIF is determined by the cells involved in posttranslational modifications rather than the amino acid sequence of the protein (6). Elucidation of the crystal structure of rhGIF, which strikingly resembles the peptide binding domain of major histocompatibility complex molecules (13), suggested to us the possibility that a substance such as a self-peptide or self-lipid may be incorporated in the groove between the two α helices during the process of posttranslational modifications and participate in the I-J epitope (13). One might speculate that Cys-60 is involved in the incorporation of the peptide/lipid, which also induces conformational changes similar to that obtained by the reaction of the same residue with a SH reagent.
Acknowledgments
This work was supported by Research Grant AI-14784 from the U.S. Department of Health and Human Services. This paper is publication no. 162 from the La Jolla Institute for Allergy and Immunology.
Footnotes
Abbreviations: rGIF, recombinant glycosylation inhibiting factor; rhGIF, recombinant human GIF; Ts cells, supressor T cells; EMTS, ethylmercurithiosalicylate; DTNB, 5,5′-dithiobis(2-nitrobenzoic acid); RGG, rabbit IgG; IgE-BF, IgE-binding factor.
References
- 1.Jardieu P, Akasaki M, Ishizaka K. J Immunol. 1987;138:1494–1501. [PubMed] [Google Scholar]
- 2.Tagaya Y, Mori A, Ishizaka K. Proc Natl Acad Sci USA. 1991;88:9117–9121. doi: 10.1073/pnas.88.20.9117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nakano T, Ishii Y, Ishizaka K. J Immunol. 1996;156:1728–1734. [PubMed] [Google Scholar]
- 4.Ishii Y, Nakano T, Ishizaka K. J Immunol. 1996;156:1735–1742. [PubMed] [Google Scholar]
- 5.Liu Y-C, Nakano T, Elly C, Ishizaka K. Proc Natl Acad Sci USA. 1994;91:11227–11231. doi: 10.1073/pnas.91.23.11227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakano T, Liu Y-C, Mikayama T, Watarai H, Taniguchi M, Ishizaka K. Proc Natl Acad Sci USA. 1995;92:9196–9200. doi: 10.1073/pnas.92.20.9196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mikayama T, Nakano T, Gomi H, Nakagawa Y, Liu Y-C, Sato M, Iwamatsu H, Ishii Y, Weiser W Y, Ishizaka K. Proc Natl Acad Sci USA. 1993;90:10056–10060. doi: 10.1073/pnas.90.21.10056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Turner J M, Brodsky M H, Irving B A, Levin S D, Perlmutter R M, Littman R R. Cell. 1990;60:755–765. doi: 10.1016/0092-8674(90)90090-2. [DOI] [PubMed] [Google Scholar]
- 9.Frankel A D, Bredt D S, Pabo C D. Science. 1988;240:70–73. doi: 10.1126/science.2832944. [DOI] [PubMed] [Google Scholar]
- 10.Green S, Chambon P. Nature (London) 1987;325:75–78. doi: 10.1038/325075a0. [DOI] [PubMed] [Google Scholar]
- 11.Glazer A N, Delange R J, Sigman D S. Chemical Modifications of Proteins. Amsterdam: North–Holland; 1975. pp. 101–120. [Google Scholar]
- 12.Iwata M, Ishizaka K. J Immunol. 1988;141:3270–3277. [PubMed] [Google Scholar]
- 13.Kato Y, Muto T, Tomura T, Tsumura H, Watarai H, Mikayama T, Ishizaka K, Kuroki R. Proc Natl Acad Sci USA. 1996;93:3007–3010. doi: 10.1073/pnas.93.7.3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sowdhamini R, Srinivasan N, Shoichet B, Santi D V, Ramakrishnan C, Balaram P. Protein Eng. 1989;3:95–103. doi: 10.1093/protein/3.2.95. [DOI] [PubMed] [Google Scholar]