Abstract
Background
Otx genes, orthologues of the Drosophila orthodenticle gene (otd), play crucial roles in vertebrate brain development. In the Xenopus eye, Xotx2 and Xotx5b promote bipolar and photoreceptor cell fates, respectively. The molecular basis of their differential action is not completely understood, though the carboxyl termini of the two proteins seem to be crucial. To define the molecular domains that make the action of these proteins so different, and to determine whether their retinal abilities are shared by Drosophila OTD, we performed an in vivo molecular dissection of their activity by transfecting retinal progenitors with several wild-type, deletion and chimeric constructs of Xotx2, Xotx5b and otd.
Results
We identified a small 8–10 amino acid divergent region, directly downstream of the homeodomain, that is crucial for the respective activities of XOTX2 and XOTX5b. In lipofection experiments, the exchange of this 'specificity box' completely switches the retinal activity of XOTX5b into that of XOTX2 and vice versa. Moreover, the insertion of this box into Drosophila OTD, which has no effect on retinal cell fate, endows it with the specific activity of either XOTX protein. Significantly, in cell transfection experiments, the diverse ability of XOTX2 and XOTX5b to synergize with NRL, a cofactor essential for vertebrate rod development, to transactivate the rhodopsin promoter is also switched depending on the box. We also show by GST-pull down that XOTX2 and XOTX5b differentially interact with NRL, though this property is not strictly dependent on the box.
Conclusion
Our data provide molecular evidence on how closely related homeodomain gene products can differentiate their functions to regulate distinct cell fates. A small 'specificity box' is both necessary and sufficient to confer on XOTX2 and XOTX5b their distinct activities in the developing frog retina and to convert the neutral orthologous OTD protein of Drosophila into a positive and specific XOTX-like retinal regulator. Relatively little is known of what gives developmental specificity to homeodomain regulators. We propose that this box is a major domain of XOTX proteins that provides them with the appropriate developmental specificity in retinal histogenesis.
Background
The vertebrate neural retina is made up of six main types of neurons (cone, rod, horizontal, bipolar, amacrine and retinal ganglion cells) plus the Müller glia cells. All these cell types are generated from a pool of multipotent retinal progenitor cells (RPCs) in a precise time schedule that is largely conserved among different vertebrates [1-4]. The molecular mechanisms driving the RPCs toward specific cell fates are under intense scrutiny and several transcription factors play a crucial role in this process. Basic helix-loop-helix (bHLH) and homeodomain factors are known to regulate the competence state of RPCs (the ability to generate one or more types of neurons), or to more directly address RPCs toward specific cell fates [5-19].
Among the homeobox genes involved in retinal cell fate regulation are the Otx genes Otx2 and Crx/Otx5. These genes are related to the orthodenticle (otd) gene of Drosophila, required for normal anterior development of the fly [20,21]. Two otx genes, Otx1 and Otx2, were initially isolated in mouse [22] and shown to be essential for correct development of the rostral brain; the Otx2-/- phenotype is especially severe, leading to complete lack of anterior structures [23-26]. This phenotype can be rescued by the Drosophila otd gene [27,28]. Conversely, the effects of otd mutation in Drosophila are rescued by either human OTX1 or OTX2 [29,30]. Finally, Otx1 and Otx2 seem interchangeable with respect to many aspects of mouse anterior development [31]. These data suggested an extensive functional conservation of the OTX/OTD class of proteins.
Crx is an otx-like gene important for the differentiation and maintenance of photoreceptors [32,33]. The CRX protein is able to bind and activate photoreceptor specific genes, such as those encoding interphotoreceptor retinoid-binding protein (IRBP), β-phosphodiesterase, arrestin and opsin [33,34]. CRX biological activity greatly depends on molecular interactions with partners such as NRL, an essential cofactor for vertebrate rod development [35]. These interactions were shown in vitro [36], and in transfection assays that demonstrate a synergy of the two factors in activating photoreceptor gene promoters [37]. Mutations in CRX are associated with diverse human eye diseases [33,38-42], and some affect CRX-NRL interaction and/or CRX transactivating ability [36,43]. In mouse, Crx function seems essential for terminal differentiation: in Crx-/- mice, photoreceptors do not develop their outer segments and display perturbed synaptogenesis [44,45]. However, though defective, photoreceptors do initially develop in Crx-/- mice, suggesting that their commitment relies on other players. In particular, results of conditional Otx2 loss-of-function in the mouse retina suggest that Otx2 controls photoreceptor initial specification and activates Crx expression in committed precursors [46].
A different picture is present in other vertebrates. In Xenopus, Xotx2 and Xotx5b (the homolog of Crx) are expressed in different patterns during retinal histogenesis: transcription of both genes starts at tailbud stage in a diffused fashion throughout the retina, but then their expression is progressively restricted and, in the mature retina, Xotx2 mRNA is found only in bipolar cells, while Xotx5b is transcribed in both photoreceptors and a subset of bipolar cells [12]. Even more dramatic is the difference in the protein expression pattern: XOTX2 protein is detected only in bipolar cells, while XOTX5b is produced only in photoreceptors, due to precise translational control through the 3' untranslated regions (UTRs) of their mRNAs [47]. Consistent with the pattern of protein distribution, lipofection of RPCs with constitutively expressed Xotx2 and Xotx5b cDNAs lacking the 3' UTR showed dramatically different effects, with Xotx2 driving cells toward bipolar cell fate, and Xotx5b toward photoreceptor cell fate [12,17,47]. Interestingly, domain-swapping experiments showed that the specific effect of either protein relies on its carboxyl terminus [12]. Because of the substantial similarity of the two proteins (XOTX2 and XOTX5b are 75% identical overall; 96% identical in the homeodomain) and of the functional conservation between Otx/otd genes in regulating early developmental events, we asked the following questions: which part of the XOTX2 or XOTX5b protein is crucial for their specific activities driving RPCs toward bipolar or photoreceptor cell type, respectively? Is Drosophila OTD able to drive RPCs toward any specific cell fate in the Xenopus retina?
To answer these questions, we performed an in vivo molecular dissection of the activity of several wild-type and mutant Xotx2 and Xotx5b constructs on retinal cell specification. We thus identified, directly downstream of the homeodomain, a small 8–10 amino acid divergent region that is both necessary and sufficient for XOTX2 and XOTX5b specific activities on cell fate; this region works as a 'specificity box' that switches the retinal activity of XOTX5b into that of XOTX2 and vice versa. Significantly, the insertion of this box into Drosophila OTD, which has no cell fate effect in the frog retina, endows it with the retinal activity of either XOTX2 or XOTX5b. We also found that the greater ability of XOTX5b, compared to XOTX2, to synergize with Xenopus NRL (XNRL) to activate the rhodopsin promoter is also switched depending on this box. We next investigated whether these results may be due to differential interactions of XOTX/OTD proteins with XNRL: we found that XOTX2 and XOTX5b differentially interact with XNRL, but also that OTD and mutant XOTX/OTD proteins are able to bind XNRL, suggesting that this domain is not essential for XOTX interactions with XNRL, though it may modulate XOTX/OTD interactions with XNRL and their overall specific actions. Our data provide in vivo and in vitro molecular evidence on how closely related homeodomain factors differentiate their functions to regulate distinct cell fates.
Results
A small 8–10 amino acid region confers specific activities on XOTX2 and XOTX5b in the Xenopus retina
Previous lipofection experiments showed that two chimeric constructs, Xotx2/engR and Xotx5b/engR, in which most of the transactivation domain of either XOTX2 or XOTX5b was replaced with the repressor domain of the Drosophila Engrailed protein, had specific effects on either bipolar cells or photoreceptor cells, respectively, in this case leading to a decrease, rather than an increase, in their frequency [12]. This observation suggested that these XOTX/engR chimeric proteins might retain a region of XOTX2 or XOTX5b crucial for their specific activities, and focused our attention on the only part of their carboxyl termini that was included in the Xotx2/engR and Xotx5b/engR constructs. This region spans amino acids 100–109 of XOTX2 and amino acids 100–107 of XOTX5b, where the two proteins differ in six residues (Figure 1).
Figure 1.
Constructs used in this study. On the left are schematics of the different constructs; on the right are their sequences in the final region of the homeodomain (HD) and directly downstream of it, with different colors shading the parental sequences of XOTX2 (red), XOTX5b (yellow) and OTD (blue). Lines are introduced for sequence alignment. The divergent region responsible for the different retinal activities of XOTX2 and XOTX5b (RS box) is shown in the blue box.
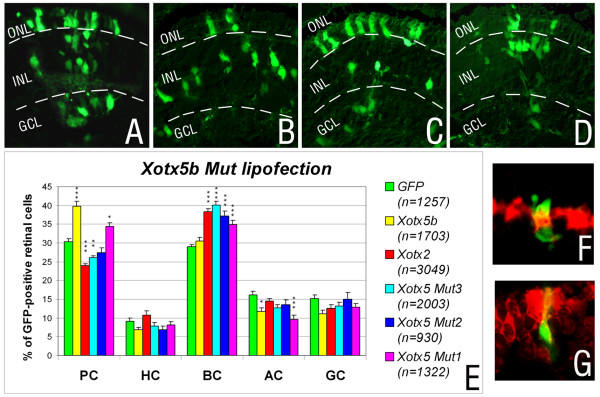
To test if these residues are crucial for the respective activities of the two factors, we changed the XOTX5b amino acid sequence in this region into the corresponding one of XOTX2, and vice versa. We first generated three sequential constructs encoding mutant forms of XOTX5b, in which two (construct Xotx5bMut1), four (Xotx5bMut2), or six (Xotx5bMut3) amino acid residues of the relevant region were changed to those of XOTX2, thereby switching this region of XOTX5b into that of XOTX2 (Figure 1). These constructs were lipofected in RPCs and their activities compared to those of wild-type Xotx2 and Xotx5b (Additional files 1 and 2). As expected [12], lipofections with wild-type Xotx5b or Xotx2 constructs promoted photoreceptor or bipolar cell fate, respectively (Figure 2a–c, e). On the other hand, unlike wild-type Xotx5b, the Xotx5bMut3 construct yielded the same effect as wild-type Xotx2, increasing bipolar cell and decreasing photoreceptor frequency (Figure 2d, e). Interestingly, lipofections of RPCs with the Xotx5bMut2 construct (four amino acid change) increased bipolar cells, but did not decrease photoreceptors; even more interestingly, the Xotx5bMut1 construct (two amino acid change) increased bipolar as well as photoreceptor cells, therefore showing the joint effects of both parental proteins (Figure 2e). Molecular markers confirmed the identity of cells lipofected with these different constructs: in particular, cells lipofected with Xotx5b and scored as photoreceptors expressed IRBP, and were thus bona fide photoreceptors and not displaced cells with a different identity (Figure 2f); on the other hand, cells transfected with Xotx5bMut3 and scored as bipolar cells expressed the bipolar marker Xvsx1 [48] (Figure 2g). Other markers were also used in this study to confirm our diagnoses of different cell types (Additional file 3).
Figure 2.
Results of in vivo lipofection of RPCs with wild-type Xotx2 and Xotx5b, and mutant Xotx5b constructs. (a-d) Sample sections are shown for control retinae lipofected with GFP+vector DNA alone (a), GFP+Xotx2 (b), GFP+Xotx5b (c) or GFP+Xotx5bMut3 (d); GCL, ganglion cell layer; INL, inner nuclear layer; ONL, outer nuclear layer. (e) Overall distribution of retinal cell types in clones lipofected with the different constructs; PC, photoreceptor cells; HC, horizontal cells; BC, bipolar cells; AC, amacrine cells; GC, ganglion cells. The proportion of each cell type is represented as an average. Error bars indicate the standard error of the mean. The experiment was repeated at least three times for all constructs. Counted cells are indicated in the histogram (n), from 15 retinae for GFP, 15 retinae for Xotx5b, 18 retinae for Xotx2, 16 retinae for Xotx5bMut3, 10 retinae for Xotx5bMut2, and 13 retinae for Xotx5bMut1. Asterisks represent significant differences between Xotx constructs and GFP, as calculated by ANOVA analysis using the Tukey-Kramer post-test (*p < 0.05, **p < 0.01, ***p < 0.001). (f, g) In situ hybridization analyses showing examples of GFP-positive (green), Xotx5-lipofected photoreceptor cell positive for IRBP probe (Fast Red detection) (f), and a Xotx5bMut3-lipofected bipolar cell expressing Xvsx1 (Fast Red detection) (g).
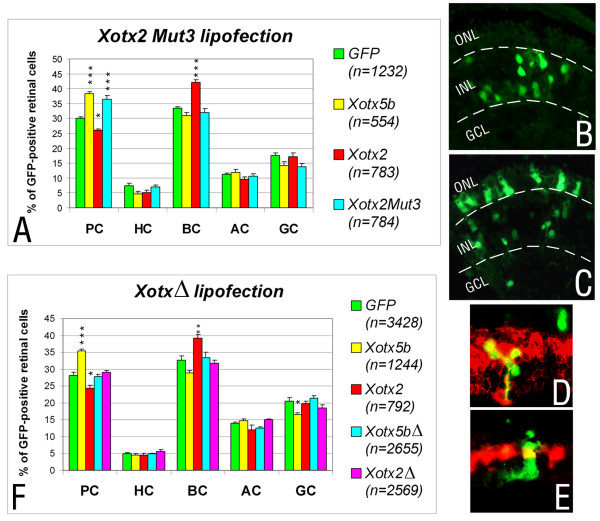
We also switched Xotx2 into Xotx5b. For this, we generated a mutant Xotx2 construct (Xotx2Mut3) in which the crucial region of XOTX2 was converted to that of XOTX5b (Figure 1). The activity of Xotx2Mut3 in lipofections was essentially identical to that of Xotx5b: instead of promoting bipolar cell fate like Xotx2, the mutant Xotx2Mut3 construct promoted photoreceptor fate (Figure 3a–e; Additional files 4 and 5). The identity of photoreceptors generated by RPCs lipofected with Xotx2Mut3 was confirmed by testing the expression of the IRBP marker (Figure 3e). We conclude that this small region works as a 'retinal specificity (RS) box' that is sufficient to confer on the XOTX2 and XOTX5b proteins their ability to drive RPCs toward specific fates.
Figure 3.
The 'specificity box' downstream of the homeodomain is necessary and sufficient for specific retinal action of XOTX2 and XOTX5b. (a-c) Results of in vivo lipofection of RPCs with wild-type Xotx2 and Xotx5b, and mutant Xotx2Mut3 constructs, showing the overall distribution of retinal cell types in clones lipofected with the different constructs, as indicated (a); PC, photoreceptor cells; HC, horizontal cells; BC, bipolar cells; AC, amacrine cells; GC, ganglion cells. The proportion of each cell type is represented as average ± standard error of the mean. Counted cells are indicated in the histogram (n), from 9 retinae for GFP, 10 retinae for Xotx5b, 9 retinae for Xotx2, and 14 retinae for Xotx2Mut3. Asterisks represent significant differences between Xotx constructs and GFP, as calculated by Tukey-Kramer test (*p < 0.05, **p <0.01, ***p <0.001). Sample sections are shown for retinae co-lipofected with GFP+Xotx2 (b) and GFP+Xotx2Mut3 (c); GCL, ganglion cell layer; INL, inner nuclear layer; ONL, outer nuclear layer. (d, e) In situ hybridization analyses showing examples of GFP-positive (green), Xotx2-lipofected bipolar cell expressing Xvsx1 (Fast Red detection) (d), and Xotx2Mut3-lipofected photoreceptor cell positive for IRBP probe (Fast Red detection) (e). (f) The RS box is required for the biological action of either XOTX2 or XOTX5b proteins: the histogram reports the overall distribution of retinal cell types in clones lipofected with the different constructs, as indicated. Counted cells are indicated in the histogram (n), from 11 retinae for GFP, 9 retinae for Xotx5b, 6 retinae for Xotx2, 8 retinae for Xotx2Δ, and 7 retinae for Xotx5Δ.
We next asked whether the RS box is required for XOTX protein activity in retinal cell fate specification, or whether XOTX proteins still possess a retinal 'default' activity without it. To test this, we generated deletion constructs (Xotx2Δ and Xotx5bΔ) by removing the RS box (Figure 1), and compared their activities to that of wild-type constructs. Deletion of the RS box completely abrogates any biological effect of either XOTX2 or XOTX5b, showing that this small region is required for their activity in the frog retina (Figure 3f; Additional files 6 and 7).
The RS box confers specific retinal activities on Drosophila OTD
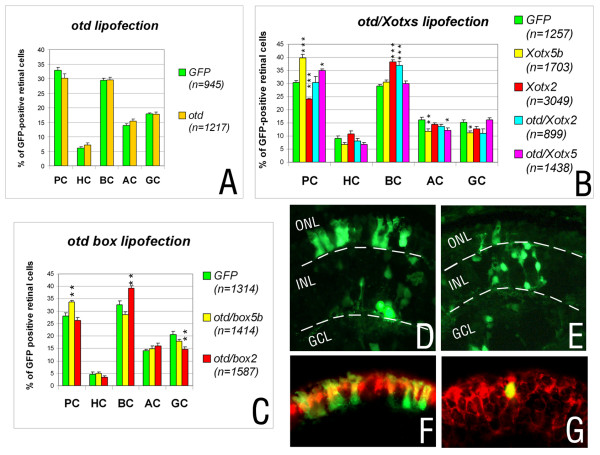
Because of the extensive functional conservation of OTX/OTD proteins in early development of the anterior region of fly and mouse embryos, we asked whether Drosophila OTD was able to direct RPCs to any specific cell fate in the Xenopus retina. In lipofections, no difference was observed in the frequency of the different retinal cell types between otd lipofected and control clones (Figure 4a; Additional file 1). The carboxyl terminus of OTD is quite divergent compared to XOTX2 or XOTX5b (it shares only 11.4% identity with XOTX2 and only 8% with XOTX5b); instead, in the same region, XOTX2 and XOTX5b are 75% identical, suggesting that a possible reason for the lack of OTD activity could be due to such a strong divergence, and possibly to absence of the box in the OTD protein (Figure 1). To test this, we first replaced the entire OTD region carboxy-terminal to the homeodomain with that of either XOTX2 or XOTX5b, and compared the activities of chimeric OTD/XOTX2 and OTD/XOTX5b (Figure 1) with those of XOTX2 and XOTX5b. Significantly, OTD/XOTX2 drives RPCs toward the bipolar cell fate, while OTD/XOTX5b increased photoreceptors (Figure 4b). However, the decrease in photoreceptor cell frequency produced by wild-type Xotx2 was not observed with the otd/Xotx2 chimeric construct (Figure 4b; Additional files 1 and 8).
Figure 4.
The RS box is sufficient to confer a specific retinal action on Drosophila OTD protein. (a) Results of lipofection of RPCs with GFP alone or with GFP+otd; counted cells were as indicated in the histogram (n), from 17 retinae for GFP, and 11 retinae for otd; PC, photoreceptor cells; HC, horizontal cells; BC, bipolar cells; AC, amacrine cells; GC, ganglion cells. (b) Results of lipofection of RPCs with GFP alone, with GFP+Xotx wild-type constructs or GFP+otd/Xotx chimeric constructs, as indicated; counted cells are indicated in the histogram (n), from 15 retinae for GFP, 18 retinae for Xotx2, 15 retinae for Xotx5b, 9 retinae for otd/Xotx2, and 11 retinae for otd/Xotx5b. (c) Results of lipofection of RPCs with GFP alone, or with GFP+otd/box2/5b chimeric constructs, as indicated. Counted cells are indicated in the histogram (n), from six retinae for GFP, nine retinae for otd/box2, and nine retinae for otd/box5b. The proportion of each cell type (a-c) is represented as average ± standard error of the mean. (d, e) Lipofected retinae with otd/box5b are enriched in photoreceptors (d), those with otd/box2 are enriched in bipolar cells (e); GCL, ganglion cell layer; INL, inner nuclear layer; ONL, outer nuclear layer. (f) An example of GFP+otd/box5b-lipofected photoreceptor cell positive for IRBP probe after in situ hybridization (Fast Red detection). (g) A GFP+otd/box2-lipofected bipolar cell expressing Xvsx1 is shown following in situ hybridization (Fast Red detection).
We then tested whether the RS box is sufficient to confer the biological activity of either XOTX2 or XOTX5b on Drosophila OTD. To do this, we generated chimeric otd/box2 and otd/box5b contructs in which the XOTX2 or XOTX5b RS box, respectively, was inserted immediately downstream of the OTD homeodomain (Figure 1), and transfected them into Xenopus RPCs. Though the carboxyl terminus of OTD is strongly divergent from those of both XOTX proteins, the specificity box enabled OTD/box2 or OTD/box5b proteins to drive RPCs toward bipolar or photoreceptor fates, respectively (Figure 4c–g). Similar to OTD/XOTX2, OTD/box2 did not lead to a significant reduction in photoreceptor cells (Figure 4c). No other effect was observed on the frequencies of the remaining cell types, with the exception of a decrease of ganglion cells with OTD/box2 (Figure 4c; Additional files 9 and 10).
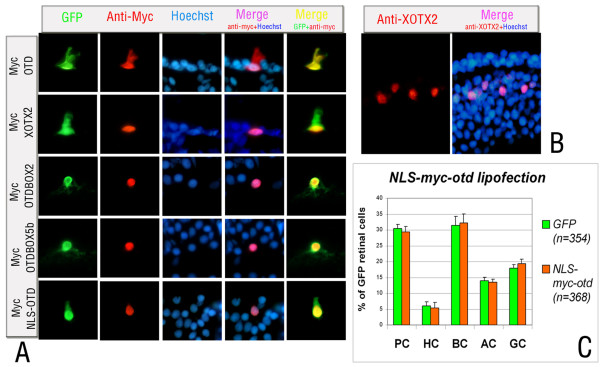
Previous work on CRX showed evidence of an additional nuclear localization signal (NLS) at the carboxyl terminus of the homeodomain. Several CRX mutations are associated with human retinal pathologies and have an effect on CRX nuclear localization [49]; one such mutation, leading to CRX mislocalization, replaces Arg98 with leucine. Because a leucine residue is present in Drosophila OTD at this position (Figure 1), we asked whether the inactivity of OTD in retinal specification was due to insufficient translocation to the nucleus, rather than to the absence of the RS box. To test this, we compared the distributions of MYC-XOTX2, MYC-XOTX5b, MYC-OTD, MYC-OTD/box2 and MYC-OTD/box5b proteins in lipofected retinal cells, detected with an anti-MYC antibody, with those of endogenous XOTX2 and XOTX5b, detected by specific antibodies [47]. Endogenous XOTX2 and XOTX5b were found only in the nuclei of bipolar and photoreceptor cells, respectively, but not in the cytoplasmic compartment of these or other retinal cells (Figure 5b; data not shown for XOTX5b). A nuclear distribution was found for MYC-XOTX2 and MYC-XOTX5b in all lipofected cells (Figure 5a; data nor shown for MYC-XOTX5b). Interestingly, while MYC-OTD was present in both nuclear and cytoplasmic compartments of lipofected cells, MYC-OTD/box2 and MYC-OTD/box5b had a clear nuclear localization (Figure 5a).
Figure 5.
The effect of the specificity box upon commitment of RPCs is not simply due to possible effects on nuclear localization. (a) The nuclear/cytoplasmic distribution of MYC-XOTX2 and MYC-OTD (as indicated at the left) is shown in lipofected retinal cells, and is compared to cytoplasmic GFP fluorescence; while MYC-XOTX2 shows an exclusively nuclear localization, MYC-OTD is partly cytoplasmic; on the contrary, MYC-OTD/box2 and MYC-OTD/box5b are targeted to the nucleus; a NLS-MYC-OTD fusion protein is forced into the nucleus (bottom row). (b) Endogenous XOTX2 protein distribution is detected by a specific antibody (red fluorescence) in the Xenopus retinal nuclei. (c) Although it forces OTD to the nucleus, a NLS-Myc-otd fusion construct does not have any effect on cell fate of RPCs. Counted cells were as indicated in the histogram (n), from six retinae for GFP, and four retinae for NLS-Myc-otd. PC, photoreceptor cells; HC, horizontal cells; BC, bipolar cells; AC, amacrine cells; GC, ganglion cells. Nuclei are counterstained with Hoechst.
While these results suggest that the RS box might be important for correct nuclear targeting of XOTX proteins, the possibility remained that potential OTD effects on cell fate did not occur due to insufficient translocation of it to the nucleus. To rule out this possibility, we lipofected a NLS-myc-otd construct (containing a nuclear localization signal) to force OTD to the nucleus, and evaluated its ability to drive RPCs to specific cell fates. As expected, the NLS-MYC-OTD protein was correctly localized to the nucleus (Figure 5a), but it did not significantly affect cell fate (Figure 5c; Additional file 11). Therefore, efficient translocation to the nuclei of retinal cells did not provide OTD with the ability to drive frog RPCs toward a precise neuronal fate; instead, OTD gained this ability when its homeodomain was directly followed by a RS box of the XOTX2 or XOTX5b type.
XOTX2 and XOTX5b differentially synergize with XNRL to transactivate the rhodopsin promoter
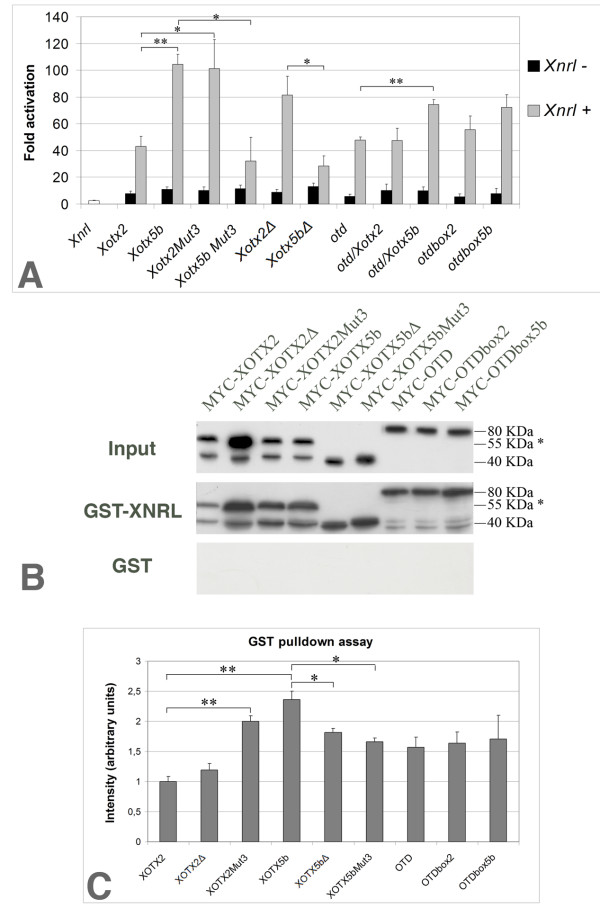
Previous work showed that CRX/XOTX5b interacts with NRL to activate the rhodopsin promoter [36,50], and that a lower level of activation is obtained when Otx2 and Nrl are co-transfected in cultured cells [37]. We therefore asked whether the Xotx mutant constructs also switched their activity in similar transfection assays. We thus monitored the ability of XOTX2 and XOTX5b (alone or together with XNRL) to activate a Xenopus rhodopsin promoter (XOP) driving green fluorescent protein (GFP) expression in HEK 293T cells, and compared it with the activities of XOTX5bMut3, XOTX2Mut3, OTD, OTD/XOTX2, OTD/XOTX5b, OTD/box2, OTD/box5b, XOTX2Δ and XOTX5bΔ. When each of these constructs was co-transfected with the XOP-GFP reporter [50] in the absence of XNRL, scarce activation of the reporter gene was detected (5- to 13-fold activation compared to the ground level given by transfection of XOP-GFP alone); the same held true for transfection of Xnrl alone (Figure 6a; Additional file 12). A significant difference (p < 0.01, Student's t-test) was observed between Xotx2+Xnrl and Xotx5b+Xnrl transfections, which elicited activation of the co-transfected reporter 43- and 105-fold over the ground level, respectively; these results are consistent with those of Peng and Chen [37].
Figure 6.
XOTX2 and XOTX5b differentially synergize with XNRL to activate the rhodopsin promoter and differentially interact in vitro with XNRL. (a) Results of rhodopsin promoter cell transfection assays with several Xotx/otd constructs (with or without Xnrl). Error bars indicate the standard error of the mean. (b) GST-pull down assays compare the interaction of MYC-XOTX/OTD fusion proteins to GST-XNRL or GST alone. The band indicated by an asterisk corresponds to a higher molecular weight (55 kDa) than the one expected for XOTX proteins (41 kDa) and may result from post-translational modification; this needs further investigation. (c) Results of two of these experiments, analyzed by Image J, were statistically processed; columns show the ratio of the retained MYC-tagged proteins relative to their respective input, normalized with respect to the MYC-XOTX2 retained/input ratio. Error bars indicate the standard deviation. The p-value was calculated by bilateral Student's t-test. Asterisks in histograms show statistically significant differences only for more relevant comparisons (*p < 0.05, **p <0.01). (see Additional files 12 and 13 for the full data set).
More significantly, Xotx2Mut3+Xnrl transfection gave similar results to Xotx5b+Xnrl (101-fold reporter activation; p = 0.035 for Xotx2Mut3+Xnrl versus Xotx2+Xnrl; p = 0.89 for Xotx2Mut3+Xnrl versus Xotx5b+Xnrl), while Xotx5bMut3+Xnrl (32-fold activation) gave similar results to Xotx2+Xnrl (Xotx5bMut3+Xnrl versus Xotx2+Xnrl, p = 0.55; Xotx5bMut3+Xnrl versus Xotx5b+Xnrl, p = 0.02). Therefore, exchanging the RS box in XOTX2 or XOTX5b leads to a switch in their ability to transactivate, together with XNRL, one of the key photoreceptor specific genes. In addition, otd, otd/Xotx2 or otd/box2, when combined with Xnrl, all gave results similar to Xotx2 (48-, 47- and 56-fold activation, respectively); a slightly stronger effect was observed with otd/Xotx5b+Xnrl (otd/Xotx5b+Xnrl versus otd+Xnrl, p = 0.003) and otd/box5+Xnrl (74- and 72-fold activation, respectively). Surprisingly, a rather strong effect was obtained with Xotx2Δ+Xnrl (82-fold activation; Xotx2Δ+Xnrl versus Xotx2+Xnrl, p = 0.047; Xotx2Δ+Xnrl versus Xotx5b+Xnrl, p = 0.22; Xotx2Δ+Xnrl versus Xotx5bΔ+Xnrl, p = 0.029), but not with Xotx5bΔ+Xnrl (28-fold activation; p = 0.014 versus Xotx5b+Xnrl). Significant differences were also found between Xotx5b+Xnrl and otd+Xnrl (p = 0.002).
In vitro interactions of XOTX/OTD proteins with XNRL
One possible way to explain the different activities of XOTX2 and XOTX5b is that the two proteins differentially interact with other key molecular players involved in retinal differentiation, such as XNRL itself. Full-length CRX, or truncated CRX forms containing the homeodomain, the Q-rich region and the basic region altogether, were shown to strongly interact with NRL, whereas the CRX homeodomain alone showed much lower interaction [36].
Because the RS box spans from the Q-rich region to part of the basic region [51], we decided to test whether XOTX2 and XOTX5b show differential interaction with XNRL. We therefore prepared a GST-XNRL fusion construct and purified the corresponding protein; this was used in GST-pull down assays against affinity purified full-length MYC-XOTX2, MYC-XOTX2Δ, MYC-XOTX2Mut3, MYC-XOTX5b, MYC-XOTX5bΔ, MYC-XOTX5bMut3, MYC-OTD, MYC-OTD/box2, and MYC-OTD/box5b. The results of these experiments are shown in Figure 6b; the pull-down results were scanned and analyzed by Image J [52], and the resulting data were plotted (Figure 6c; Additional file 13). We found that MYC-XOTX5b interacts with GST-XNRL about 2.4-fold more compared to MYC-XOTX2 (p < 0.01, Student's t-test); significant differences were also observed compared to MYC-XOTX2Δ, MYC-XOTX5bΔ, MYC-XOTX5bMut3, MYC-OTD and MYC-OTD/box2, but not compared to MYC-XOTX2Mut3, or MYC-OTD/box5b. Therefore, XOTX5b interacts more strongly with XNRL, but other XOTX/OTD proteins also interact in vitro with XNRL, even without the box.
Discussion
We have identified a small, divergent region that confers specific retinal activities to XOTX2 and XOTX5b. This RS box lies directly carboxy-terminal to the homeodomain, extending for 8–10 amino acids from the the poly-Q tail to embrace part of the basic region as identified in CRX [51]. Remarkably, this divergent region is necessary and sufficient to confer on these XOTX proteins their specific cell fate specification activity in the frog retina. First, deletion of the box completely abrogated any cell fate activity of both XOTX2 and XOTX5b. Furthermore, exchanging the sequence of the XOTX5b RS box into that of the XOTX2 box (construct Xotx5bMut3) and vice versa (construct Xotx2Mut3) completely switched the biological activities of the two proteins. Two other Xotx5b mutant constructs showed interesting intermediate effects: Xotx5bMut2 pushed RPCs toward a bipolar cell fate (like Xotx2), but had no significant effect on decreasing photoreceptor cell frequency (unlike Xotx2); Xotx5bMut1 showed the activities of both Xotx2 and Xotx5, as it was able to increase both bipolar and photoreceptor cells (though in the latter case with significantly lower efficiency than Xotx5b). These data show that the first two changes in the XOTX5b amino acid sequence (S100N, T101G) are sufficient to endow XOTX5bMut1 with a great part of the XOTX2 ability to promote bipolar fate; in fact, there was no statistical difference between Xotx2 and Xotx5bMut1 (p > 0.05; or Xotx5bMut2 or Xotx5bMut3, p > 0.05) in their efficiency to promote bipolar cells. This suggests that Asn102 and/or Gly103 are particularly important residues for this aspect of XOTX2 action.
Mutant and wild-type Xotx5b constructs also showed graded effects on photoreceptor commitment: Xotx5bMut1 promoted, rather then repressed, photoreceptors, similar to Xotx5b; Xotx5bMut2 showed no effect on photoreceptor frequency; and Xotx5bMut3 led to significantly fewer photoreceptors compared to GFP controls. In particular, Xotx5bMut1 photoreceptor-promoting activity was significantly lower than that of Xotx5b; furthermore, constructs Xotx5bMut1 and Xotx5bMut2 yielded significantly different effects (p < 0.001), whereas no significant difference occurred between constructs Xotx5bMut2 and Xotx5bMut3 (p > 0.05; or these two and Xotx2, p > 0.05). These results suggest that the changing of the first two amino acids may not have completely compromised the photoreceptor promoting activity of XOTX5b, while the next specific mutagenetic changes led to its abrogation and a reversal of its effect. The six residues may, therefore, have additive roles in determining XOTX2 repressive effect on photoreceptor fate.
We show that wild-type OTD does not mimic XOTX2 or XOTX5b in the frog retina. However, replacement of the OTD carboxyl terminus with that of either XOTX protein, or even the simple insertion of either the XOTX2 or XOTX5b specificity box into OTD, provides it with the activity of XOTX2 or XOTX5b. This is remarkable since OTD lacks some of the functional domains important for the transactivating ability of CRX/OTX proteins, such as the OTX tail and the WSP domain [43,51]. Therefore, the RS box is sufficient to promote specific cell fates in a rather more divergent context than that of vertebrate OTX proteins. This is not due to mere effects of the box on cytoplasmic-nuclear trafficking, because forcing OTD to the nucleus using a NLS does not have any effect on RPC fate and because the effect of OTD/box2 and OTD/box5b specifically depends on the type of RS box. Therefore, we suggest that while the carboxy-terminal domain of OTD mimics, to a certain extent, the transactivating activity of the XOTX2 and XOTX5b carboxyl termini, OTD, in the absence of the RS box, fails to properly target the gene sets that address RPCs to their fates.
However, not all XOTX2 or XOTX5b retinal functions depend on the RS box. Unlike XOTX2, OTD/XOTX2 is unable to repress photoreceptors. This is different from the effect shown by the Xotx5b/Xotx2 chimeric construct, which retains Xotx2 anti-photoreceptor activity [12], suggesting that some features of XOTX retinal activity may also depend on the amino terminus. XOTX5b and XOTX2 amino-terminal regions (excluding the homeodomain) are about 73% identical, while the OTD amino terminus is only about 15% identical to that of XOTX2. It is possible that the amino terminus of XOTX5b may better match the carboxyl terminus of XOTX2 (and vice versa) than the amino terminus of OTD, thus allowing the exploitation of the full spectrum of protein activities. Such activity of the amino terminus could be due to possible interactions with other parts of the XOTX protein at either the intramolecular level, for example, to allow proper folding of the protein, or at the intermolecular level, for example, with other XOTX monomers [53] or other molecular partners.
How can the RS box modulate the activity of XOTX2 and XOTX5b proteins? Two possibilities, not mutually exclusive, are that the box refines the DNA binding abilities of XOTX proteins towards different sets of promoters, or that it modulates interactions with other molecular partners. While the overall effects on cell fate of wild-type and mutant XOTX2 and XOTX5b strongly suggest that different sets of genes are indeed activated depending on the type of RS box, we also show that XOTX2 and XOTX5b differentially synergize with XNRL in the regulation of the rhodopsin promoter, and that this ability is switched by the RS box. Some activation is also observed when XNRL is transfected together with OTD/XOTX5b or OTD/box5b (although this is significant only in the case of OTD/XOTX5b). Instead, XOTX5bΔ, OTD, OTD/XOTX2 and OTD/box2 do not appear to synergize strongly with XNRL on the rhodopsin promoter. While these results are quite consistent with the in vivo photoreceptor promoting activity of these constructs, we found, unexpectedly, that XOTX2Δ+XNRL also activates the rhodopsin promoter. This result does not seem completely consistent with the lipofection results, where Xotx2Δ does not promote photoreceptor fate. However, the in vivo cell fate specification activity of XOTX proteins presumably occurs through the activation of entire sets of genes, and may be more complex that what can be measured from their activity on a single promoter. XOTX2Δ may have some intrinsic ability to act on the rhodopsin promoter together with XNRL; this may normally be impaired by the RS box in XOTX2, and becomes unmasked when the box is removed. On the other hand, XOTX5bΔ does not have this intrinsic ability, and XOTX5b specifically requires the RS box to synergize with XNRL. In this respect, the box would have a negative regulatory role in XOTX2, and a positive one in XOTX5b.
We also investigated whether the difference in the synergy of XOTX5b and XOTX2 with XNRL on the rhodopsin promoter may be due to differences in their interaction with XNRL. We found that MYC-XOTX5b had a significantly greater affinity toward GST-XNRL than MYC-XOTX2, MYC-XOTX5bMut3 and MYC-OTD/box2 (all with an XOTX2-type box) or MYC-XOTX2Δ, MYC-XOTX5bΔ, and MYC-OTD (all lacking a box); instead, MYC-XOTX2Mut3 and MYC-OTD/box5b (with a XOTX5b-type box) were not significantly different from XOTX5b in this respect. Therefore, while the box is not an essential determinant for XOTX/OTD versus XNRL interaction, it seems to modulate this interaction in a way that is quite consistent with the roles of XOTX2 and XOTX5b in frog retinogenesis [12] and with the results on rhodopsin promoter activation [37] (and present data).
On the whole, our results show that XOTX proteins are pivotal in regulating retinal cell fate. Although their effects on cell fate may appear limited, and not all transfected progenitors are turned into bipolar cells or photoreceptors, they are statistically significant. Failure to address all transfected cells to a single and specific fate may be due to differences in their time of cell cycle exit, which may underlie the diverse competence of progenitors; it may be significant, in this respect, that the effect of Xotx5b in enhancing photoreceptor fate is potentiated when co-transfected with X-gadd45γ, which promotes cell cycle exit [47]. Besides, retinal cell fate commitment and differentiation are, at the molecular level, the result of a multifactorial process [6,16,17], and, therefore, other factors may contribute to the action of either XOTX2 or XOTX5b in addressing retinal cells to their proper neuronal fate.
Conclusion
We have provided in vivo and in vitro evidence for the different biochemical activities of XOTX/OTD proteins in the Xenopus retina, due to the presence/absence of the RS box. We suggest that the RS box allows XOTX2 and XOTX5b proteins to appropriately target gene sets involved in bipolar and photoreceptor cell specification, respectively. This is particularly significant since OTX/CRX/OTD proteins are able to bind in vitro to the same consensus sequence, TAATCC/T [33,53,54], and yet they have significantly different effects in the Xenopus retina; this is reminiscent of many Hox gene products, which also have largely overlapping DNA binding abilities but perform very specific and different developmental functions (reviewed in [55,56]). While still relatively little is known about what confers in vivo targeting specificity on homeodomain containing factors, our data show that the RS box of XOTX2 and XOTX5b is an essential and major domain of their functioning in vivo and is involved in providing such specificity in the developing frog retina.
Methods
Xenopus laevis embryos
Xenopus embryos were obtained and staged as previously described [16]. All protocols involving the use of animals were approved by the Bioethical Committee of Pisa University.
DNA constructs
The main constructs used in this study are shown in Figure 1. pCS2Xotx2 and pCS2Xotx5b wild-type constructs were described in Viczian et al. [12]. pCS2Xotx5bMut1, pCS2Xotx5bMut2 and pCS2Xotx5bMut3 were generated by in vitro mutagenesis from pCS2Xotx5b, converting the initial STGQAKPR sequence of amino acids 100-107 of XOTX5b to generate mutant constructs as shown in Figure 1. Similarly, pCS2Xotx2Mut3 was obtained from pCS2Xotx2. pCS2otd was obtained by PCR cloning of the otd coding region plus 50 nucleotides (nt) of the 5' UTR and 24 nt of the 3' UTR into pCS2+; fragments were amplified from an otd plasmid (kindly provided by Dr Antonio Simeone), and cloned into the EcoRI site of pCS2+. The chimeric pCS2otd/Xotx2 is an in-frame fusion encoding amino acids 1–96 of OTD and amino acids 62–288 of XOTX2, plus 50 nt of the 5' UTR of otd and 4 nt of the 3' UTR of Xotx2; the chimeric pCS2otd/Xotx5b is an in-frame fusion encoding amino acids 1–96 of OTD and amino acids 62–290 of XOTX5b, plus 50 nt of the 5' UTR of otd and 25 nt of the 3' UTR of Xotx5b. The pCS2Xotx2Δ and pCS2Xotx5bΔ deletion constructs correspond to pCS2Xotx2 and pCS2Xotx5b, respectively, except that the regions encoding amino acids 100–109 of XOTX2 and amino acids 100–107 of XOTX5b were removed by site directed mutagenesis. pCS2otd/box2 and pCS2otd/box5b correspond to pCS2otd, but have an insertion encoding amino acids 100–109 of XOTX2 and 100–107 of XOTX5b, respectively, replacing amino acids 132–137 of OTD.
pCS2Myc-Xotx2, pCS2Myc-Xotx5b, pCS2Myc-Xotx2Mut3, pCS2Myc-Xotx5bMut3, pCS2Myc-Xotx2Δ, pCS2Myc-Xotx5bΔ, pCS2Myc-otd, pCS2Myc-otd/box2, pCS2Myc-otd/box5b, and pCS2-NLS-Myc-otd were prepared by PCR cloning from the parental Xotx2, Xotx5b, otd or chimeric plasmids into pCS2Myc and pCS2-NLS-Myc vectors.
Xnrl full-length cDNA was cloned by RT-PCR from stage 42 Xenopus embryo RNA; the GST-Xnrl fusion construct was obtained by in frame PCR cloning of the Xnrl coding region into the pYEX vector (a modified pGEX 2TK, a kind gift of Dr Luciana Dente). All constructs were verified by sequencing.
Lipofections
Lipofections were performed at stages 17–18 as described in Poggi et al. [16]. At stage 42, embryos were fixed in 4% paraformaldehyde for 1 h at room temperature, sunk in 20% sucrose overnight at 4°C and cryostat sectioned (12 μm). Samples were rehydrated with two washes of 1× phosphate-buffered saline (PBS) for 5 minutes, incubated in 1 μg/ml Hoechst to label nuclei and mounted in Aqua Polymount (Polysciences, Inc., Warrington, PA, U.S.A). Lipofected cells were scored by GFP fluorescence and assigned to the different cell types on the basis of their position within layers and their morphology; their identity was confirmed by molecular marker analysis (Additional file 3). Statistical analysis on cell frequencies was performed by means of one-way ANOVA and Tukey-Kramer multiple comparison test.
In situ hybridization, immunostaining and immunofluorescence
In situ hybridization on sections was performed as previously described [57]; immunostaining on sections as described in [12,47]. Probes used for in situ hybridizations were: XIRBP for photoreceptors [58]; Xhermes for ganglion cells [59]; Xprox1 for horizontal cells [11]; and Xvsx1 for bipolar cells [48]. To identify amacrine cells we used anti-5-HT, anti-GABA and anti-Tyrosine Hydroxylase, all purchased from DiaSorin (Saluggia, Italy). The anti-XOTX2 and anti-XOTX5b antibodies were described in Decembrini et al. [47].
GST-XNRL protein production and purification
GST-fusion proteins were expressed in Escherichia coli BL21 upon transformation with appropriate constructs. Cultures were grown to mid-log phase (A600 = 0.7) in LB medium at 37°C, induced with 1.0 mM isopropyl thio-β-D-galactopiranoside, and grown for an additional 4 h at 32°C. Culture (50 ml) was centrifuged at 4,000 rpm for 15 minutes, resuspended in ice cold PBS and lysed on ice. After addition of lysozyme (200 μg/ml), 10 mM DTT (in AcONa 10 mM pH 5.2), protease inhibitor mix (2 mM AEBSF, 1 mM EDTA, 130 μM bestatin, 14 μM E-64, 1 μM leupeptin, 0.3 μM aprotinin; final concentrations; Sigma (S. Louis, MO, U.S.A.)), the mixture was left on ice for 30 minutes. Then 1% (v/v) Triton X-100, 10 mM MgCl2, and 100 μg/ml DNase (final concentrations) were added; the mixture was left on ice for further 30 minutes and then centrifuged at 4°C, 14,000 rpm for 20 minutes.
Glutathione Sepharose 4B resin (100 μl) (Amersham, GE Healthcare, Little Chalfont, Buckinghamshire, U.K.) was used for each experiment and control. Resin was washed 3 times with ice cold PBS and centrifuged at 2,500 rpm for 1 minute following each wash. BL21 extract was then incubated with the resin for 1 h at 4°C on a shaker. Then three washings were carried out as above. Finally, the resin was soaked in a solution of 3% (w/v) bovine serum albumin in PBS to achieve blocking, and left at 4°C overnight.
Cell transfection and pull-down assay
HEK 293T cells were cultured in Dulbecco's modified Eagle's medium (Gibco/Invitrogen, Grand Island, NY, U.S.A.) supplemented with 10% (v/v) fetal bovine serum (GIBCO). Transfections were performed using Lipofectamine 2000 (Invitrogen, Grand Island, NY, U.S.A.). Following a 48 h incubation at 37°C and 5% CO2, cells were washed with ice-cold PBS and lysed with 100 μl ice-cold lysis buffer (1% (v/v) Triton X-100, 1 mM phenylmethylsulfonyl fluoride, 50 mM HEPES pH 7.5, 150 mM NaCl, 1% glycerol, 1.5 mM MgCl2, 5 mM EGTA, 1 mM Na3VO4 and protease inhibitor cocktail (Sigma)). After 30 minutes incubation on ice, lysates were cleared by centrifugation for 40 minutes at 14,000 g and 4°C. After protein quantification of the extracts (Bradford assay), Myc fusion proteins were purified on anti-cMyc antibody agarose beads (Clontech n. 631208, Clontech, Mountain View, CA, U.S.A.), quantified again and subsequently incubated with Glutathione-Sepharose-bound GST-XNRL or GST alone in the binding buffer (0.1% Triton X-100, 50 mM HEPES pH 7.5, 150 mM NaCl, 1% glycerol, 1.5 mM MgCl2, 5 mM EGTA) overnight at 4°C on a shaker; then washed three times and finally denatured with loading buffer for 5 minutes at 95°C.
Western blotting
Protein samples were loaded onto a 12% polyacrylamide gel for size separation. Subsequently, proteins were transferred to Immobilon-P Tranfer membrane (Millipore, Billerica, MA, U.S.A.) by electroblotting for 1–2 h. Blots were blocked for 1 h using 5% nonfat dry milk in TBS-T (10 mM Tris/HCl, pH 8.0, 150 mM NaCl, 0.05% (v/v) Tween-20 (Sigma)). Monoclonal primary anti-MYC antibody (Sigma) (dilution 1:500) and secondary anti mouse IgG (peroxidase conjugate; Sigma; 1:10,000) were used to detect MYC-tagged proteins. Filters were incubated for 1 h at room temperature for each antibody, and then washed three times with TBS-T to remove excess antibody. The SuperSignal West Pico Chemiluminescent Substrate (Pierce, Rockford, IL, U.S.A.) was used to visualize immunoreactive bands by exposure to Amersham Hyperfilm. Samples from two independent experiments were analyzed.
Cell transfection and reporter assays
HEK 293T cells were co-transfected in 24-well plates with a total of 600 ng of DNA using Lipofectamine. XOP-GFP construct (400 ng) [50] was added to each well, along with various combinations of 100 ng of pCS2Xotx2, pCS2Xotx2Mut3, pCS2Xotx2Δ, pCS2Xotx5b, pCS2Xotx5bMut3, pCS2Xotx5bΔ, pCS2otd, pCS2otd/Xotx2, pCS2otd/Xotx5b, pCS2otd/box2, pCS2otd/box5b, pCS2Xnrl, or empty pYEX expression constructs. GFP fluorescence was analyzed using flow cytometry. Fold activation was assumed as the ratio of the volume of fluorescence between each sample and the basal activation sample (XOP-GFP transfection alone), where the fluorescent volume is the fraction of GFP positive cells in the population multiplied by the mean fluorescence intensity [60]. Samples from at least three independent experiments were analyzed.
Competing interests
The author(s) declare that they have no competing interests.
Authors' contributions
MO performed most of the experiments, designed and generated many of the constructs used in this study and analyzed the data. FC contributed to conceive some of the experiments, performed some of the immunostaining experiments, analyzed the lipofection results and contributed to the writing of the paper. YL designed and prepared the mutant Xotx5b constructs. R-QE participated in the design of lipofection experiments and of some of the constructs. GB and RV conceived the study, participated in the design of most of the experiments and wrote the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Raw data of the lipofection experiments with the Xotx5bMut1, Xotx5bMut2, Xotx5bMut3, otd and otd/Xotx constructs. Raw data of the lipofection experiments with the Xotx5bMut1, Xotx5bMut2, and Xotx5bMut3 constructs (summarized in Figure 2e), with the otd construct (summarized in Figure 4a) and with the otd/Xotx constructs (summarized in Figure 4b).
Results of the statistical ANOVA analysis on neuronal cell type frequency in lipofection experiments with Xotx5bMut1, Xotx5bMut2, and Xotx5bMut3 constructs. Results of the statistical ANOVA analysis on neuronal cell type frequency in lipofection experiments with Xotx5bMut1, Xotx5bMut2, and Xotx5bMut3 constructs (Figure 2e).
Expression of retinal markers used in this study to confirm neuron cell identities of lipofected cells. Lipofected GFP-positive cells (green) can be identified by using specific probes (all detected with Fast Red): (a) an IRBP probe identifies photoreceptors by in situ hybridization; (b) in situ hybridization with Xprox1 identifies horizontal cells (red); (c) in situ hybridization with Xvsx1 identifies bipolar cells (red); (d) a specific antibody identifies GABAergic amacrine cells (red); (e) a specific antibody identifies 5-HT positive amacrine cells (red); (f) a specific antibody identifies tyrosine hydroxylase (TH)-positive amacrine cells (red); (g) in situ hybridization with Xhermes identifies ganglion cells. Hoechst staining (in blue) identifies cell nuclei.
Raw data of the lipofection experiments with the Xotx2Mut3 construct. Raw data of the lipofection experiments with the Xotx2Mut3 construct (summarized in Figure 3a).
Results of the statistical ANOVA analysis on neuronal cell type frequency in lipofection experiments with the Xotx2Mut3 construct. Results of the statistical ANOVA analysis on neuronal cell type frequency in lipofection experiments with the Xotx2Mut3 construct (Figure 3a).
Raw data of the lipofection experiments with the Xotx2Δ and Xotx5bΔ constructs. Raw data of the lipofection experiments with the Xotx2Δ and Xotx5bΔ constructs (summarized in Figure 3f).
Results of the statistical ANOVA analysis on neuronal cell type frequency in lipofection experiments with the Xotx deletion constructs. Results of the statistical ANOVA analysis on neuronal cell type frequency in lipofection experiments with the Xotx deletion constructs (Figure 3f).
Results of the statistical ANOVA analysis on neuronal cell type frequency in lipofection experiments with the otd/Xotx constructs. Results of the statistical ANOVA analysis on neuronal cell type frequency in lipofection experiments with the otd/Xotx constructs (Figure 4b).
Raw data of the lipofection experiments with the otd/box2 and otd/box5b constructs. Raw data of the lipofection experiments with the otd/box2 and otd/box5b constructs (summarized in Figure 4c).
Results of the statistical ANOVA analysis on neuronal cell type frequency in lipofection experiments with the otd/box2 and otd/box5b constructs. Results of the statistical ANOVA analysis on neuronal cell type frequency in lipofection experiments with the otd/box2 and otd/box5b constructs (Figure 4c).
Raw data of the lipofection experiments with the NLS-Myc-otd construct. Raw data of the lipofection experiments with the NLS-Myc-otd construct (summarized in Figure 5c).
Raw data of the rhodopsin promoter transactivation assays. Raw data of the rhodopsin promoter transactivation assays summarized in Figure 6a.
Raw data of the GST-pull down experiments. Raw data of the GST-pull down experiments summarized in Figure 6b, c.
Acknowledgments
Acknowledgements
We thank Massimiliano Andreazzoli, Simona Casarosa, Luciana Dente and Bill Harris for comments on the manuscript; Massimiliano Andreazzoli, Luciana Dente, Daniel Kessler, Barry Knox, Paul Krieg, Antonio Simeone and Andrea Viczian for plasmids; Elena Landi, Manuela Barilari and Marco Mainardi for advice on the biochemical work; Elisa Zabogli and Mauro Pistello for help with FACS. We thank Donatella De Matienzo and Marzia Fabbri for great technical assistance, and Salvatore Di Maria for frog care. This work was supported by EEC grant QLRT-2000-01460, Contributo Ministero Affari Esteri Cooperazione Italia-Cina RP (NFNS 30370453), Telethon Grant GPP04268, Cofinanziamento PRIN-Università di Pisa, FIRB Neuroscienze, Centro di Eccellenza AmbiSEN to GB; MO is a recipient of a Borsa di Perfezionamento della Scuola Normale Superiore di Pisa.
Contributor Information
Marco Onorati, Email: m.onorati@sns.it.
Federico Cremisi, Email: fcremisi@biologia.unipi.it.
Yang Liu, Email: yang_liu@dfci.harvard.edu.
Rong-Qiao He, Email: herq@sun5.ibp.ac.cn.
Giuseppina Barsacchi, Email: gbarsacchi@biologia.unipi.it.
Robert Vignali, Email: rvignali@biologia.unipi.it.
References
- Turner DL, Cepko CL. A common progenitor for neurons and glia persists in rat retina late in development. Nature. 1987;328:131–136. doi: 10.1038/328131a0. [DOI] [PubMed] [Google Scholar]
- Wetts R, Fraser SE. Multipotent precursors can give rise to all major cell types of the frog retina. Science. 1988;239:1142–1145. doi: 10.1126/science.2449732. [DOI] [PubMed] [Google Scholar]
- Harris WA. Cellular diversification in the vertebrate retina. Curr Opin Genet Dev. 1997;7:651–658. doi: 10.1016/S0959-437X(97)80013-5. [DOI] [PubMed] [Google Scholar]
- Livesey FJ, Cepko CL. Vertebrate neural cell-fate determination: lessons from the retina. Nat Rev Neurosci. 2001;2:109–118. doi: 10.1038/35053522. [DOI] [PubMed] [Google Scholar]
- Brown NL, Patel S, Brzezinski J, Glaser T. Math5 is required for retinal ganglion cell and optic nerve formation. Development. 2001;128:2497–2508. doi: 10.1242/dev.128.13.2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatakeyama J, Tomita K, Inoue T, Kageyama R. Roles of homeobox and bHLH genes in specification of a retinal cell type. Development. 2001;128:1313–1322. doi: 10.1242/dev.128.8.1313. [DOI] [PubMed] [Google Scholar]
- Kay JN, Finger-Baier KC, Roeser T, Staub W, Baier H. Retinal ganglion cell genesis requires lakritz, a zebrafish atonal homolog. Neuron. 2001;30:725–736. doi: 10.1016/S0896-6273(01)00312-9. [DOI] [PubMed] [Google Scholar]
- Marquardt T, Ashery-Padan R, Andrejewski N, Scardigli R, Guillemot F, Gruss P. Pax6 is required for the multipotent state of retinal progenitor cells. Cell. 2001;105:43–55. doi: 10.1016/S0092-8674(01)00295-1. [DOI] [PubMed] [Google Scholar]
- Inoue T, Hojo M, Bessho Y, Tano Y, Lee JE, Kageyama R. Math3 and NeuroD regulate amacrine cell fate specification in the retina. Development. 2002;129:831–842. doi: 10.1242/dev.129.4.831. [DOI] [PubMed] [Google Scholar]
- Moore KB, Schneider ML, Vetter ML. Posttranslational mechanisms control the timing of bHLH function and regulate retinal cell fate. Neuron. 2002;34:183–195. doi: 10.1016/S0896-6273(02)00666-9. [DOI] [PubMed] [Google Scholar]
- Dyer MA, Livesey FJ, Cepko CL, Oliver G. Prox1 function controls progenitor cell proliferation and horizontal cell genesis in the mammalian retina. Nat Genet. 2003;34:53–58. doi: 10.1038/ng1144. [DOI] [PubMed] [Google Scholar]
- Viczian AS, Vignali R, Zuber ME, Barsacchi G, Harris WA. XOtx5b and XOtx2 regulate photoreceptor and bipolar fates in the Xenopus retina. Development. 2003;130:1281–1294. doi: 10.1242/dev.00343. [DOI] [PubMed] [Google Scholar]
- Chow RL, Volgyi B, Szilard RK, Ng D, McKerlie C, Bloomfield SA, Birch DG, McInnes RR. Control of late off-center cone bipolar cell differentiation and visual signaling by the homeobox gene Vsx1. Proc Natl Acad Sci USA. 2004;101:1754–1759. doi: 10.1073/pnas.0306520101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramblett DE, Pennesi ME, Wu SM, Tsai MJ. The transcription factor Bhlhb4 is required for rod bipolar cell maturation. Neuron. 2004;43:779–793. doi: 10.1016/j.neuron.2004.08.032. [DOI] [PubMed] [Google Scholar]
- Ohtoshi A, Wang SW, Maeda H, Saszik SM, Frishman LJ, Klein WH, Behringer RR. Regulation of retinal cone bipolar cell differentiation and photopic vision by the CVC homeobox gene Vsx1. Curr Biol. 2004;14:530–536. doi: 10.1016/j.cub.2004.02.027. [DOI] [PubMed] [Google Scholar]
- Poggi L, Vottari T, Barsacchi G, Wittbrodt J, Vignali R. The homeobox gene Xbh1 cooperates with proneural genes to specify ganglion cell fate within the Xenopus neural retina. Development. 2004;131:2305–2315. doi: 10.1242/dev.01099. [DOI] [PubMed] [Google Scholar]
- Wang JC, Harris WA. The role of combinational coding by homeodomain and bHLH transcription factors in retinal cell fate specification. Dev Biol. 2005;285:101–115. doi: 10.1016/j.ydbio.2005.05.041. [DOI] [PubMed] [Google Scholar]
- Hatakeyama J, Kageyama R. Retinal cell fate determination and bHLH factors. Semin Cell Dev Biol. 2004;15:83–89. doi: 10.1016/j.semcdb.2003.09.005. [DOI] [PubMed] [Google Scholar]
- Mu X, Klein WH. A gene regulatory hierarchy for retinal ganglion cell specification and differentiation. Semin Cell Dev Biol. 2004;15:115–123. doi: 10.1016/j.semcdb.2003.09.009. [DOI] [PubMed] [Google Scholar]
- Cohen SM, Jurgens G. Mediation of Drosophila head development by gap-like segmentation genes. Nature. 1990;346:482–485. doi: 10.1038/346482a0. [DOI] [PubMed] [Google Scholar]
- Finkelstein R, Smouse D, Capaci TM, Spradling AC, Perrimon N. The orthodenticle gene encodes a novel homeo domain protein involved in the development of the Drosophila nervous system and ocellar visual structures. Genes Dev. 1990;4:1516–1527. doi: 10.1101/gad.4.9.1516. [DOI] [PubMed] [Google Scholar]
- Simeone A, Acampora D, Gulisano M, Stornaiuolo A, Boncinelli E. Nested expression domains of four homeobox genes in developing rostral brain. Nature. 1992;358:687–690. doi: 10.1038/358687a0. [DOI] [PubMed] [Google Scholar]
- Acampora D, Mazan S, Lallemand Y, Avantaggiato V, Maury M, Simeone A, Brulet P. Forebrain and midbrain regions are deleted in Otx2-/- mutants due to a defective anterior neuroectoderm specification during gastrulation. Development. 1995;121:3279–3290. doi: 10.1242/dev.121.10.3279. [DOI] [PubMed] [Google Scholar]
- Acampora D, Mazan S, Avantaggiato V, Barone P, Tuorto F, Lallemand Y, Brulet P, Simeone A. Epilepsy and brain abnormalities in mice lacking the Otx1 gene. Nat Genet. 1996;14:218–222. doi: 10.1038/ng1096-218. [DOI] [PubMed] [Google Scholar]
- Ang SL, Jin O, Rhinn M, Daigle N, Stevenson L, Rossant J. A targeted mouse Otx2 mutation leads to severe defects in gastrulation and formation of axial mesoderm and to deletion of rostral brain. Development. 1996;122:243–252. doi: 10.1242/dev.122.1.243. [DOI] [PubMed] [Google Scholar]
- Matsuo I, Kuratani S, Kimura C, Takeda N, Aizawa S. Mouse Otx2 functions in the formation and patterning of rostral head. Genes Dev. 1995;9:2646–2658. doi: 10.1101/gad.9.21.2646. [DOI] [PubMed] [Google Scholar]
- Acampora D, Avantaggiato V, Tuorto F, Barone P, Reichert H, Finkelstein R, Simeone A. Murine Otx1 and Drosophila otd genes share conserved genetic functions required in invertebrate and vertebrate brain development. Development. 1998;125:1691–1702. doi: 10.1242/dev.125.9.1691. [DOI] [PubMed] [Google Scholar]
- Acampora D, Boyl PP, Signore M, Martinez-Barbera JP, Ilengo C, Puelles E, Annino A, Reichert H, Corte G, Simeone A. OTD/OTX2 functional equivalence depends on 5' and 3' UTR-mediated control of Otx2 mRNA for nucleo-cytoplasmic export and epiblast-restricted translation. Development. 2001;128:4801–4813. doi: 10.1242/dev.128.23.4801. [DOI] [PubMed] [Google Scholar]
- Leuzinger S, Hirth F, Gerlich D, Acampora D, Simeone A, Gehring WJ, Finkelstein R, Furukubo-Tokunaga K, Reichert H. Equivalence of the fly orthodenticle gene and the human OTX genes in embryonic brain development of Drosophila. Development. 1998;125:1703–1710. doi: 10.1242/dev.125.9.1703. [DOI] [PubMed] [Google Scholar]
- Nagao T, Leuzinger S, Acampora D, Simeone A, Finkelstein R, Reichert H, Furukubo-Tokunaga K. Developmental rescue of Drosophila cephalic defects by the human Otx genes. Proc Natl Acad Sci USA. 1998;95:3737–3742. doi: 10.1073/pnas.95.7.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acampora D, Annino A, Puelles E, Alfano I, Tuorto F, Simeone A. OTX1 compensates for OTX2 requirement in regionalisation of anterior neuroectoderm. Gene Expr Patterns. 2003;3:497–501. doi: 10.1016/S1567-133X(03)00056-5. [DOI] [PubMed] [Google Scholar]
- Freund CL, Gregory-Evans CY, Furukawa T, Papaioannou M, Looser J, Ploder L, Bellingham J, Ng D, Herbrick JA, Duncan A, et al. Cone-rod dystrophy due to mutations in a novel photoreceptor-specific homeobox gene (CRX) essential for maintenance of the photoreceptor. Cell. 1997;91:543–553. doi: 10.1016/S0092-8674(00)80440-7. [DOI] [PubMed] [Google Scholar]
- Furukawa T, Morrow EM, Cepko CL. Crx, a novel otx-like homeobox gene, shows photoreceptor-specific expression and regulates photoreceptor differentiation. Cell. 1997;91:531–541. doi: 10.1016/S0092-8674(00)80439-0. [DOI] [PubMed] [Google Scholar]
- Chen S, Wang QL, Nie Z, Sun H, Lennon G, Copeland NG, Gilbert DJ, Jenkins NA, Zack DJ. Crx, a novel Otx-like paired-homeodomain protein, binds to and transactivates photoreceptor cell-specific genes. Neuron. 1997;19:1017–1030. doi: 10.1016/S0896-6273(00)80394-3. [DOI] [PubMed] [Google Scholar]
- Mears AJ, Kondo M, Swain PK, Takada Y, Bush RA, Saunders TL, Sieving PA, Swaroop A. Nrl is required for rod photoreceptor development. Nat Genet. 2001;29:447–452. doi: 10.1038/ng774. [DOI] [PubMed] [Google Scholar]
- Mitton KP, Swain PK, Chen S, Xu S, Zack DJ, Swaroop A. The leucine zipper of NRL interacts with the CRX homeodomain. A possible mechanism of transcriptional synergy in rhodopsin regulation. J Biol Chem. 2000;275:29794–29799. doi: 10.1074/jbc.M003658200. [DOI] [PubMed] [Google Scholar]
- Peng GH, Chen S. Chromatin immunoprecipitation identifies photoreceptor transcription factor targets in mouse models of retinal degeneration: new findings and challenges. Vis Neurosci. 2005;22:575–586. doi: 10.1017/S0952523805225063. [DOI] [PubMed] [Google Scholar]
- Freund CL, Wang QL, Chen S, Muskat BL, Wiles CD, Sheffield VC, Jacobson SG, McInnes RR, Zack DJ, Stone EM. De novo mutations in the CRX homeobox gene associated with Leber congenital amaurosis. Nat Genet. 1998;18:311–312. doi: 10.1038/ng0498-311. [DOI] [PubMed] [Google Scholar]
- Swain PK, Chen S, Wang QL, Affatigato LM, Coats CL, Brady KD, Fishman GA, Jacobson SG, Swaroop A, Stone E, et al. Mutations in the cone-rod homeobox gene are associated with the cone-rod dystrophy photoreceptor degeneration. Neuron. 1997;19:1329–1336. doi: 10.1016/S0896-6273(00)80423-7. [DOI] [PubMed] [Google Scholar]
- Sohocki MM, Sullivan LS, Mintz-Hittner HA, Birch D, Heckenlively JR, Freund CL, McInnes RR, Daiger SP. A range of clinical phenotypes associated with mutations in CRX, a photoreceptor transcription-factor gene. Am J Hum Genet. 1998;63:1307–1315. doi: 10.1086/302101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzekov RT, Liu Y, Sohocki MM, Zack DJ, Daiger SP, Heckenlively JR, Birch DG. Autosomal dominant retinal degeneration and bone loss in patients with a 12-bp deletion in the CRX gene. Invest Ophthalmol Vis Sci. 2001;42:1319–1327. [PMC free article] [PubMed] [Google Scholar]
- Cremers FP, van den Hurk JA, den Hollander AI. Molecular genetics of Leber congenital amaurosis. Hum Mol Genet. 2002;11:1169–1176. doi: 10.1093/hmg/11.10.1169. [DOI] [PubMed] [Google Scholar]
- Chen S, Wang QL, Xu S, Liu I, Li LY, Wang Y, Zack DJ. Functional analysis of cone-rod homeobox (CRX) mutations associated with retinal dystrophy. Hum Mol Genet. 2002;11:873–884. doi: 10.1093/hmg/11.8.873. [DOI] [PubMed] [Google Scholar]
- Furukawa T, Morrow EM, Li T, Davis FC, Cepko CL. Retinopathy and attenuated circadian entrainment in Crx-deficient mice. Nat Genet. 1999;23:466–470. doi: 10.1038/70591. [DOI] [PubMed] [Google Scholar]
- Morrow EM, Furukawa T, Raviola E, Cepko CL. Synaptogenesis and outer segment formation are perturbed in the neural retina of Crx mutant mice. BMC Neurosci. 2005;6:5. doi: 10.1186/1471-2202-6-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida A, Furukawa A, Koike C, Tano Y, Aizawa S, Matsuo I, Furukawa T. Otx2 homeobox gene controls retinal photoreceptor cell fate and pineal gland development. Nat Neurosci. 2003;6:1255–1263. doi: 10.1038/nn1155. [DOI] [PubMed] [Google Scholar]
- Decembrini S, Andreazzoli M, Vignali R, Barsacchi G, Cremisi F. Timing the generation of distinct retinal cells by homeobox proteins. PLoS Biol. 2006;4:e272. doi: 10.1371/journal.pbio.0040272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Autilia S, Decembrini S, Casarosa S, He R-Q, Barsacchi G, Cremisi F, Andreazzoli M. Cloning and developmental expression of the Xenopus homeobox gene Xvsx1. Dev Gen Evol. 2006;216:1562–71. doi: 10.1007/s00427-006-0109-0. [DOI] [PubMed] [Google Scholar]
- Fei Y, Hughes TE. Nuclear trafficking of photoreceptor protein crx: the targeting sequence and pathologic implications. Invest Ophthalmol Vis Sci. 2000;41:2849–2856. [PubMed] [Google Scholar]
- Whitaker SL, Knox BE. Conserved transcriptional activators of the Xenopus rhodopsin gene. J Biol Chem. 2004;279:49010–49018. doi: 10.1074/jbc.M406080200. [DOI] [PubMed] [Google Scholar]
- Chau KY, Chen S, Zack DJ, Ono SJ. Functional domains of the cone-rod homeobox (CRX) transcription factor. J Biol Chem. 2000;275:37264–37270. doi: 10.1074/jbc.M002763200. [DOI] [PubMed] [Google Scholar]
- http://rsb.info.nih.gov/ij/
- Briata P, Ilengo C, Bobola N, Corte G. Binding properties of the human homeodomain protein OTX2 to a DNA target sequence. FEBS Lett. 1999;445:160–164. doi: 10.1016/S0014-5793(99)00113-1. [DOI] [PubMed] [Google Scholar]
- Wilson DS, Sheng G, Jun S, Desplan C. Conservation and diversification in homeodomain-DNA interactions: a comparative genetic analysis. Proc Natl Acad Sci USA. 1996;93:6886–6891. doi: 10.1073/pnas.93.14.6886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahaffey JW. Assisting Hox proteins in controlling body form: are there new lessons from flies (and mammals)? Curr Opin Genet Dev. 2005;15:422–429. doi: 10.1016/j.gde.2005.06.009. [DOI] [PubMed] [Google Scholar]
- Moens CB, Selleri L. Hox cofactors in vertebrate development. Dev Biol. 2006;291:193–206. doi: 10.1016/j.ydbio.2005.10.032. [DOI] [PubMed] [Google Scholar]
- Casarosa S, Amato MA, Andreazzoli M, Gestri G, Barsacchi G, Cremisi F. Xrx1 controls proliferation and multipotency of retinal progenitors. Mol Cell Neurosci. 2003;22:25–36. doi: 10.1016/S1044-7431(02)00025-8. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Fernandez F, Kittredge KL, Rayborn ME, Hollyfield JG, Landers RA, Saha M, Grainger RM. Interphotoreceptor retinoid-binding protein (IRBP), a major 124 kDa glycoprotein in the interphotoreceptor matrix of Xenopus laevis. Characterization, molecular cloning and biosynthesis. J Cell Sci. 1993;105:7–21. doi: 10.1242/jcs.105.1.7. [DOI] [PubMed] [Google Scholar]
- Gerber WV, Yatskievych TA, Antin PB, Correia KM, Conlon RA, Krieg PA. The RNA-binding protein gene, hermes, is expressed at high levels in the developing heart. Mech Dev. 1999;80:77–86. doi: 10.1016/S0925-4773(98)00195-6. [DOI] [PubMed] [Google Scholar]
- Soboleski MR, Oaks J, Halford WP. Green fluorescent protein is a quantitative reporter of gene expression in individual eukaryotic cells. Faseb J. 2005;19:440–442. doi: 10.1096/fj.04-3180fje. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Raw data of the lipofection experiments with the Xotx5bMut1, Xotx5bMut2, Xotx5bMut3, otd and otd/Xotx constructs. Raw data of the lipofection experiments with the Xotx5bMut1, Xotx5bMut2, and Xotx5bMut3 constructs (summarized in Figure 2e), with the otd construct (summarized in Figure 4a) and with the otd/Xotx constructs (summarized in Figure 4b).
Results of the statistical ANOVA analysis on neuronal cell type frequency in lipofection experiments with Xotx5bMut1, Xotx5bMut2, and Xotx5bMut3 constructs. Results of the statistical ANOVA analysis on neuronal cell type frequency in lipofection experiments with Xotx5bMut1, Xotx5bMut2, and Xotx5bMut3 constructs (Figure 2e).
Expression of retinal markers used in this study to confirm neuron cell identities of lipofected cells. Lipofected GFP-positive cells (green) can be identified by using specific probes (all detected with Fast Red): (a) an IRBP probe identifies photoreceptors by in situ hybridization; (b) in situ hybridization with Xprox1 identifies horizontal cells (red); (c) in situ hybridization with Xvsx1 identifies bipolar cells (red); (d) a specific antibody identifies GABAergic amacrine cells (red); (e) a specific antibody identifies 5-HT positive amacrine cells (red); (f) a specific antibody identifies tyrosine hydroxylase (TH)-positive amacrine cells (red); (g) in situ hybridization with Xhermes identifies ganglion cells. Hoechst staining (in blue) identifies cell nuclei.
Raw data of the lipofection experiments with the Xotx2Mut3 construct. Raw data of the lipofection experiments with the Xotx2Mut3 construct (summarized in Figure 3a).
Results of the statistical ANOVA analysis on neuronal cell type frequency in lipofection experiments with the Xotx2Mut3 construct. Results of the statistical ANOVA analysis on neuronal cell type frequency in lipofection experiments with the Xotx2Mut3 construct (Figure 3a).
Raw data of the lipofection experiments with the Xotx2Δ and Xotx5bΔ constructs. Raw data of the lipofection experiments with the Xotx2Δ and Xotx5bΔ constructs (summarized in Figure 3f).
Results of the statistical ANOVA analysis on neuronal cell type frequency in lipofection experiments with the Xotx deletion constructs. Results of the statistical ANOVA analysis on neuronal cell type frequency in lipofection experiments with the Xotx deletion constructs (Figure 3f).
Results of the statistical ANOVA analysis on neuronal cell type frequency in lipofection experiments with the otd/Xotx constructs. Results of the statistical ANOVA analysis on neuronal cell type frequency in lipofection experiments with the otd/Xotx constructs (Figure 4b).
Raw data of the lipofection experiments with the otd/box2 and otd/box5b constructs. Raw data of the lipofection experiments with the otd/box2 and otd/box5b constructs (summarized in Figure 4c).
Results of the statistical ANOVA analysis on neuronal cell type frequency in lipofection experiments with the otd/box2 and otd/box5b constructs. Results of the statistical ANOVA analysis on neuronal cell type frequency in lipofection experiments with the otd/box2 and otd/box5b constructs (Figure 4c).
Raw data of the lipofection experiments with the NLS-Myc-otd construct. Raw data of the lipofection experiments with the NLS-Myc-otd construct (summarized in Figure 5c).
Raw data of the rhodopsin promoter transactivation assays. Raw data of the rhodopsin promoter transactivation assays summarized in Figure 6a.
Raw data of the GST-pull down experiments. Raw data of the GST-pull down experiments summarized in Figure 6b, c.