Abstract
Background
Glucocorticoids (GCs) are widely used anti-inflammatory drugs. While useful in clinical practice, patients taking GCs often suffer from skeletal side effects including growth retardation in children and adolescents, and decreased bone quality in adults. On a physiological level, GCs have been implicated in the regulation of chondrogenesis and osteoblast differentiation, as well as maintaining homeostasis in cartilage and bone. We identified the glucocorticoid receptor (GR) as a potential regulator of chondrocyte hypertrophy in a microarray screen of primary limb bud mesenchyme micromass cultures. Some targets of GC regulation in chondrogenesis are known, but the global effects of pharmacological GC doses on chondrocyte gene expression have not been comprehensively evaluated.
Results
This study systematically identifies a spectrum of GC target genes in embryonic growth plate chondrocytes treated with a synthetic GR agonist, dexamethasone (DEX), at 6 and 24 hrs. Conventional analysis of this data set and gene set enrichment analysis (GSEA) was performed. Transcripts associated with metabolism were enriched in the DEX condition along with extracellular matrix genes. In contrast, a subset of growth factors and cytokines were negatively correlated with DEX treatment. Comparing DEX-induced gene expression data to developmental changes in gene expression in micromass cultures revealed an additional layer of complexity in which DEX maintains the expression of certain chondrocyte marker genes while inhibiting factors that promote vascularization and ultimately ossification of the cartilaginous template.
Conclusion
Together, these results provide insight into the mechanisms and major molecular classes functioning downstream of DEX in primary chondrocytes. In addition, comparison of our data with microarray studies of DEX treatment in other cell types demonstrated that the majority of DEX effects are tissue-specific. This study provides novel insights into the effects of pharmacological GC on chondrocyte gene transcription and establishes the foundation for subsequent functional studies.
Background
Cartilage provides a scaffold for the deposition of osteoblast precursors and ultimately the development of long bones. This process, termed endochondral ossification, describes a coordinated developmental series that involves commitment of mesenchymal precursor cells to the chondrogenic lineage and subsequent alternating phases of proliferation and differentiation, which culminate in the replacement of the cartilage by bone tissue [1-4]. In the first phase of this process, multipotent mesenchymal progenitors condense and initiate expression of the pro-chondrogenic Sox family members 9, 5 and 6 [5,6]. A subset of cells at the center of these aggregates differentiates into chondrocytes. Newly formed chondrocytes secrete an extracellular matrix rich in type II collagen (Col2a1), proliferate and ultimately terminally differentiate into hypertrophic chondrocytes [7]. Chondrocyte hypertrophy precedes the end of the chondrocyte life cycle by apoptosis and is accompanied by vascularization of the hypertrophic template and mineralization of the cartilaginous extracellular matrix [8-12]. Concomitantly, osteoclasts degrade the calcified cartilage extracellular matrix, making way for the invasion and deposition of an osteoprogenitor population that form the primary ossification center [13].
These events take place in a region called the growth plate that illustrates the organization of different phases of cartilage development into distinct zones. The resting zone delineates newly differentiated chondrocytes with low mitotic activity and the cellular reserve for subsequent stages of chondrocyte differentiation. Proliferative zone chondrocytes exhibit higher mitotic activity resulting in distinct columns containing cells reminiscent of stacked coins. The hypertrophic zone demarcates terminally differentiated chondrocytes which are identified by high cytoplasm to nuclear ratio and the expression of type X collagen (Col10a1) [14-16]. Terminally differentiated chondrocytes are fated for programmed cell death after which primary ossification occurs by way of vascularization of the remaining cartilaginous matrix and the deposition of osteoprogenitor cells [17-19].
Glucocorticoids (GC) are among various endocrine molecules including growth hormone (GH) and thyroid hormone (TH) known to regulate linear growth [20-23]. Regulation of linear growth follows the paradigm in which steroid hormones affect target tissue through both local and systemic mechanisms [24-27]. Indirect effects occur through modulation of other endocrine systems such as the GH/IGF-I axis. Generally, GC decrease IGF-I, GH receptor and IGF receptor 1 expression and also abrogate the release of GH from the pituitary [20,28,29]. Direct regulation of growth occurs through GC receptor (GR)-mediated gene transcription in chondrocytes [24,30,31].
GC functions are primarily mediated by the glucocorticoid receptor (GR) that is encoded by the Nr3c1 gene. The GR is ubiquitously expressed in mammalian tissues, including the growth plate, and is essential for life [31-36]. Many studies have examined GC regulation of the skeleton and have led to various theories on potential modes of GC function in cartilage [37-40]. The specific function of the receptor in terms of its transcriptional regulation in cartilage, however, remains enigmatic.
While endogenous GCs have been shown to promote the differentiation of both chondrocytes and osteoblasts, exogenous GCs in pharmacological doses which are also widely used in clinical practice to treat inflammatory disorders [41-46]. Their have different effects. Indeed, their utility in treating various diseases is, however, limited by numerous side effects such as growth failure and decreased bone quality [47]. GC-target genes including C-type natriuretic peptide and VEGF have been identified in chondrocytes [28,48,49]; however, the cartilage-specific transcriptional consequences of high-GC-doses in the growth plate have not been studied comprehensively.
Work in our laboratory identified GR amongst factors that were up-regulated during chondrocyte maturation [50] Thus, to comprehensively understand the transcriptional effects of pharmacological GC doses in growth plate, we completed a genomic screen of gene expression changes in chondrocytes derived from E15.5 day old mouse embryos. Primary monolayer chondrocytes were treated with a synthetic GC, dexamethasone (DEX), and RNA was isolated for microarray analysis. We complemented traditional microarray analysis methods with the gene set enrichment algorithm to correlate the behaviour of specific molecular classes with DEX treatment [51,52].
Results and Discussion
Microarray screen of dexamethasone-treated primary chondrocyte monolayers
We identified the GR as a candidate for the regulation of chondrocyte hypertrophy in a previous expression profiling screen using primary micromass cultures [50]. The Nr3c1 probe set which encodes the GR was up-regulated 4-fold from day 3 to day 15 of micromass culture (Figure 1A, top panel). Confirmation of the GR expression profile with qRT-PCR showed an approximately 8-fold increase over the same time course (Figure 1A, bottom panel). Studies in our laboratory and others have implicated GCs in chondrocyte differentiation and growth plate function [25,26,47,48,53,54]. In addition, our cell counting experiments revealed that DEX consistently decreases cell numbers after 24 hrs (Figure 1B), in agreement with other studies that show increased apoptosis [38,55] and reduced proliferation [56] in response to GCs. We therefore aimed at extending this analysis to examine pharmacological effects of GCs on growth plate chondrocytes by systematically identifying downstream effector genes of DEX. Primary chondrocytes derived from the long bones of 15.5 day old embryonic mice were treated with DEX or the vehicle control, and total RNA was isolated after 6 and 24 hrs of culture, respectively.
Figure 1.
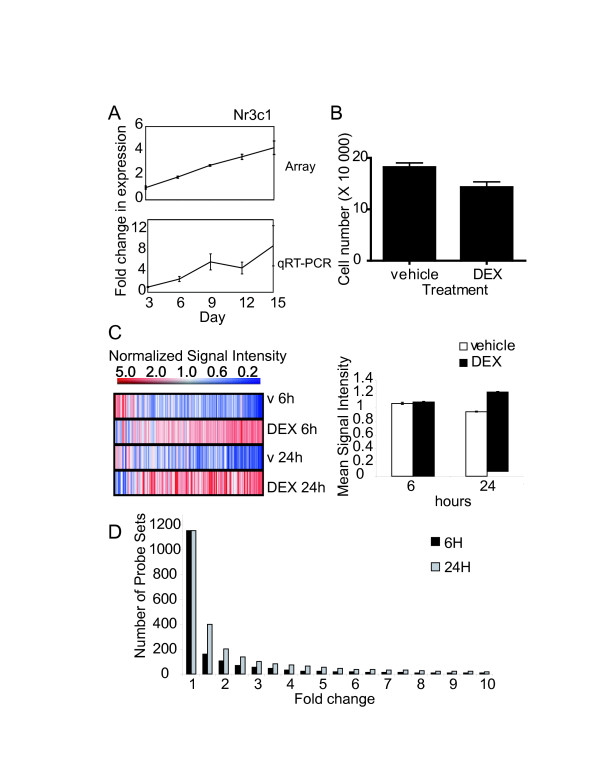
Gene expression changes in DEX-treated primary chondrocytes. Microarray and quantitative RT-PCR expression profiles of the Glucocorticoid receptor (Nr3c1) in primary mesenchymal micromass cultures (A). Primary chondrocytes are plated in high density monolayers and treated with DEX or vehicle for 24 hrs and counted with a hemocytometer (B). Ordered list of global microarray data set derived from the hybridization of RNA isolated from primary chondrocytes treated with 10-7 M DEX and the vehicle (v) control (C, left panel). One-Way ANOVA testing for significantly expressed probe sets between DEX-treated samples and the vehicle control resulted in a list of 1158 transcripts. Mean normalized signal intensities for all 1158 probe sets are shown (C, right panel). Fold change filtering of these transcripts reveal that the majority of probe sets vary in the range of 1 to 2-fold (D).
Gene expression was evaluated using Affymetrix MOE 430 2.0 mouse genome chips using three independent cell isolations. We first analyzed gene expression using conventional analysis functions in GeneSpring GX*. After pre-processing the data set using the GC-RMA algorithm and eliminating probe sets showing expression levels close to background, 22 091 probe sets remained, reducing the data set by 48% (Table 1). Significance testing with one-Way ANOVA analysis identified probe sets differentially expressed between DEX and vehicle-treated cultures over the entire time course (Figure 1C, left panel). The resulting list contained 1158 probe sets, which is 2% of the data set's original size. Approximately 70% of significantly changed probe sets exhibited upregulation in response to DEX treatment. This data set was further subdivided by using 1.5-, 5- and 10- fold change filters which generated lists of 162, 21 and 7 probe sets for the 6 hr time point and 399, 53 and 19 probe sets for the 24 hr time point, respectively (Table 1). Examination of the overall differences between the mean normalized signal intensities associated with each condition showed minimal changes in gene expression (Figure 1C, right panel), indicating that GC treatment affects the expression of only a small subset of all expressed genes in this system. A distribution of fold differences between 6 and 24 hrs showed that the majority of gene expression changes did not exceed 2-fold (Figure 1D). In each case, both time points exhibited the same overall trends in gene expression, but, as expected, the 24 hr time point consistently showed a higher proportion of probe sets altered by DEX treatment.
Table 1.
Microarray analysis of DEX-treated primary chondrocyte monolayers.
| Specifications | Probe sets at 6 hrs | Probe sets at 24 hrs |
| Total number of probe sets | 45101 | 45101 |
| Significantly expressed | 22091 | 22091 |
| Differentially expressed | 1158 | 1158 |
| 1.5-fold changed | 162 | 399 |
| 5-fold changed | 21 | 53 |
| 10-fold changed | 7 | 33 |
| 1.5-fold up-regulated | 141 | 342 |
| 5-fold up-regulated | 20 | 50 |
| 10-fold up-regulated | 7 | 19 |
| 1.5-fold down-regulated | 21 | 57 |
| 5-fold down-regulated | 1 | 3 |
| 10-fold down-regulated | 0 | 0 |
Probe set validation
To confirm the accuracy of the microarrays in identifying biologically significant differences, we selected a variety of expressed transcripts for qRT-PCR analysis (Figure 2A). Transcripts that either belonged to a functional class implicated in cartilage development or exhibited marked changes with DEX treatment were chosen. Markers exhibiting marginal changes in gene expression were also selected for control purposes. Specifically, we evaluated the expression patterns of Indian hedgehog (Ihh), Tissue inhibitor of matrix metalloproteinase 4 (Timp4), Cyclin-dependent kinase inhibitor 1C (Cdkn1c), which contains a GC response element in its promoter [57], Integrin beta like 1 protein (Itgbl1), GC receptor (Nr3c1), Integrin beta 1 (Itgb1) and Kruppel-like factor 15 (Klf15) over 0, 6, 12, and 24 hrs of culture with or without DEX treatment. Transcripts for Klf15 were up-regulated from 0 to 6 hrs while Ihh, Timp4, Cdkn1c and Itgbl1 all increased after the 6 hr time point. Nr3c1, which encodes the GR, was not affected by DEX-treatment at both 6 and 24 hrs, but does contain a putative GRE [58]. Transcripts such as Itgb1 that exhibited less than 1.5-fold change in our arrays were also confirmed with qRT-PCR, providing further evidence that the microarray data represented authentic gene expression data. Interestingly, the fold change difference varied according to the experimental method. In cases such as Timp4 and to a lesser extent Cdkn1c, qtPCR data showed higher fold change increases with the DEX treatment than in microarrays. In contrast, the expression pattern for Klf15 exhibits a higher fold-change difference in the microarrays compared to the control. While data normalization using the RMA algorithm provides excellent estimates of reliable signal intensities, other methods such as the M.A.S. 5.0 algorithm are known to outperform RMA in its ability to accurately estimate fold change differences in transcript levels [59].
Figure 2.
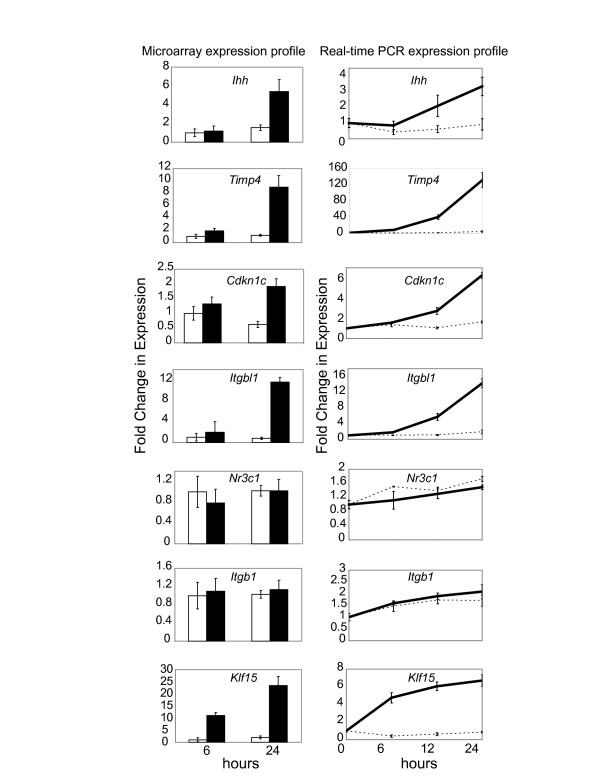
Identification of significantly expressed probe sets and subsequent validation with real-time RT-PCR. Expression profiles for selected transcripts in vehicle- or DEX-treated chondrocytes are confirmed with real-time RT-PCR at 0, 6, 12 and 24 hr time points. Indian hedgehog (Ihh), tissue inhibitor of matrix metalloproteinase 4 (Timp4), cyclin-dependent kinase inhibitor 1C (Cdkn1c, p57), integrin beta like 1 protein (Itgbl1), glucocorticoid receptor (Nr3c1), integrin beta 1 (Itgb1) and kruppel-like factor 15 (Klf15) microarray data are shown on the left at the 6 and 24 hr time points and corresponding real-time expression values are shown on the right. P-values less than 0.01 are deemed significant. Specifically, Ihh, Timp4, Itgbl1 and Klf15 exhibit significant differences between the 6 and 24 hr time point and between treatments. Dotted lines indicate the control and solid lines denote DEX treatment.
GSEA to identify the effects of dexamethasone on gene expression in chondrocytes
Traditional microarray analysis methods are useful for the identification of probe sets exhibiting transcriptional responses to DEX-treatment, but are limited in certain capacities. Alternate statistical methods such as ANOVA testing produced transcript lists that, while effectively reducing the dimensionality or sample size of the data set, increased the rate of false negative data thus hampering our ability to generate meaningful hypotheses from the data (Figure 1). Also, the overall effect of DEX treatment on gene expression was modest, which may have reduced the significance of biologically relevant genes because their signal intensities were close to background levels. Accordingly, we did not have a clear concept of the central pathways and biological categories affected by DEX treatment. Similarly, Gene Ontology annotations were not sufficiently robust to detect differences in the representation of specific molecular categories (data not shown). We therefore implemented GSEA [52], an algorithm that is designed to effectively evaluate the effect of a specific experimental condition on known biological pathways and functional categories. These analyses show whether a given treatment (e.g. DEX stimulation) results in enrichment of genes sets involved in the regulation of a specific phenotype (see materials and methods for details).
We created a gene set consisting of 77 gene lists representing different tissue types, functional categories and pathways derived from other microarray studies in the literature (Table 2). We drew conclusions from the top gene sets that had a false discovery rate (FDR) less than 25% and a p-value less than 0.001, both of which are acceptable cut-offs for the identification of biologically relevant probe sets. This cut-off, although relatively high, was optimized to reduce the occurrence of false negative data in data sets interrogating a small number of gene sets. Additionally, the FDR compensates for the inherent lack of coherence microarray data sets exhibit between gene expression and specific experimental conditions [52]. Enriched gene sets were identified in both DEX and vehicle data (Table 3). Specifically, the highest statistical confidence and correlation with the DEX phenotype was assigned to metabolism and extracellular matrix, which contained 196 and 228 genes, respectively (Figure 3, left panels, Table 4 and 5). In each case, the expression of genes positively correlated with the DEX phenotype at the 24 hr time point exceeded the number of genes at the 6 hr time point (Figure 3, right panels). Metabolic genes included aldehyde and alcohol dehydrogenases (Table 4), among others, and were identified in accordance with previously documented roles for GC in various metabolic processes and tissues [60,61]. Closer examination of the genes contributing to the enrichment scores for the ECM gene set revealed that Dentin matrix protein 1 (Dmp1) was the top ranking gene (Table 5). DMP1 belongs to the SIBLING family of matrix molecules and has been linked to chondrocyte differentiation. Dmp1 knockout mice display disordered postnatal chondrogenesis, among other skeletal abnormalities [62]. Interestingly, integrin binding sialoprotein (Ibsp) [63-66]), another SIBLING family member, and osteocalcin (Bglap2) both contain putative GRE sequences, but did not contribute to the enrichment score for this category [63,66]. They did, however, belong to the core group of genes that were enriched when a micromass culture gene set was used to interrogate the DEX data (Figure 4).
Table 2.
Gene sets used in GSEA.
| Category name | Number of genes | Category name | Number of genes |
| Adipose | 70 | Nucleus_3 | 510 |
| Apoptosis | 39 | Fkbp | 33 |
| Bone | 116 | 3vs15_1.5x_1 | 497 |
| Cartilage | 28 | 3vs15_1.5x_2 | 497 |
| Catalytic | 245 | 3vs15_1.5x_3 | 497 |
| Chaperone | 81 | 3vs15_1.5x_4 | 497 |
| Chemokine | 31 | 3vs15_1.5x_5 | 76 |
| Chromatin/Hdacs | 24 | Igf | 48 |
| Cyclin | 225 | Cart_2 | 299 |
| Cytokine | 127 | Cart_3 | 352 |
| 1_Dnabind | 500 | Liver_1 | 260 |
| 2_Dnabind | 448 | Liver_2 | 260 |
| Ecm | 228 | Blood | 111 |
| Electron_Transp | 40 | Protease_1 | 269 |
| Gf Receptor | 327 | Protease_2 | 269 |
| Gluconeogen | 31 | Phosphatase | 473 |
| Growth Factor | 106 | Dusp | 20 |
| Gtpase Activator | 46 | Kinase_1 | 499 |
| Gtpase Activ | 73 | Kinase_2 | 499 |
| Heparin Bind | 37 | Kinase_3 | 227 |
| Hormone | 75 | Integrin_Rel | 173 |
| Muscle | 198 | Brain_Rel | 379 |
| Neg_Apoptosis | 50 | Hepatocyte | 19 |
| Oncogene | 154 | Obl_Oclast | 16 |
| Pos_Apoptosis | 79 | Interleukinrelated | 175 |
| Related_Apoptosis | 311 | Rgs_Related | 44 |
| Structure | 151 | Caspase_Related | 47 |
| Sugar_Bind | 104 | Creb_Atf3 | 32 |
| Tf_Activ | 56 | Nuclear Receptor | 138 |
| Tf_Repress | 55 | Nuc_Hormone_Receptor | 55 |
| Tgfb | 45 | Mapkrelated | 267 |
| Tnf_Receptor | 69 | Membrane | 260 |
| Tumor Suppressor | 48 | Metabolism | 196 |
| Wnt | 53 | Nucleus_1 | 494 |
| Actin_Cytoskel | 38 | Nucleus_2 | 494 |
| Angiogen | 57 | Pzhorton.Farnum | 413 |
| Bmprelated | 62 | Hzhorton.Farnum | 407 |
| Cytoplasm | 411 | ||
| Erk_Related | 40 | ||
| Fgf_Related | 64 |
Table 3.
GSEA of DEX-treated primary chondrocytes.
| Gene set name | Size | ES | NES | NOM p-val | FDR q-val |
| Metabolism | 196 | 0.471 | 1.935 | <0.001 | 0.016 |
| Extracellualr Matrix | 228 | 0.451 | 1.878 | <0.001 | 0.016 |
| Fkbp | 33 | 0.559 | 1.696 | 0.011 | 0.054 |
| Integrin_Related | 173 | 0.407 | 1.643 | <0.001 | 0.001 |
| Angiogenesis | 57 | 0.479 | 1.610 | 0.012 | 0.065 |
| Kinase_1 | 499 | 0.343 | 1.549 | <0.001 | 0.092 |
| Tumor Suppressor | 48 | 0.457 | 1.492 | 0.037 | 0.126 |
| Catalytic | 245 | 0.337 | 1.420 | 0.008 | 0.172 |
| Hepatocyte | 19 | 0.529 | 1.406 | 0.104 | 0.161 |
| D3 Vs D15_2 | 497 | 0.304 | 1.368 | 0.004 | 0.194 |
| Igf | 48 | 0.412 | 1.348 | 0.093 | 0.208 |
| Cyclin | 224 | 0.322 | 1.344 | 0.028 | 0.199 |
| Actin_Cytoskel | 38 | 0.426 | 1.325 | 0.124 | 0.213 |
| Structure | 151 | 0.332 | 1.312 | 0.053 | 0.219 |
| Cytoplasm | 411 | 0.292 | 1.300 | 0.023 | 0.224 |
| Adipose | 70 | 0.368 | 1.285 | 0.116 | 0.232 |
| Gtpase Activity | 73 | 0.363 | 1.280 | 0.113 | 0.230 |
| Cartilage | 28 | 0.432 | 1.262 | 0.169 | 0.246 |
| Chemokine | 31 | -0.779 | -2.40 | <0.001 | 0 |
| Cytokine | 127 | -0.579 | -2.31 | <0.001 | 0 |
| Growth Factor | 106 | -0.517 | -2.01 | <0.001 | 7.698E-04 |
| Interleukinrelated | 175 | -0.469 | -1.98 | <0.001 | 9.475E-04 |
| Bone | 16 | -0.577 | -1.51 | 0.051 | 8.945E-02 |
| Creb_Atf3 | 30 | -0.469 | -1.43 | 0.065 | 1.300E-01 |
| Dusp | 20 | -0.508 | -1.40 | 0.102 | 1.418E-01 |
| Blood | 111 | -0.351 | -1.37 | 0.037 | 1.425E-01 |
| 3vs15_1.5x_3 | 496 | -0.288 | -1.35 | 0.002 | 1.518E-01 |
| Protease_2 | 268 | -0.306 | -1.35 | 0.015 | 1.411E-01 |
| Nuc_Hormone_Receptor | 55 | -0.381 | -1.32 | 0.086 | 1.570E-01 |
| Tf_Repress | 55 | -0.380 | -1.32 | 0.091 | 1.498E-01 |
| 3vs15_1.5x_4 | 497 | -0.272 | -1.28 | 0.011 | 1.817E-01 |
| Erk_Related | 40 | -0.385 | -1.25 | 0.157 | 2.169E-01 |
ES, enrichment score
NES, normalized enrichment score
FDR q-val, false discovery rate and multiple testing corrections (q-value)
NOM p-val; the uncorrected p-value
Figure 3.
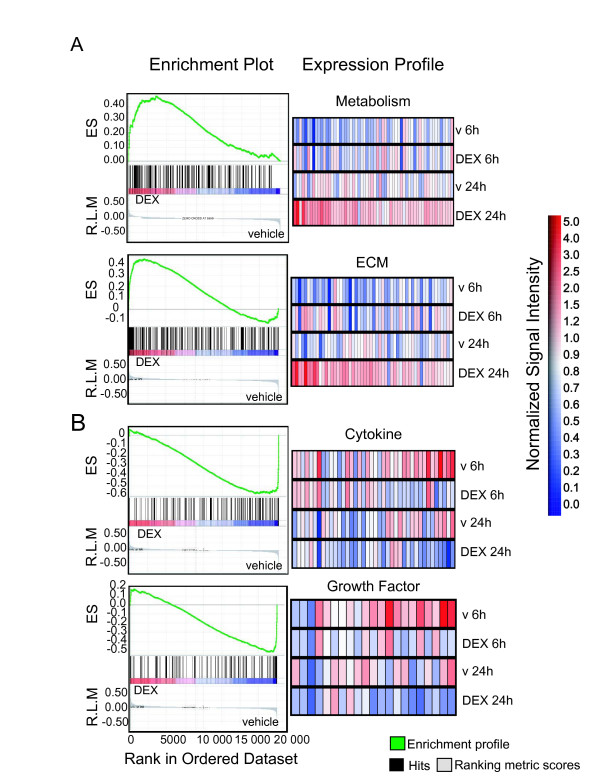
Enrichment plots for statistically significant gene sets identified by GSEA. User-defined gene sets enriched with the DEX or vehicle conditions are depicted. Black bars illustrate the position of probe sets belonging to metabolic, extracellular matrix (A), cytokine and growth factor (B) gene sets in the context of all probes on the DEX array. The running enrichment score (RES) plotted as a function of the position within the ranked list of array probes is shown in green. The ranked list metric shown in gray illustrates the correlation between the signal to noise values of all individually ranked genes according and the class labels (experimental conditions). Metabolic and ECM genes are overrepresented in the left side of the enrichment plot indicating correlation to differential expression in DEX-treated chondrocytes. In contrast, cytokines and growth factor genes are enriched in the right side of the plots and correspond to the vehicle control. Significantly enriched data sets are defined according to GSEA default settings i.e., a p < 0.001 and a false discovery rate (FDR) < 0.25. Individual expression profiles for probe sets contributing to the normalized enrichment score are shown in the right panel. R.L.M = ranked list metric, E.S. = enrichment score.
Table 4.
Metabolic transcripts enriched in DEX-treated chondrocytes. I.
| HUGO symbol | Rank | RMS* | RES** | HUGO symbol | Rank | RMS* | RES** |
| Aldh1a1 | 26 | 0.417 | 0.053 | Slc27a4 | 1616 | 0.058 | 0.426 |
| Eya2 | 40 | 0.355 | 0.099 | Ltbp2 | 1721 | 0.056 | 0.428 |
| Vcl | 106 | 0.228 | 0.125 | Hsd17b1 | 1783 | 0.055 | 0.432 |
| Adhfe1 | 116 | 0.222 | 0.154 | P4ha2 | 1783 | 0.055 | 0.432 |
| Ids | 123 | 0.212 | 0.181 | Mut | 1850 | 0.053 | 0.443 |
| Cbr3 | 133 | 0.204 | 0.207 | Pde3a | 2195 | 0.048 | 0.432 |
| Aldh6a1 | 202 | 0.165 | 0.225 | Sulf2 | 2200 | 0.048 | 0.438 |
| Bcat2 | 224 | 0.157 | 0.245 | Prep | 2316 | 0.046 | 0.438 |
| Pmm1 | 278 | 0.145 | 0.261 | Plod3 | 2387 | 0.045 | 0.441 |
| Pcx | 553 | 0.105 | 0.261 | 1110013G13RIK | 2510 | 0.043 | 0.440 |
| Fthfd | 554 | 0.105 | 0.275 | Pld1 | 2669 | 0.041 | 0.437 |
| Atp1a1 | 560 | 0.104 | 0.288 | Au041707 | 2721 | 0.040 | 0.440 |
| Gstm1 | 619 | 0.099 | 0.298 | Decr1 | 2837 | 0.039 | 0.439 |
| Gstm2 | 742 | 0.088 | 0.303 | Gstm5 | 2872 | 0.038 | 0.443 |
| 1700061G19RIK | 787 | 0.086 | 0.312 | Bckdha | 2932 | 0.038 | 0.445 |
| Slc38a4 | 833 | 0.084 | 0.321 | Atp11a | 2951 | 0.038 | 0.449 |
| Pyp | 847 | 0.083 | 0.331 | Gstp1 | 2967 | 0.037 | 0.453 |
| Aacs | 901 | 0.080 | 0.339 | Dhrs7 | 3014 | 0.037 | 0.455 |
| Plod1 | 934 | 0.079 | 0.348 | Cbr2 | 3147 | 0.035 | 0.453 |
| Acas2 | 983 | 0.077 | 0.355 | Echdc3 | 3152 | 0.035 | 0.458 |
| Auh | 1068 | 0.074 | 0.361 | Acy3 | 3254 | 0.035 | 0.457 |
| Gcat | 1109 | 0.072 | 0.368 | Dhrs1 | 3483 | 0.032 | 0.450 |
| Dhrs8 | 1184 | 0.070 | 0.373 | Itgb1 | 3527 | 0.032 | 0.452 |
| Egln3 | 1232 | 0.068 | 0.380 | 4933406E20RIK | 3553 | 0.031 | 0.454 |
| Mthfs | 1268 | 0.067 | 0.387 | Plod2 | 3574 | 0.031 | 0.458 |
| Mvk | 1298 | 0.066 | 0.394 | Pmm2 | 3582 | 0.031 | |
| Aup1 | 1325 | 0.065 | 0.401 | Ugp2 | 3583 | 0.031 | |
| Spr | 1456 | 0.062 | 0.403 | Gnpat | 3633 | 0.031 | |
| Sc5dl | 1462 | 0.062 | 0.411 | 1110003P22RIK | 3636 | 0.031 | |
| 1300018J18RIK | 1516 | 0.061 | 0.416 | Dbt | 3710 | 0.030 | |
| Agpat3 | 1524 | 0.061 | 0.423 |
Rank = position of genes in the context of the ranked list of array genes
RMS = the ranked metric score
RES = the running enrichment score
Table 5.
ECM-related transcripts enriched in DEX-treated chondrocytes.
| HUGO symbol | Rank | RMS | RES | HUGO symbol | Rank | RMS* | RES** |
| Dmp1 | 18 | 0.470 | 0.036 | Matn4 | 882 | 0.081 | 0.420 |
| Omd | 27 | 0.409 | 0.068 | Lama3 | 886 | 0.081 | 0.427 |
| Itga5 | 38 | 0.358 | 0.095 | Nyx | 992 | 0.077 | 0.427 |
| Adamts1 | 57 | 0.305 | 0.118 | Lamb2 | 1082 | 0.073 | 0.429 |
| Timp4 | 61 | 0.296 | 0.141 | Bsg | 1100 | 0.072 | 0.433 |
| Col4a1 | 86 | 0.268 | 0.161 | Fbn2 | 1242 | 0.068 | 0.432 |
| Col4a2 | 98 | 0.247 | 0.180 | Ntn4 | 1245 | 0.068 | 0.437 |
| Adam12 | 112 | 0.225 | 0.197 | 5730577E14RIK | 1381 | 0.064 | 0.435 |
| Prelp | 139 | 0.200 | 0.211 | Col6a2 | 1405 | 0.064 | 0.439 |
| Postn | 142 | 0.195 | 0.227 | Ntn3 | 1415 | 0.063 | 0.443 |
| Chad | 176 | 0.174 | 0.239 | Tgfb2 | 1531 | 0.060 | 0.442 |
| Mgp | 195 | 0.168 | 0.251 | Mia1 | 1575 | 0.059 | 0.445 |
| Col1a1 | 232 | 0.154 | 0.261 | Mmp14 | 1803 | 0.054 | 0.438 |
| Mfap5 | 233 | 0.153 | 0.273 | Col15a1 | 1845 | 0.053 | 0.440 |
| Col10a1 | 266 | 0.146 | 0.283 | Ctgf | 1882 | 0.052 | 0.442 |
| Smoc2 | 279 | 0.145 | 0.294 | Col6a1 | 1942 | 0.052 | 0.443 |
| Aspn | 294 | 0.141 | 0.304 | Gpld1 | 1946 | 0.051 | 0.447 |
| Col4a5 | 367 | 0.128 | 0.310 | Emid2 | 2043 | 0.050 | 0.446 |
| Adamts15 | 385 | 0.126 | 0.319 | Col7a1 | 2047 | 0.050 | 0.450 |
| Tgfb1 | 394 | 0.125 | 0.329 | Adam10 | 2107 | 0.049 | 0.451 |
| Sparcl1 | 440 | 0.119 | 0.336 | Col9a2 | 605 | 0.100 | 0.370 |
| Adam17 | 483 | 0.112 | 0.343 | Matn3 | 610 | 0.099 | 0.377 |
| Lama5 | 508 | 0.110 | 0.350 | Col11a2 | 636 | 0.097 | 0.384 |
| Lamc1 | 517 | 0.109 | 0.358 | Hapln1 | 650 | 0.096 | 0.391 |
| Spock2 | 581 | 0.102 | 0.363 | Lama2 | 685 | 0.092 | 0.396 |
| Lama1 | 688 | 0.092 | 0.403 | Gpc3 | 796 | 0.086 | 0.412 |
| Ltbp4 | 704 | 0.091 | 0.410 | Lama4 | 827 | 0.084 | 0.417 |
*RMS = the ranked metric score
**RES = the running enrichment score
Figure 4.
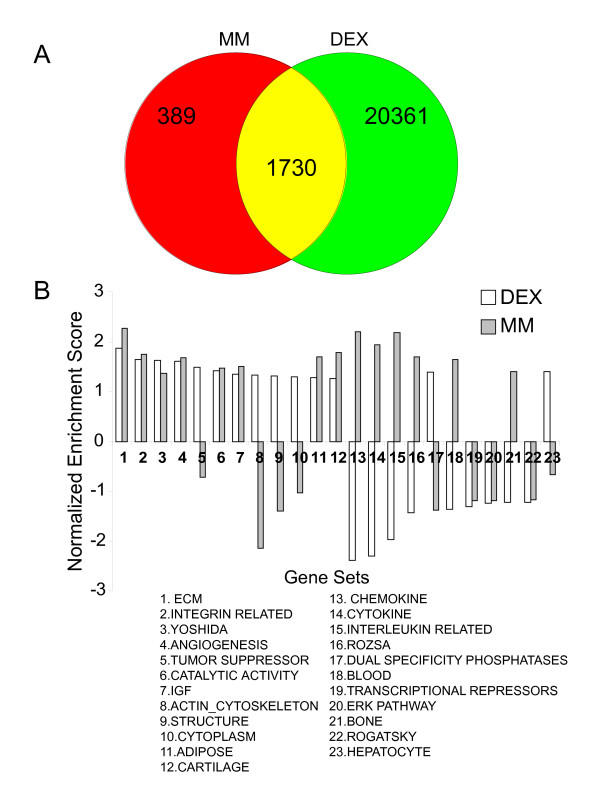
Comparison of DEX-treated primary chondrocytes to a time course of chondrocyte differentiation in micromass culture. The Venn diagram depicts probe sets that are common between the list of 2119 probe sets differentially expressed between days 3 and 15 of micromass culture and the list of 22 091 significantly expressed probe sets in primary chondrocyte monolayer cultures (A). The matrix of 77 user-defined gene sets are used to interrogate microarray data from days 15 and day 3 of micromass culture. Normalized enrichment scores (NES) generated from this analysis are then compared to NES scores derived from the DEX study to evaluate similarities in the regulation of different groups of genes in chondrocytes (B). Positive enrichment scores (ES) indicate gene sets that are enriched and up-regulated in DEX-treated chondrocytes or d15 of micromass culture. Negative ES indicate gene set enrichment and down-regulation in the DEX-treatment or up-regulation in the day 3 samples of the micromass (MM) culture data set.
Osteomodulin, an additional matrix molecule shown to be structurally similar to IBSP [67], ranked second in the list of enriched ECM genes. Additional ECM molecules expressed in terminally differentiated chondrocytes such as collagen 10 (Col10a1) and osteonectin (Spock1) were identified, suggesting that this molecular classification is important for transmitting GC signaling in the growth plate.
Interestingly, the normalized enrichment scores for factors down-regulated by DEX treatment were higher than those positively correlated with DEX, but contained fewer probe sets contributing to the scores. Gene sets composed of 127 and 106 genes associated with cytokine and growth factor activity, respectively, were negatively correlated with DEX treatment (Figure 4, Table 6, 7). In other studies, cytokines such as Il-8 and GROα were found to promote the hypertrophy of osteoarthritic cartilage, and excess interleukins 1β(IL-1β), interleukin 6 (IL-6) and Tumor Necrosis Factor alpha (TNF-α) cause growth failure in children [68-70]. Our studies identified three members of the GP-130 family of cytokines, namely interleukins -11,-6 (Il11, Il6) and leukemia inhibitory factor (Lif), as part of the core enrichment group for cytokines (Table 6). Transgenic mice overexpressing Il-6 exhibit growth retardation, and LIF is thought to regulate the rate at which terminally differentiated cartilage is calcified and vascularized [71,72].
Table 6.
Cytokine transcripts enriched in vehicle-treated chondrocytes. I.
| HUGO gene symbol | Rank | RMS | RES |
| Cklfsf2b | 16971 | -0.0424 | -0.574 |
| Il7 | 16981 | -0.0425 | -0.569 |
| Il1f9 | 17007 | -0.0427 | -0.566 |
| Grn | 17130 | -0.0439 | -0.567 |
| Il1f6 | 17153 | -0.0442 | -0.563 |
| Ifna2 | 17418 | -0.0468 | -0.571 |
| Tslp | 17503 | -0.0477 | -0.570 |
| Il17 | 17568 | -0.0483 | -0.568 |
| A730028g07rik | 17606 | -0.0486 | -0.564 |
| Cxcl11 | 17634 | -0.0490 | -0.560 |
| Ctf1 | 17857 | -0.0519 | -0.565 |
| Lta | 17864 | -0.0519 | -0.559 |
| Il1a | 18018 | -0.0539 | -0.561 |
| Ccl20 | 18038 | -0.0542 | -0.556 |
| Ccl17 | 18334 | -0.0584 | -0.564 |
| Ccl12 | 18384 | -0.0592 | -0.560 |
| Cklf | 18618 | -0.0639 | -0.564 |
| Ifna11 | 18855 | -0.0688 | -0.568 |
| Cklfsf6 | 18874 | -0.0693 | -0.561 |
| Il15 | 18955 | -0.0719 | -0.557 |
| Ltb | 19146 | -0.0779 | -0.558 |
| Ccl3 | 19220 | -0.0814 | -0.552 |
| Tnfsf9 | 19228 | -0.0816 | -0.543 |
| Cx3cl1 | 19523 | -0.0975 | -0.547 |
| Gdf15 | 19660 | -0.1100 | -0.541 |
| Bmp5 | 19775 | -0.1238 | -0.533 |
| Cxcl14 | 19798 | -0.1289 | -0.519 |
| Cxcl1 | 19928 | -0.1698 | -0.507 |
| Cxcl10 | 19951 | -0.1807 | -0.487 |
| Ccl7 | 19956 | -0.1849 | -0.466 |
| Gdf5 | 19973 | -0.2066 | -0.444 |
| Cxcl12 | 19978 | -0.2104 | -0.420 |
| Areg | 19983 | -0.2189 | -0.395 |
| Cxcl2 | 19996 | -0.2421 | -0.369 |
| Ppbp | 20014 | -0.2944 | -0.336 |
| Lif | 20024 | -0.3296 | -0.299 |
| Ccl2 | 20030 | -0.3589 | -0.258 |
| Il11 | 20035 | -0.4036 | -0.213 |
| Cxcl5 | 20039 | -0.5406 | -0.152 |
| Tnfsf11 | 20041 | -0.5835 | -0.085 |
| Il6 | 20043 | -0.7529 |
Rank = position of genes in the context of the ranked list of array genes
RMS = the ranked metric score
RES = the running enrichment score
Table 7.
Growth factor transcripts vehicle in DEX-treated chondrocytes.
| HUGO gene symbol | Rank | RMS | RES |
| Fgf21 | 18968 | -0.073 | -0.508 |
| Nrg3 | 19132 | -0.077 | -0.506 |
| Fgf5 | 19190 | -0.080 | -0.499 |
| Ereg | 19507 | -0.096 | -0.502 |
| Fgf7 | 19581 | -0.102 | -0.493 |
| Gdf15 | 19660 | -0.110 | -0.483 |
| Igf1 | 19679 | -0.111 | -0.469 |
| Bmp5 | 19775 | -0.124 | -0.458 |
| Nov | 19848 | -0.144 | -0.443 |
| Vegf | 19877 | -0.150 | -0.425 |
| Ptn | 19885 | -0.153 | -0.406 |
| Cxcl1 | 19928 | -0.170 | -0.386 |
| Bdnf | 19939 | -0.176 | -0.364 |
| Inhba | 19971 | -0.204 | -0.340 |
| Gdf5 | 19973 | -0.207 | -0.313 |
| Cxcl12 | 19978 | -0.210 | -0.287 |
| Areg | 19983 | -0.219 | -0.259 |
| Hbegf | 20006 | -0.264 | -0.226 |
| Ngfb | 20013 | -0.287 | -0.189 |
| Lif | 20024 | -0.330 | -0.148 |
| Il11 | 20035 | -0.404 | -0.096 |
| Il6 | 20043 | -0.753 | 0.000 |
Rank = position of genes in the context of the ranked list of array genes
RMS = the ranked metric score
RES = the running enrichment score
This group also contained the gene encoding Tumor necrosis factor (ligand) superfamily, member 11 (Tnfsf11, RANKL), which has been localized to mature chondrocytes and is thought to promote degradation of the calcified cartilage ECM and ultimately endochondral ossification through activation of osteoclasts [73-75]. It is important to note that several independent gene sets connected to inflammation such as cytokines, chemokines and interleukins exhibit some overlap and showed similar enrichment patterns, which provides additional confirmation that DEX is indeed downregulating inflammatory molecules in chondrocytes. GC have been previously reported to down-regulate the expression of VEGF, one of the central growth factors involved in vascularization of calcified cartilage matrix [49], in agreement with our data (Table 7). Since some of these factors, such as RANKL, VEGF and LIF, promote normal tissue remodeling processes during endochondral ossification, our data suggest that DEX prevents the replacement of hypertrophic cartilage by bone. GC have been shown to delay chondrocyte maturation while retaining their capacity to re-engage in their developmental program [21]. This could account for upregulation of genes typically associated with the chondrocyte phenotype, such as ECM genes and the coordinated downregulation of factors that promote the transition from cartilage into bone.
Identification of cartilage-specific dexamethasone-effects
Identification of cartilage-specific gene sets affected by DEX treatment provided further insight into the complex nature of GC functions in cartilage. We knew from other studies that DEX effects on chondrogenic differentiation are dependent on cell source, experimental system and DEX concentration [40,42,76-78]. We aimed to systematically characterize the effects of DEX on growth plate chondrocytes. To ensure that our DEX data set was expressing bona fide cartilage markers, we compared the DEX data to our previously generated micromass culture data set [50]. We compared all expressed probe sets in the DEX array to probe sets exhibiting a minimum 1.5-fold change in expression between days 3 and 15 of micromass cultures that encompass the various stages of the chondrocyte life cycle. Day 3 of micromass culture likely coincides with the onset of the cartilage developmental program and early chondrogenesis. After 15 days of culture, the cell population is comprised primarily of terminally differentiated chondrocytes and thus corresponds mostly to the hypertrophic zone of the growth plate [50,79], although small numbers of other cells are present at all stages. Out of the 2119 probe sets displaying at least 1.5-fold changes in expression in the micromass culture data set (a probe set list generated from the pair-wise comparison of day 3 versus day 15 of micromass culture), 1730 were also expressed in the DEX array. This shows that our primary chondrocyte monolayers do exhibit prototypical chondrocyte gene expression patterns in both the presence and absence of DEX treatment.
To complete more robust classification of the data in which we could correlate chondrocyte gene expression to the DEX phenotype, we created a gene set from this list of 2119 probe sets (Table 8, 9). The micromass derived gene list was enriched in this study; however, the list was found to correlate both positively and negatively with different aspects of the DEX phenotype. We therefore proceeded to evaluate both the micromass (MM) data set and the DEX data set using GSEA analysis and the previously created gene sets. If both the micromass time course and the DEX data sets show the same enrichment pattern, we would have evidence to suggest that pharmacological DEX doses promote chondrocyte differentiation. Normalized enrichment scores for gene sets common to both culture methods were therefore compared to identify differences and similarities between DEX-treated chondrocytes and the chondrocyte phenotype (Figure 4B).
Table 8.
Micromass culture-derived gene sets are enriched in DEX-treated primary chondrocytes (d3 vs d15_2). I.
| HUGO gene symbol | Rank | RMS | RES |
| Itgbl1 | 32 | 0.391 | 0.015 |
| Adrb2 | 54 | 0.308 | 0.026 |
| Bst1 | 80 | 0.271 | 0.036 |
| Gpx3 | 83 | 0.269 | 0.047 |
| Myocd | 90 | 0.259 | 0.058 |
| Grk5 | 105 | 0.229 | 0.066 |
| Ids | 123 | 0.212 | 0.074 |
| Ms4a6b | 140 | 0.200 | 0.082 |
| 1810057c19rik | 146 | 0.193 | 0.090 |
| Igfbp2 | 149 | 0.190 | 0.097 |
| Zfp36 | 218 | 0.159 | 0.100 |
| Serpina3n | 222 | 0.158 | 0.107 |
| P2ry6 | 225 | 0.157 | 0.113 |
| Adm | 228 | 0.156 | 0.120 |
| Crym | 277 | 0.145 | 0.123 |
| Ppap2a | 303 | 0.139 | 0.128 |
| Pycard | 307 | 0.138 | 0.133 |
| Kcns1 | 320 | 0.134 | 0.138 |
| Cd80 | 321 | 0.134 | 0.144 |
| Trim24 | 330 | 0.133 | 0.149 |
| C1qtnf6 | 339 | 0.131 | 0.154 |
| A330049m08rik | 377 | 0.127 | 0.157 |
| Adamts15 | 385 | 0.126 | 0.162 |
| Elovl4 | 398 | 0.124 | 0.167 |
| C1qa | 402 | 0.124 | 0.172 |
| Sox9 | 434 | 0.119 | 0.175 |
| Htra3 | 455 | 0.116 | 0.179 |
| Adam17 | 483 | 0.112 | 0.182 |
| Mgll | 493 | 0.112 | 0.186 |
| Ibsp | 507 | 0.110 | 0.190 |
| C1qb | 511 | 0.109 | 0.194 |
| Bambi | 516 | 0.109 | 0.199 |
| Anxa4 | 551 | 0.105 | 0.201 |
| Cd109 | 555 | 0.105 | 0.206 |
| Nrk | 559 | 0.104 | 0.210 |
| Gstm1 | 619 | 0.099 | 0.211 |
| Asb4 | 634 | 0.097 | 0.214 |
| Pygl | 654 | 0.095 | 0.217 |
| Rasl11b | 655 | 0.095 | 0.221 |
| Cdc42ep4 | 674 | 0.093 | 0.224 |
| Slc9a3r2 | 683 | 0.092 | 0.227 |
| Lama1 | 688 | 0.092 | 0.231 |
| Bb146404 | 707 | 0.091 | 0.234 |
| Ai194308 | 724 | 0.090 | 0.237 |
| Smn1 | 752 | 0.088 | 0.239 |
| Alcam | 772 | 0.087 | 0.242 |
| Cst3 | 790 | 0.086 | 0.244 |
| Pyp | 847 | 0.083 | 0.245 |
| 2700017m01rik | 870 | 0.082 | 0.247 |
| Fgfr3 | 884 | 0.081 | 0.250 |
| Mrpl34 | 912 | 0.080 | 0.252 |
| C9orf46 | 972 | 0.077 | 0.252 |
| Maf | 981 | 0.077 | 0.255 |
| 8430420c20rik | 1028 | 0.075 | 0.255 |
| Gfm2 | 1030 | 0.075 | 0.259 |
| Anxa6 | 1041 | 0.075 | 0.261 |
| Isg20 | 1064 | 0.074 | 0.263 |
| Auh | 1068 | 0.074 | 0.266 |
| Bsg | 1100 | 0.072 | 0.267 |
| Peg3 | 1179 | 0.070 | 0.266 |
| Adam23 | 1208 | 0.069 | 0.268 |
| Ezh1 | 1213 | 0.069 | 0.270 |
| 2810022l02rik | 1214 | 0.069 | 0.273 |
| 0610011i04rik | 1248 | 0.068 | 0.274 |
| Pbx2 | 1257 | 0.067 | 0.277 |
| Jup | 1291 | 0.066 | 0.278 |
| Zcwcc2 | 1301 | 0.066 | 0.280 |
| Whsc2 | 1317 | 0.066 | 0.282 |
| 2410004l22rik | 1344 | 0.065 | 0.283 |
| Lmnb2 | 1388 | 0.064 | 0.284 |
| Fndc1 | 1435 | 0.063 | 0.284 |
| Rarres2 | 1460 | 0.062 | 0.285 |
| Tap2 | 1512 | 0.061 | 0.285 |
| Ctbs | 1559 | 0.060 | 0.285 |
| Jdp2 | 1574 | 0.059 | 0.287 |
| Hck | 1712 | 0.056 | 0.282 |
| 5031400m07rik | 1792 | 0.054 | 0.281 |
| Pkn1 | 1839 | 0.053 | 0.280 |
| Dag1 | 1929 | 0.052 | 0.278 |
| Fth1 | 1976 | 0.051 | 0.278 |
| 1110001e17rik | 1979 | 0.051 | 0.280 |
| Rbp4 | 1984 | 0.051 | 0.282 |
| Pdcd6ip | 2044 | 0.050 | 0.281 |
| Siat7d | 2050 | 0.050 | 0.283 |
| Kcnd2 | 2074 | 0.050 | 0.284 |
| 2310004k06rik | 2076 | 0.050 | 0.286 |
| D19ertd678e | 2106 | 0.049 | 0.286 |
| Npdc1 | 2114 | 0.049 | 0.288 |
| Fts | 2116 | 0.049 | 0.290 |
| Prickle1 | 2123 | 0.049 | 0.291 |
| 1110037f02rik | 2171 | 0.048 | 0.291 |
| Cdc42se1 | 2246 | 0.047 | 0.289 |
| Chpt1 | 2261 | 0.047 | 0.290 |
| Wwp2 | 2341 | 0.045 | 0.288 |
| Dact1 | 2363 | 0.045 | 0.289 |
| Rragd | 2380 | 0.045 | 0.290 |
| Irf5 | 2406 | 0.044 | 0.291 |
| Nrbf2 | 2414 | 0.044 | 0.292 |
| Cox4i2 | 2436 | 0.044 | 0.293 |
| Bmp7 | 2456 | 0.044 | 0.294 |
| 1810008a18rik | 2517 | 0.043 | 0.292 |
| Asph | 2533 | 0.043 | 0.293 |
| Stat2 | 2550 | 0.042 | 0.294 |
| Hoxa11 | 2560 | 0.042 | 0.296 |
| Bax | 2599 | 0.042 | 0.295 |
| Sspn | 2611 | 0.042 | 0.297 |
| Ifngr2 | 2612 | 0.042 | 0.298 |
| Glrx1 | 2672 | 0.041 | 0.297 |
| Gba | 2739 | 0.040 | 0.295 |
| Fzd2 | 2759 | 0.040 | 0.296 |
| Crtap | 2772 | 0.040 | 0.297 |
| Slc1a5 | 2786 | 0.040 | 0.298 |
| Slco3a1 | 2831 | 0.039 | 0.297 |
| 3110040n11rik | 2833 | 0.039 | 0.299 |
| Tep1 | 2845 | 0.039 | 0.300 |
| Fastk | 2860 | 0.039 | 0.301 |
| Tmed3 | 2869 | 0.038 | 0.302 |
| Ephb4 | 2876 | 0.038 | 0.303 |
| Asah2 | 2908 | 0.038 | 0.303 |
| Pold4 | 2989 | 0.037 | 0.301 |
| 1110001a07rik | 2995 | 0.037 | 0.302 |
| Pcp4 | 3010 | 0.037 | 0.303 |
| Mab21l2 | 3025 | 0.037 | 0.304 |
Rank = position of genes in the context of the ranked list of array genes
RMS = the ranked metric score
RES = the running enrichment score
Table 9.
Micromass culture-derived transcripts enriched in vehicle-treated primary chondrocytes (d3 vs d15_3/4). I.
| HUGO gene symbol | Rank | RMS | RES |
| Rabggtb | 16734 | -0.040 | -0.271 |
| Ube2e2 | 16759 | -0.041 | -0.270 |
| Cd68 | 16769 | -0.041 | -0.269 |
| H2-T23 | 16830 | -0.041 | -0.270 |
| Derl1 | 16834 | -0.041 | -0.268 |
| Smarcc1 | 16853 | -0.041 | -0.267 |
| Srxn1 | 16856 | -0.041 | -0.266 |
| Klf10 | 16868 | -0.042 | -0.264 |
| Zfhx1b | 16879 | -0.042 | -0.263 |
| H2afy3 | 16929 | -0.042 | -0.264 |
| Wisp2 | 16973 | -0.042 | -0.264 |
| Tbl1xr1 | 16976 | -0.042 | -0.262 |
| Ppp1r3c | 16979 | -0.042 | -0.260 |
| D11lgp2e | 17036 | -0.043 | -0.261 |
| Smpdl3b | 17079 | -0.043 | -0.262 |
| Dock2 | 17125 | -0.044 | -0.262 |
| Purb | 17127 | -0.044 | -0.260 |
| Grn | 17130 | -0.044 | -0.258 |
| 1110035l05rik | 17139 | -0.044 | -0.257 |
| Kiaa1008 | 17185 | -0.045 | -0.257 |
| E430025l02rik | 17195 | -0.045 | -0.256 |
| Timm8a | 17293 | -0.046 | -0.259 |
| C130006e23 | 17307 | -0.046 | -0.257 |
| Rbm10 | 17319 | -0.046 | -0.256 |
| A230103n10rik | 17347 | -0.046 | -0.255 |
| Cd151 | 17401 | -0.047 | -0.256 |
| Srf | 17409 | -0.047 | -0.254 |
| Cacna1s | 17507 | -0.048 | -0.257 |
| Ythdf1 | 17529 | -0.048 | -0.256 |
| Ppp2r1b | 17539 | -0.048 | -0.254 |
| Tead2 | 17545 | -0.048 | -0.252 |
| Igsf7 | 17590 | -0.049 | -0.252 |
| Per3 | 17604 | -0.049 | -0.251 |
| G1p2 | 17739 | -0.050 | -0.256 |
| Slco2a1 | 17786 | -0.051 | -0.256 |
| Coq7 | 17918 | -0.053 | -0.260 |
| Rarb | 17940 | -0.053 | -0.259 |
| Lcp1 | 17954 | -0.053 | -0.257 |
| Dnaja1 | 17987 | -0.053 | -0.256 |
| Thoc3 | 17993 | -0.054 | -0.254 |
| Cd44 | 18041 | -0.054 | -0.254 |
| Slc41a1 | 18171 | -0.056 | -0.258 |
| Kif11 | 18232 | -0.057 | -0.259 |
| Hspa5bp1 | 18235 | -0.057 | -0.257 |
| Ncf4 | 18290 | -0.058 | -0.257 |
| Bub1b | 18292 | -0.058 | -0.254 |
| Cap2 | 18295 | -0.058 | -0.252 |
| Aig1 | 18340 | -0.059 | -0.251 |
| Rfc3 | 18361 | -0.059 | -0.250 |
| Stmn1 | 18396 | -0.060 | -0.249 |
| 9130213b05rik | 18408 | -0.060 | -0.247 |
| Tyms-Ps | 18432 | -0.060 | -0.245 |
| Timp3 | 18513 | -0.062 | -0.247 |
| Tiparp | 18564 | -0.063 | -0.247 |
| Thbs4 | 18627 | -0.064 | -0.247 |
| Wasf1 | 18652 | -0.064 | -0.245 |
| Nupr1 | 18686 | -0.065 | -0.244 |
| Ezh2 | 18706 | -0.066 | -0.242 |
| Fbxl14 | 18709 | -0.066 | -0.239 |
| Prim1 | 18780 | -0.067 | -0.240 |
| Insig2 | 18805 | -0.068 | -0.238 |
| B3gnt5 | 18858 | -0.069 | -0.238 |
| Fam60a | 18963 | -0.072 | -0.240 |
| H2-M3 | 18972 | -0.073 | -0.237 |
| Gja7 | 18974 | -0.073 | -0.234 |
| Bex2 | 18987 | -0.073 | -0.231 |
| Tk1 | 19043 | -0.074 | -0.231 |
| 1200015n20rik | 19109 | -0.076 | -0.231 |
| Clecsf5 | 19114 | -0.077 | -0.228 |
| Ms4a7 | 19141 | -0.078 | -0.226 |
| Cdca5 | 19163 | -0.079 | -0.223 |
| C730042f17rik | 19180 | -0.079 | -0.220 |
| Trim25 | 19194 | -0.080 | -0.218 |
| Efnb2 | 19207 | -0.081 | -0.215 |
| Apex1 | 19236 | -0.082 | -0.212 |
| Ddah2 | 19243 | -0.082 | -0.209 |
| Bub1 | 19262 | -0.083 | -0.206 |
| Nup43 | 19263 | -0.083 | -0.203 |
| Rdh10 | 19270 | -0.083 | -0.199 |
| 2610201a13rik | 19330 | -0.086 | -0.199 |
| Rp2h | 19406 | -0.089 | -0.198 |
| Tnni1 | 19407 | -0.089 | -0.195 |
| Myog | 19423 | -0.091 | -0.191 |
| Osmr | 19486 | -0.095 | -0.190 |
| Mmp9 | 19524 | -0.097 | -0.188 |
| Tnnt1 | 19525 | -0.098 | -0.184 |
| Fhod3 | 19528 | -0.098 | -0.179 |
| D930038m13rik | 19537 | -0.099 | -0.175 |
| Nes | 19567 | -0.101 | -0.172 |
| Sbk1 | 19571 | -0.102 | -0.168 |
| Dusp9 | 19594 | -0.103 | -0.165 |
| Akr1b8 | 19622 | -0.106 | -0.161 |
| Pdgfrb | 19663 | -0.110 | -0.158 |
| Tfrc | 19667 | -0.111 | -0.154 |
| Moxd1 | 19670 | -0.111 | -0.149 |
| 1810008k03rik | 19681 | -0.112 | -0.145 |
| Cpeb1 | 19710 | -0.115 | -0.141 |
| 6720475j19rik | 19716 | -0.116 | -0.136 |
| Ripk4 | 19718 | -0.116 | -0.131 |
| Itga6 | 19756 | -0.121 | -0.127 |
| Bmp5 | 19775 | -0.124 | -0.123 |
| Lhx9 | 19776 | -0.124 | -0.117 |
| Pkp2 | 19797 | -0.129 | -0.113 |
| Chrna1 | 19808 | -0.131 | -0.108 |
| Bhlhb2 | 19837 | -0.142 | -0.103 |
| Gp49a | 19847 | -0.144 | -0.097 |
| Clecsf10 | 19893 | -0.155 | -0.092 |
| Gch1 | 19902 | -0.159 | -0.086 |
| D0h4s114 | 19908 | -0.161 | -0.079 |
| Cxcl1 | 19928 | -0.170 | -0.072 |
| Ch25h | 19946 | -0.178 | -0.065 |
| Mkrn3 | 19988 | -0.228 | -0.057 |
| Ptprc | 20016 | -0.297 | -0.046 |
| Car6 | 20017 | -0.298 | -0.032 |
| Nr1d2 | 20031 | -0.368 | -0.017 |
| Evi2a | 20033 | -0.393 | 0.001 |
| Plxnc1 | 18075 | -0.055 | -0.286 |
| Cilp2 | 18106 | -0.055 | -0.285 |
| Brca1 | 18148 | -0.056 | -0.285 |
| Litaf | 18149 | -0.056 | -0.283 |
| Bc027246 | 18154 | -0.056 | -0.281 |
| 6820424l24rik | 18268 | -0.057 | -0.285 |
| Hrb | 18272 | -0.057 | -0.283 |
| Nnat | 18303 | -0.058 | -0.282 |
| P2ry12 | 18329 | -0.058 | -0.282 |
| Cdca4 | 18343 | -0.059 | -0.280 |
| 6030404e16rik | 18367 | -0.059 | -0.279 |
| Tfec | 18429 | -0.060 | -0.280 |
| Nfe2l2 | 18440 | -0.060 | -0.278 |
| Gtf2h2 | 18467 | -0.061 | -0.277 |
| 4930469p12rik | 18504 | -0.062 | -0.277 |
| Cul4b | 18535 | -0.062 | -0.276 |
| H2afy2 | 18547 | -0.063 | -0.274 |
| 1190002n15rik | 18582 | -0.063 | -0.274 |
| B430218l07rik | 18591 | -0.063 | -0.272 |
| Rgs18 | 18607 | -0.064 | -0.270 |
| Frk | 18631 | -0.064 | -0.269 |
| Slc6a9 | 18633 | -0.064 | -0.267 |
| Tgfbr2 | 18687 | -0.065 | -0.267 |
| Tia1 | 18802 | -0.068 | -0.270 |
| Lgr5 | 18844 | -0.068 | -0.270 |
| Sgpp1 | 18909 | -0.071 | -0.271 |
| Matn2 | 18924 | -0.071 | -0.269 |
| Sox11 | 18931 | -0.071 | -0.266 |
| Hus1 | 18980 | -0.073 | -0.266 |
| D930015e06rik | 19028 | -0.074 | -0.266 |
| Apob48r | 19032 | -0.074 | -0.263 |
| Av344025 | 19045 | -0.074 | -0.261 |
| Eno2 | 19047 | -0.074 | -0.258 |
| 2610024e20rik | 19053 | -0.075 | -0.256 |
| Chd1l | 19093 | -0.076 | -0.255 |
| Emr1 | 19145 | -0.078 | -0.255 |
| Rgs4 | 19200 | -0.081 | -0.254 |
| D030028o16rik | 19211 | -0.081 | -0.252 |
| Kif2c | 19216 | -0.081 | -0.249 |
| Ccl3 | 19220 | -0.081 | -0.246 |
| Trim30 | 19232 | -0.082 | -0.244 |
| Qrsl1 | 19242 | -0.082 | -0.241 |
| Nr3c1 | 19281 | -0.083 | -0.240 |
| Trip13 | 19282 | -0.084 | -0.237 |
| Dna2l | 19317 | -0.085 | -0.236 |
| Tcf8 | 19335 | -0.086 | -0.233 |
| Clecsf8 | 19341 | -0.086 | -0.230 |
| Lyzs | 19422 | -0.090 | -0.231 |
| Palmd | 19475 | -0.095 | -0.230 |
| Tjp2 | 19487 | -0.095 | -0.227 |
| D430019h16rik | 19493 | -0.096 | -0.224 |
| Sesn3 | 19501 | -0.096 | -0.221 |
| Ereg | 19507 | -0.096 | -0.218 |
| Cx3cl1 | 19523 | -0.097 | -0.215 |
| Fzd6 | 19529 | -0.098 | -0.211 |
| Sod3 | 19564 | -0.101 | -0.209 |
| Tnnt2 | 19580 | -0.102 | -0.206 |
| Satb1 | 19599 | -0.104 | -0.203 |
| Cd14 | 19606 | -0.104 | -0.200 |
| Gbp2 | 19607 | -0.104 | -0.196 |
| Tgfbi | 19609 | -0.105 | -0.192 |
| Chek1 | 19652 | -0.109 | -0.190 |
| Tm4sf1 | 19653 | -0.109 | -0.186 |
| Igf1 | 19679 | -0.111 | -0.183 |
| Enpp1 | 19695 | -0.113 | -0.180 |
| Slc15a3 | 19704 | -0.114 | -0.176 |
| Pdpn | 19725 | -0.117 | -0.173 |
| Dkk1 | 19747 | -0.119 | -0.169 |
| Slk | 19759 | -0.121 | -0.166 |
| Ankrd1 | 19794 | -0.128 | -0.163 |
| Trp53bp1 | 19801 | -0.129 | -0.158 |
| C79407 | 19804 | -0.130 | -0.153 |
| 2210010l05rik | 19809 | -0.131 | -0.149 |
| Eps8 | 19815 | -0.133 | -0.144 |
| Dkk2 | 19862 | -0.147 | -0.141 |
| Arhgap18 | 19863 | -0.147 | -0.136 |
| Twist2 | 19878 | -0.151 | -0.131 |
| Pcdha8 | 19915 | -0.164 | -0.126 |
| Il4r | 19926 | -0.169 | -0.121 |
| Mdm1 | 19931 | -0.172 | -0.115 |
| Phlda1 | 19957 | -0.188 | -0.109 |
| Bhlhb5 | 19960 | -0.192 | -0.102 |
| C130076o07rik | 19964 | -0.196 | -0.095 |
| 5830411e10rik | 19974 | -0.207 | -0.088 |
| Ptpre | 19989 | -0.228 | -0.080 |
| Trib3 | 19990 | -0.235 | -0.071 |
| 9230117n10rik | 19994 | -0.241 | -0.062 |
| Pcdhb7 | 19998 | -0.249 | -0.053 |
| Mmp3 | 20001 | -0.252 | -0.044 |
| Cd34 | 20009 | -0.274 | -0.034 |
| Thbd | 20022 | -0.310 | -0.023 |
| A830016g23rik | 20023 | -0.323 | -0.011 |
| Ahr | 20028 | -0.336 | 0.001 |
Rank = position of genes in the context of the ranked list of array genes
RMS = the ranked metric score
RES = the running enrichment score
Four different patterns were observed when comparing DEX treatment and micromass differentiation data sets for gene enrichments scores (Figure 4B). First, similar gene sets were indeed enriched in both day 15 micromass and DEX-treated monolayer cultures, and core genes contributing to the normalized enrichment scores were similarly overlapping between the two data sets in results with low FDR. For example, ECM genes were enriched with both DEX treatment and the day 15 micromass phenotype. Other gene sets following this enrichment pattern included genes involved in integrin function, angiogenesis, catalytic activity, IGF related, adipocyte and cartilage, all of which have a precedent for being involved in chondrocyte maturation [28,49,80,81]. The enrichment of angiogenic transcripts with DEX treatment was unexpected since DEX was shown to have anti-angiogenic roles in cartilage; however, upon closer examination of the genes contributing to the enrichment score, Vegf, which is thought to be a central angiogenic factor in endochondral ossification [82], was excluded from the core enrichment genes and had the lowest correlation with the DEX phenotype in that gene set. In contrast, Vegf was enriched in the growth factor data set which positively correlated with the vehicle control and not DEX treatment (Table 7).
Gene sets associated with the actin cytoskeleton, tumour suppressors, structure, cytoplasmic genes, hepatocyte markers and dual specificity phosphatases (DUSPs) were enriched in the DEX data set and the phenotype positively correlated with day 3 of micromass culture. The identification of DUSPs was particularly interesting since DEX has been shown to induce genes encoding for these proteins [77,83,84]. DUSPS counteract the activation of MAP kinase pathways, known regulators of chondrocyte differentiation [85], and are thought to mediate DEX's anti-inflammatory functions and to influence hepatic gluconeogenesis [83,86,87].
Additional comparisons identified genes that show enrichment in day 15 micromass cultures and downregulation with DEX treatment. These include the previously identified chemokines, cytokines and interleukins. A final trend in similarly enriched gene sets identified lists that were negatively correlated both with the DEX phenotype and day 15 of micromass cultures. Only transcriptional repressors and molecules involved in the extracellular signal-regulated kinase (ERK) pathway were identified. This pattern is consistent with DEX's anti-proliferative functions, as another study showed that DEX decreases ERK phosphorylation and thus cell cycle progression in a pre-osteoblast cell line [77]. Altogether this analysis shows that DEX regulation of growth plate chondrocyte differentiation is multifaceted. The patterns identified here are in agreement with a dual role of DEX in maintenance of the cartilage phenotype and delay in the cartilage-to-bone transition, as we suggested above.
We also wanted to determine whether DEX target genes identified in the current study were similar to DEX-responsive genes identified in alternate studies, in different cell types [88]. Out of a total of twelve microarray studies evaluating the transcriptional effects of DEX treatment on a specific tissue or cell type, only ten genes were common to at least three of the chosen DEX studies. Specifically, bone morphogenetic protein 2 (Bmp2), delta sleep inducing factor 1 (Dsip1), beta-2 microglobulin (B2m), neuroepithelial cell transforming gene 1 (Net1), TNFAIP3 interacting protein 1 (Tnip1), bone marrow stromal cell antigen 2 (Bst2), B-cell leukemia/lymphoma 6 (Bcl6), nuclear factor of kappa light chain gene enhancer in B-cells inhibitor, alpha (Nfkbia), FK506 binding protein 5 (Fkbp5) and B-cell translocation gene 1, anti-proliferative (Btg1) were identified. It therefore appears that while DEX affects similar functional categories across various species, tissue types and experimental conditions, the individual genes that respond to DEX treatment are variable. These results also reinforce the paradigm that GC regulation is inextricably linked to its physiological context [88-99].
Analyses of GC response elements in dexamethasone target genes in chondrocytes
Classical genomic GC action is thought to be mediated by a cytoplasmic GR that modifies transcription upon binding its cognate ligand and translocating to the nucleus. In the nucleus, the GR binds a GRE sequence. GR can both activate and repress transcription, depending on the GRE variant present in the regulatory regions of GC target genes [100]. Binding to composite GREs involves homodimerization of the GR to bind a non-palindromic consensus sequence comprised of two GR binding sites and is generally associated with transcriptional activation. In some instances, however, GR can function to block access or activity of transcription factors within promoter regions of certain genes, thus impeding transcription [101]. GR are also able to bind a modified GRE consisting of composite GRE half-sites, termed negative GREs, since they have documented roles in transcriptional repression. Variations on the genomic functions of GC include transcriptional regulation at the level of protein-protein interactions between the GR and other transcription factors, co-activators or co-repressors. In addition to the GRE-dependent roles, the GR is capable of interacting with other co-activators and repressors to influence transcription indirectly [102,103].
The 100 most highly expressed probe sets with greatest enrichment in the DEX or vehicle-treated chondrocytes are shown in Figure 5. Probe sets identified in this analysis included both known cartilage markers and established DEX target genes such as Vegf, Ibsp, Bglap2 and Fkbp5 [49,63-66,104,105]. We examined the proximal promoter regions of three separate gene lists, the top 100 DEX-responsive transcripts generated by GSEA analysis (Figure 5), the 22 091 probe sets deemed expressed in primary chondrocyte cultures and the 1158 transcripts deemed differentially expressed between DEX and vehicle treated cultures by one-Way ANOVA. Specifically, we searched the 9990 base pairs upstream regulatory regions in this list for the composite GRE consensus sequence. We identified putative GRE sequences in many genes, including Fkbp5, pyruvate dehydrogenasekinase (Pdk4), RANKL (Tnfsf11), Interleukin 6 (Il6) and prostaglandin I2 synthase (Ptgis) (bold in Figure 5). However, the majority of DEX-regulated probe sets such as prostaglandin-endoperoxide synthase 2 (Ptgs2), phosphodiesterase 4A (Pde4), Vegf, Period homolog 1 (Per1) and Krüppel like factor 15 (Klf15) do not appear to contain a GRE in the first 10 kilobases and may by regulated by DEX via a GRE-independent mechanism, through a GRE that deviates from the consensus GRE sequence or through GREs at other locations in the gene.
Figure 5.
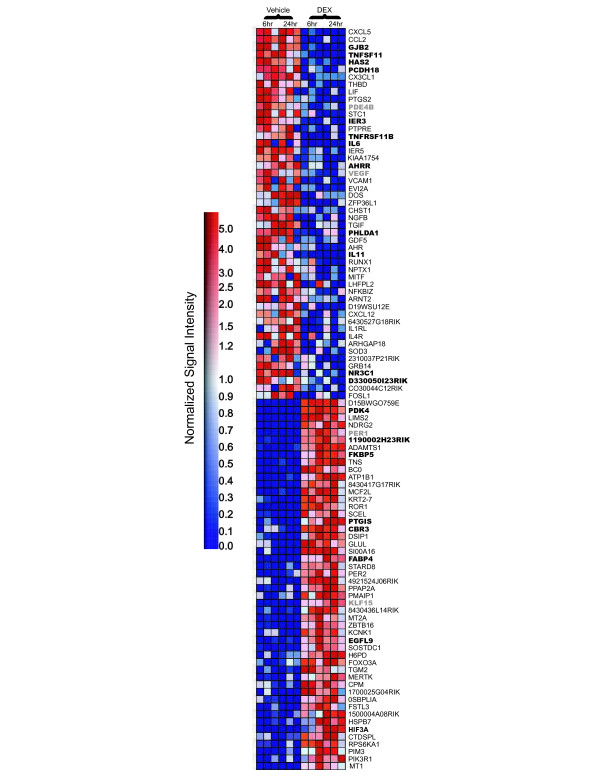
Heat map of top 100 probe sets determined by GSEA analysis. GSEA-derived heat maps of the top 100 differentially expressed probe sets enriched with DEX or the vehicle control are shown (B). Expression profiles for all experimental replicates are shown for each time point. Genes containing a putative GRE are shown in bold, and examples of genes that do not contain GREs but have been documented as targets of DEX regulation are depicted by bold gray lettering. Signal intensities are illustrated by varying shades of red (up-regulation) and blue (down-regulation).
Examination of all lists generated similar results in that approximately 16–20% of all probes contained the consensus GRE. Consequently, we cannot exclude the presence of less conventional GRE loci in the transcripts, or the presence of GREs that deviate from the consensus sequence or are located outside the queried sequence. Since many of the genes affected at the 6 hr time point encode transcription factors, it is likely that a large proportion of the genes that only change after 24 hrs are regulated indirectly by DEX, through altered expression of these transcription factors and other regulatory proteins (e.g. phosphatases and cytokines, as discussed above).
Functional analysis is required to unequivocally evaluate the contribution of GRE-dependent mechanisms to GC regulation in chondrocytes. In addition to the genomic functions of GC, non-genomic modes of GC regulation have been documented. Non-genomic mechanisms are thought to occur through specific and non-specific mechanisms. Specific non-genomic GC regulation occurs through the classical GR and its cytoplasmic heteroprotein complex or non-classical GRs such as membrane GR [106-109]. Conversely, non-specific non-genomic mechanisms rely on the physiochemical properties of GC and the phospholipid bilayer (Buttgereit and Scheffold, 2002). Further, studies in which candidate molecules are selected and characterized in depth are imperative to discern the specific regulatory mechanisms occurring in chondrocytes.
Conclusion
This study elucidates the downstream transcriptional impact of pharmacological GC exposure on developing chondrocytes. We have identified a small subset of transcripts containing putative GREs in cartilage, but it appears that GRE-independent or indirect mechanisms of GC regulation also contribute to GC regulation in primary chondrocyte monolayer cultures. In addition, traditional microarray analysis methods and gene class testing point to a dual role for pharmacological GC doses in chondrocytes. DEX acts in a gene class-specific manner in cartilage in which it promotes the expression of ECM and metabolic transcripts necessary for maintaining the chondrocyte phenotype while simultaneously downregulating cytokines and growth factors which stimulate the cartilage to bone transition. Understanding the implications of gene expression changes and integrating them into the network of molecules controlling cartilage development continues to be challenging, but robust analytical methods will prove to be useful in constructing the networks of gene interactions and understanding the complex nature of GC signaling in the skeleton. The ultimate objective of this study will be to translate these findings into more efficacious therapeutic GCs.
Methods
Animals and Materials
Timed-pregnant CD1 mice were purchased from Charles River Laboratories at embryonic day E15.5 mice (E15.5). Dexamethasone was obtained from Calbiochem and reconstituted in Dimethyl sulfoxide (DMSO, vehicle) according to the manufacturer's instructions. Cell culture materials and general chemicals were obtained from Invitrogen, Sigma or VWR unless otherwise stated.
Primary cell culture and dexamethasone-treatment
Tibiae, femurs and humeri were isolated from E15.5 mouse embryos and placed in α-MEM media (Invitrogen) containing 0.2% Bovine Serum Albumin (BSA), 1 mM β-glycerophosphate, 0.05 mg/ml ascorbic acid and penicillin/streptomycin and incubated at 37°C in a humidified 5% CO2 incubator overnight. The following morning media was removed and the bones placed in 4 ml of 0.25% trypsin-EDTA (Invitrogen) for 15 min at 37°C. Trypsin was subsequently replaced with 1 mg/ml collagenase P (Roche) in DMEM/10% fetal bovine serum (Invitrogen), and cells were incubated at 37°C with rotation at 100 rpm for 90 min. Following digestion, the cell suspension was centrifuged for 5 min at 1000 rpm, and the collagenase containing supernatant was decanted. Chondrocytes were resuspended in media containing 2:3 DMEM:F12, 10% fetal bovine serum, 0.5 mM L-glutamine, and penicillin/streptomycin (25 units/ml). Cells were seeded in 6-well NUNC plates at a density of 2.5 × 104 cells per ml and incubated overnight. Primary monolayer chondrocytes were treated with 10-7 M dexamethasone (DEX) or the DMSO control (vehicle) diluted in fresh media supplemented with 0.25 mM ascorbic acid (Sigma) and 1 mM β-glycerophosphate (Sigma) and incubated for up to 24 hrs. Micromass cultures were completed as previously described [50].
Cell counting studies
Chondrocytes were isolated and seeded in 24-well NUNC plates (Nunc Inc.) at a density of 16 000 cells/cm2. Cells were cultured, treated and enzymatically digested as described with some modifications. Collagenase digestion occurred for 5 minutes followed by mechanical digestion to liberate cells from the ECM. Cells were counted with a hemocytometer in triplicate with a minimum of 3 individual wells per treatment and three independent cell isolations.
RNA isolations and quantitative real-time PCR
All RNA protocols were completed as previously outlined [50]. Total RNA was isolated at 6 hrs and 24 hrs after treatment using the RNeasy mini extraction kit (Qiagen) according to the manufacturer's instructions. RNA quantity and integrity was assessed using the Bioanalyzer 2000 system (Agilent). Quantitative real-time polymerase chain reaction (qRT-PCR) amplification was completed using the ABI Prism 7900 Sequence Detection System (Applied Biosystems). Triplicate reactions were executed for each sample of each of three independent trials. The TaqMan one-step master mix kit (Applied Biosystems) with gene-specific target primers and probes were used for amplification. The collagen X (Col10a1) probe and primer set (forward primer 5'-ACGCCTACGATGTACACGTATGA-3', reverse primer 5'-ACTCCCTGAAGCCTGATCCA-3', 6-FAM-5'-AGTACAGCAAAGGCTAC-MGBNFQ) was designed with PrimerDesigner 2.0 software (Applied Biosystems) [79]. TaqMan GAPDH control reagents for house-keeping gene glyceraldehyde-3-phosphate dehydrogenase (Gapdh, forward primer 5'-GAAGGTGAAGGTCGGAGTC; reverse primer 5'-GAAGATGGTGATGGGATTTC; probe JOE-CAAGCTTCCCGTTCTCAGCC-TAMRA) was used as an internal amplification control. Probes for Indian hedgehog (Ihh), Tissue inhibitor of matrix metalloproteinase 4 (Timp4), Cyclin-dependent kinase inhibitor 1C (Cdkn1c, p57), Integrin beta like 1 protein (Itgbl1), GC receptor (Nr3c1), Integrin beta 1 (Itgb1) and Kruppel-like factor 15 (Klf15) were assayed using the TaqMan® gene expression assays in accordance with the manufacturers directions. Amplified transcripts were quantified using the standard curve method, and the relative transcript abundance was determined by calculating the quotient of the gene of interest and equivalent Gapdh values.
Microarray analysis
Total RNA was extracted from control and DEX-treated cultures at 6 hr and 24 hr following treatment, in three independent experiments. RNA integrity and quantity was assessed using the Agilent 2000 Bioanalyzer system, and RNA samples were subsequently hybridized to the MOE 430 2.0 mouse chip from Affymetrix© containing 45 101 probe sets as described [50]. Bioanalysis, microarray hybridization, scanning and preliminary MAS 5.0 normalizations were completed at the London Regional Genomics Facility. Data were deposited in the GEO database (NCBI; accession number GSE7683).
Data normalization
Microarray data were pre-processed using the GC-RMA algorithm in Genespring GX*. Expression values were further filtered by retaining only those probe sets with expression values of at least 50 in at least 25% of all conditions, thus generating a list of 22 091 probe sets. To assess differential gene expression between treatments at both the 6 and 24 hr time points, a Welch ANOVA test with a p-value cut-off of 0.01 and a 5% false discovery rate (FDR) reduced the data to 1158 probe sets. Subsequent 1.5-, 5- and 10-fold change filters produced lists of 162, 21 and 7 probe sets for the 6 hr time point and 399, 53 and 19 probe sets for the 24 hr time point, respectively.
The same data set was normalized in parallel using Robust Multichip Analysis using RMAEXPRESS software v.0.4.1 developed by B. Bolstad, University of California, Berkeley [110]. Background adjustment and quantile normalization parameters were selected for data processing. Logarithmically transformed expression values were used to implement Gene Set Enrichment Analysis (GSEA).
Gene set enrichment analysis (GSEA)
The GSEA algorithm was implemented with GSEA v2.0 software [51,52]. Ranked expression lists were derived from RMAEXPRESS and GeneSpring GX® 7.3.1. Briefly, the GSEA algorithm ranks all array genes according to their expression under each experimental condition. The resulting ranked metric score (RMS) is therefore a function of the correlation between a gene's signal intensity, the experimental conditions in question and all other genes in the data set. An enrichment score (ES) is then calculated for an a priori gene list or gene set that is associated with a particular molecular classification. In our analysis, gene sets were created from different functional groupings, molecular classifications, tissues, and other microarray screens. A Ranked enrichment score (RES) which determines the extent to which a given gene from a gene set is represented at the extremes of the ranked gene list is then calculated. Specifically, this value is obtained by walking along the ranked list using a cumulative sum statistic which increases when a member of a particular gene set is found in the ranked gene list and is coordinately penalized when it does not appear in the gene set. A null distribution of ES is subsequently generated by permutation filtering to evaluate the statistical significance of the observed RES values. Permutation filtering randomly assigns the experimental conditions or class labels (i.e., DEX versus vehicle) to the different microarray samples. After this procedure has been repeated for each gene set, the ES are normalized (NES) to account for differences in gene set size. The false discovery rate (FDR) is then calculated relative to the NES values to determine the false-positive rate. Significant FDR and p-values were less than 25% and 0.001, respectively in accordance with GSEA recommendations.
Gene set creation
Gene sets were generated using the probe set search tool and the molecular function class of Gene Ontology annotations in GeneSpring GX. Additional gene sets were created using lists from pairwise comparisons between day 3 and 15 of a previously generated micromass data set (James et al., 2005), and publications that identified DEX target genes in other cartilage array screens, other tissue types and experimental systems. A total of 2119 probe sets showing a minimum 1.5-fold change in gene expression were used in the analysis. Probe set redundancy was eliminated in all gene sets using the CollapseDataset function in the GSEA program. All probe set identifiers were assimilated to the Human Genome Organization (HUGO) annotations. Probe sets lacking corresponding HUGO annotations were excluded from the analysis. Default parameters were used to execute the analysis and median values taken to represent the range of duplicated probe sets for a given gene. A total of 77 user-defined gene sets were generated from GeneSpring derived Gene Ontology annotations for various molecular classifications and probe sets of differentially expressed genes between days 3 and 15 of micromass culture (James et al., 2005).
Glucocorticoid response element (GRE) analysis
Putative GRE were identified with the GenespringGX mouse genome9999 application which allows sequences up to 9999 bp upstream of the transcriptional start sites of all annotated MOE4302.0 transcripts to be interrogated for transcription factor binding sites. The GR consensus sequence GGTACAnnntgttCT [111] was queried from 10 bp to 10 000 bp upstream of the transcriptional start sites of available probe sets. The GRE consensus sequence was screened against 10 748 probe sets derived from the list of 22 091 reliably expressed probe sets exhibiting homology to upstream regulatory regions annotated in the program. Only exact matches were retained for subsequent analyses out a total of 1,073,741,824 tests.
Abbreviations
DEX: Dexamethasone; GSEA: gene set enrichment analysis; RES: ranked enrichment score; RMS: ranked metric score, ES: enrichment scores; NES: normalized enrichment score, SOM: self-organizing maps; FDR: false discovery rate; GR: glucocorticoid receptor
Competing interests
The author(s) declare that they have no competing interests.
Authors' contributions
CGJ completed cell culture experiments, data analysis, real-time PCR and drafted the manuscript. VU completed cell culture experiments. JT and TMU contributed to the design of the study and the writing of the manuscript. FB conceived of the study and contributed to the writing of the manuscript. All authors read and approved the final manuscript.
Acknowledgments
Acknowledgements
CGJ is supported by a doctoral award from the Canadian Institutes of Health Research (CIHR) and previously by an Ontario Graduate Scholarship in Science and Technology. V.U. is the recipient of a graduate scholarship from the Canadian Arthritis Network. FB is the recipient of a Canada Research Chair. Operating funds for these studies were provided by the CIHR, the Canadian Arthritis Network and the Hospital for Sick Children Foundation to FB.
Contributor Information
Claudine G James, Email: cjames9@uwo.ca.
Veronica Ulici, Email: vulici@uwo.ca.
Jan Tuckermann, Email: jan@fli-leibniz.de.
T Michael Underhill, Email: tunderhi@interchange.ubc.ca.
Frank Beier, Email: fbeier@uwo.ca.
References
- Cancedda R, Descalzi Cancedda F, Castagnola P. Chondrocyte differentiation. Int Rev Cytol. 1995;159:265–358. doi: 10.1016/s0074-7696(08)62109-9. [DOI] [PubMed] [Google Scholar]
- Karsenty G, Wagner EF. Reaching a genetic and molecular understanding of skeletal development. Dev Cell. 2002;2:389–406. doi: 10.1016/S1534-5807(02)00157-0. [DOI] [PubMed] [Google Scholar]
- Eames BF, de la Fuente L, Helms JA. Molecular ontogeny of the skeleton. Birth Defects Res Part C Embryo Today. 2003;69:93–101. doi: 10.1002/bdrc.10016. [DOI] [PubMed] [Google Scholar]
- Cancedda R, Castagnola P, Cancedda FD, Dozin B, Quarto R. Developmental control of chondrogenesis and osteogenesis. Int J Dev Biol. 2000;44:707–714. [PubMed] [Google Scholar]
- Ikeda T, Kamekura S, Mabuchi A, Kou I, Seki S, Takato T, Nakamura K, Kawaguchi H, Ikegawa S, Chung UI. The combination of SOX5, SOX6, and SOX9 (the SOX trio) provides signals sufficient for induction of permanent cartilage. Arthritis Rheum. 2004;50:3561–3573. doi: 10.1002/art.20611. [DOI] [PubMed] [Google Scholar]
- Lefebvre V, Behringer RR, de Crombrugghe B. L-Sox5, Sox6 and Sox9 control essential steps of the chondrocyte differentiation pathway. Osteoarthritis Cartilage. 2001;9:S69–75. doi: 10.1053/joca.2001.0447. [DOI] [PubMed] [Google Scholar]
- Lefebvre V, Huang W, Harley VR, Goodfellow PN, de Crombrugghe B. SOX9 is a potent activator of the chondrocyte-specific enhancer of the pro alpha1(II) collagen gene. Mol Cell Biol. 1997;17:2336–2346. doi: 10.1128/mcb.17.4.2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafsson E, Aszodi A, Ortega N, Hunziker EB, Denker HW, Werb Z, Fassler R. Role of collagen type II and perlecan in skeletal development. Ann N Y Acad Sci. 2003;995:140–150. doi: 10.1111/j.1749-6632.2003.tb03217.x. [DOI] [PubMed] [Google Scholar]
- Li SW, Prockop DJ, Helminen H, Fassler R, Lapvetelainen T, Kiraly K, Peltarri A, Arokoski J, Lui H, Arita M, et al. Transgenic mice with targeted inactivation of the Col2 alpha 1 gene for collagen II develop a skeleton with membranous and periosteal bone but no endochondral bone. Genes Dev. 1995;9:2821–2830. doi: 10.1101/gad.9.22.2821. [DOI] [PubMed] [Google Scholar]
- O'Keefe RJ, Puzas JE, Loveys L, Hicks DG, Rosier RN. Analysis of type II and type X collagen synthesis in cultured growth plate chondrocytes by in situ hybridization: rapid induction of type X collagen in culture. J Bone Miner Res. 1994;9:1713–1722. doi: 10.1002/jbmr.5650091107. [DOI] [PubMed] [Google Scholar]
- Bell DM, Leung KK, Wheatley SC, Ng LJ, Zhou S, Ling KW, Sham MH, Koopman P, Tam PP, Cheah KS. SOX9 directly regulates the type-II collagen gene. Nat Genet. 1997;16:174–178. doi: 10.1038/ng0697-174. [DOI] [PubMed] [Google Scholar]
- Ng LJ, Wheatley S, Muscat GE, Conway-Campbell J, Bowles J, Wright E, Bell DM, Tam PP, Cheah KS, Koopman P. SOX9 binds DNA, activates transcription, and coexpresses with type II collagen during chondrogenesis in the mouse. Dev Biol. 1997;183:108–121. doi: 10.1006/dbio.1996.8487. [DOI] [PubMed] [Google Scholar]
- Kronenberg HM. Developmental regulation of the growth plate. Nature. 2003;423:332–336. doi: 10.1038/nature01657. [DOI] [PubMed] [Google Scholar]
- Kirsch T, Ishikawa Y, Mwale F, Wuthier RE. Roles of the nucleational core complex and collagens (types II and X) in calcification of growth plate cartilage matrix vesicles. J Biol Chem. 1994;269:20103–20109. [PubMed] [Google Scholar]
- Kirsch T, Wuthier RE. Stimulation of calcification of growth plate cartilage matrix vesicles by binding to type II and X collagens. J Biol Chem. 1994;269:11462–11469. [PubMed] [Google Scholar]
- Apte SS, Olsen BR. Characterization of the mouse type X collagen gene. Matrix. 1993;13:165–179. doi: 10.1016/s0934-8832(11)80075-2. [DOI] [PubMed] [Google Scholar]
- Ohyama K, Farquharson C, Whitehead CC, Shapiro IM. Further observations on programmed cell death in the epiphyseal growth plate: comparison of normal and dyschondroplastic epiphyses. J Bone Miner Res. 1997;12:1647–1656. doi: 10.1359/jbmr.1997.12.10.1647. [DOI] [PubMed] [Google Scholar]
- Gibson G, Lin DL, Roque M. Apoptosis of terminally differentiated chondrocytes in culture. Exp Cell Res. 1997;233:372–382. doi: 10.1006/excr.1997.3576. [DOI] [PubMed] [Google Scholar]
- Zenmyo M, Komiya S, Kawabata R, Sasaguri Y, Inoue A, Morimatsu M. Morphological and biochemical evidence for apoptosis in the terminal hypertrophic chondrocytes of the growth plate. J Pathol. 1996;180:430–433. doi: 10.1002/(SICI)1096-9896(199612)180:4<430::AID-PATH691>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- Itagane Y, Inada H, Fujita K, Isshiki G. Interactions between steroid hormones and insulin-like growth factor-I in rabbit chondrocytes. Endocrinology. 1991;128:1419–1424. doi: 10.1210/endo-128-3-1419. [DOI] [PubMed] [Google Scholar]
- Siebler T, Robson H, Shalet SM, Williams GR. Dexamethasone inhibits and thyroid hormone promotes differentiation of mouse chondrogenic ATDC5 cells. Bone. 2002;31:457–464. doi: 10.1016/S8756-3282(02)00855-4. [DOI] [PubMed] [Google Scholar]
- Robson H, Siebler T, Shalet SM, Williams GR. Interactions between GH, IGF-I, glucocorticoids, and thyroid hormones during skeletal growth. Pediatr Res. 2002;52:137–147. doi: 10.1203/00006450-200208000-00003. [DOI] [PubMed] [Google Scholar]
- Wang J, Zhou J, Cheng CM, Kopchick JJ, Bondy CA. Evidence supporting dual, IGF-I-independent and IGF-I-dependent, roles for GH in promoting longitudinal bone growth. J Endocrinol. 2004;180:247–255. doi: 10.1677/joe.0.1800247. [DOI] [PubMed] [Google Scholar]
- Baron J, Huang Z, Oerter KE, Bacher JD, Cutler GB., Jr Dexamethasone acts locally to inhibit longitudinal bone growth in rabbits. Am J Physiol. 1992;263:E489–492. doi: 10.1152/ajpendo.1992.263.3.E489. [DOI] [PubMed] [Google Scholar]
- Jux C, Leiber K, Hugel U, Blum W, Ohlsson C, Klaus G, Mehls O. Dexamethasone impairs growth hormone (GH)-stimulated growth by suppression of local insulin-like growth factor (IGF)-I production and expression of GH- and IGF-I-receptor in cultured rat chondrocytes. Endocrinology. 1998;139:3296–3305. doi: 10.1210/en.139.7.3296. [DOI] [PubMed] [Google Scholar]
- Hochberg Z. Mechanisms of steroid impairment of growth. Horm Res. 2002;58:33–38. doi: 10.1159/000064764. [DOI] [PubMed] [Google Scholar]
- Raisz LG, Kream BE. Hormonal control of skeletal growth. Annu Rev Physiol. 1981;43:225–238. doi: 10.1146/annurev.ph.43.030181.001301. [DOI] [PubMed] [Google Scholar]
- Smink JJ, Buchholz IM, Hamers N, van Tilburg CM, Christis C, Sakkers RJ, de Meer K, van Buul-Offers SC, Koedam JA. Short-term glucocorticoid treatment of piglets causes changes in growth plate morphology and angiogenesis. Osteoarthritis Cartilage. 2003;11:864–871. doi: 10.1016/S1063-4584(03)00187-0. [DOI] [PubMed] [Google Scholar]
- Devesa J, Barros MG, Gondar M, Tresguerres JA, Arce V. Regulation of hypothalamic somatostatin by glucocorticoids. J Steroid Biochem Mol Biol. 1995;53:277–282. doi: 10.1016/0960-0760(95)00065-8. [DOI] [PubMed] [Google Scholar]
- Silvestrini G, Ballanti P, Patacchioli FR, Mocetti P, Di Grezia R, Wedard BM, Angelucci L, Bonucci E. Evaluation of apoptosis and the glucocorticoid receptor in the cartilage growth plate and metaphyseal bone cells of rats after high-dose treatment with corticosterone. Bone. 2000;26:33–42. doi: 10.1016/S8756-3282(99)00245-8. [DOI] [PubMed] [Google Scholar]
- van der Eerden BC, Karperien M, Wit JM. Systemic and local regulation of the growth plate. Endocr Rev. 2003;24:782–801. doi: 10.1210/er.2002-0033. [DOI] [PubMed] [Google Scholar]
- Tronche F, Kellendonk C, Reichardt HM, Schutz G. Genetic dissection of glucocorticoid receptor function in mice. Curr Opin Genet Dev. 1998;8:532–538. doi: 10.1016/S0959-437X(98)80007-5. [DOI] [PubMed] [Google Scholar]
- Finotto S, Krieglstein K, Schober A, Deimling F, Lindner K, Bruhl B, Beier K, Metz J, Garcia-Arraras JE, Roig-Lopez JL, et al. Analysis of mice carrying targeted mutations of the glucocorticoid receptor gene argues against an essential role of glucocorticoid signalling for generating adrenal chromaffin cells. Development. 1999;126:2935–2944. doi: 10.1242/dev.126.13.2935. [DOI] [PubMed] [Google Scholar]
- Benoyahu D, Akavia UD, Socher R, Shur I. Gene expression in skeletal tissues: application of laser capture microdissection. J Microsc. 2005;220:1–8. doi: 10.1111/j.1365-2818.2005.01511.x. [DOI] [PubMed] [Google Scholar]
- Speirs HJ, Seckl JR, Brown RW. Ontogeny of glucocorticoid receptor and 11beta-hydroxysteroid dehydrogenase type-1 gene expression identifies potential critical periods of glucocorticoid susceptibility during development. J Endocrinol. 2004;181:105–116. doi: 10.1677/joe.0.1810105. [DOI] [PubMed] [Google Scholar]
- Cole TJ, Blendy JA, Monaghan AP, Krieglstein K, Schmid W, Aguzzi A, Fantuzzi G, Hummler E, Unsicker K, Schutz G. Targeted disruption of the glucocorticoid receptor gene blocks adrenergic chromaffin cell development and severely retards lung maturation. Genes Dev. 1995;9:1608–1621. doi: 10.1101/gad.9.13.1608. [DOI] [PubMed] [Google Scholar]
- Grigoriadis AE, Aubin JE, Heersche JN. Effects of dexamethasone and vitamin D3 on cartilage differentiation in a clonal chondrogenic cell population. Endocrinology. 1989;125:2103–2110. doi: 10.1210/endo-125-4-2103. [DOI] [PubMed] [Google Scholar]
- Mushtaq T, Farquharson C, Seawright E, Ahmed SF. Glucocorticoid effects on chondrogenesis, differentiation and apoptosis in the murine ATDC5 chondrocyte cell line. J Endocrinol. 2002;175:705–713. doi: 10.1677/joe.0.1750705. [DOI] [PubMed] [Google Scholar]
- Nadra R, Menuelle P, Chevallier S, Berdal A. Regulation by glucocorticoids of cell differentiation and insulin-like growth factor binding protein production in cultured fetal rat nasal chondrocytes. J Cell Biochem. 2003;88:911–922. doi: 10.1002/jcb.10396. [DOI] [PubMed] [Google Scholar]
- Abu EO, Horner A, Kusec V, Triffitt JT, Compston JE. The localization of the functional glucocorticoid receptor alpha in human bone. J Clin Endocrinol Metab. 2000;85:883–889. doi: 10.1210/jc.85.2.883. [DOI] [PubMed] [Google Scholar]
- Shalhoub V, Conlon D, Tassinari M, Quinn C, Partridge N, Stein GS, Lian JB. Glucocorticoids promote development of the osteoblast phenotype by selectively modulating expression of cell growth and differentiation associated genes. J Cell Biochem. 1992;50:425–440. doi: 10.1002/jcb.240500411. [DOI] [PubMed] [Google Scholar]
- Boden SD, Hair G, Titus L, Racine M, McCuaig K, Wozney JM, Nanes MS. Glucocorticoid-induced differentiation of fetal rat calvarial osteoblasts is mediated by bone morphogenetic protein-6. Endocrinology. 1997;138:2820–2828. doi: 10.1210/en.138.7.2820. [DOI] [PubMed] [Google Scholar]
- Boden SD, McCuaig K, Hair G, Racine M, Titus L, Wozney JM, Nanes MS. Differential effects and glucocorticoid potentiation of bone morphogenetic protein action during rat osteoblast differentiation in vitro. Endocrinology. 1996;137:3401–3407. doi: 10.1210/en.137.8.3401. [DOI] [PubMed] [Google Scholar]
- Kato Y, Gospodarowicz D. Stimulation by glucocorticoid of the synthesis of cartilage-matrix proteoglycans produced by rabbit costal chondrocytes in vitro. J Biol Chem. 1985;260:2364–2373. [PubMed] [Google Scholar]
- Sekiya I, Koopman P, Tsuji K, Mertin S, Harley V, Yamada Y, Shinomiya K, Nifuji A, Noda M. Dexamethasone enhances SOX9 expression in chondrocytes. J Endocrinol. 2001;169:573–579. doi: 10.1677/joe.0.1690573. [DOI] [PubMed] [Google Scholar]
- Bianchi ML. Glucorticoids and bone: some general remarks and some special observations in pediatric patients. Calcif Tissue Int. 2002;70:384–390. doi: 10.1007/s00223-001-0043-0. [DOI] [PubMed] [Google Scholar]
- De Luca F. Impaired growth plate chondrogenesis in children with chronic illnesses. Pediatr Res. 2006;59:625–629. doi: 10.1203/01.pdr.0000214966.60416.1b. [DOI] [PubMed] [Google Scholar]
- Agoston H, Baybayan L, Beier F. Dexamethasone stimulates expression of C-type Natriuretic Peptide in chondrocytes. BMC Musculoskelet Disord. 2006;7:87. doi: 10.1186/1471-2474-7-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koedam JA, Smink JJ, van Buul-Offers SC. Glucocorticoids inhibit vascular endothelial growth factor expression in growth plate chondrocytes. Mol Cell Endocrinol. 2002;197:35–44. doi: 10.1016/S0303-7207(02)00276-9. [DOI] [PubMed] [Google Scholar]
- James CG, Appleton CT, Ulici V, Underhill TM, Beier F. Microarray analyses of gene expression during chondrocyte differentiation identifies novel regulators of hypertrophy. Mol Biol Cell. 2005;16:5316–5333. doi: 10.1091/mbc.E05-01-0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstrale M, Laurila E, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derfoul A, Perkins GL, Hall DJ, Tuan RS. Glucocorticoids promote chondrogenic differentiation of adult human mesenchymal stem cells by enhancing expression of cartilage extracellular matrix genes. Stem Cells. 2006;24:1487–1495. doi: 10.1634/stemcells.2005-0415. [DOI] [PubMed] [Google Scholar]
- Robson H, Anderson E, Eden OB, Isaksson O, Shalet S. Chemotherapeutic agents used in the treatment of childhood malignancies have direct effects on growth plate chondrocyte proliferation. J Endocrinol. 1998;157:225–235. doi: 10.1677/joe.0.1570225. [DOI] [PubMed] [Google Scholar]
- Chrysis D, Zaman F, Chagin AS, M T, Savendahl L. Dexamethasone induces apoptosis in proliferative chondrocytes through activation of caspases and suppression of the Akt-(PI3K) signaling pathway. Endocrinology. 2004 doi: 10.1210/en.2004-1152. [DOI] [PubMed] [Google Scholar]
- Klaus G, Jux C, Fernandez P, Rodriguez J, Himmele R, Mehls O. Suppression of growth plate chondrocyte proliferation by corticosteroids. Pediatr Nephrol. 2000;14:612–615. doi: 10.1007/s004670000344. [DOI] [PubMed] [Google Scholar]
- Alheim K, Corness J, Samuelsson MK, Bladh LG, Murata T, Nilsson T, Okret S. Identification of a functional glucocorticoid response element in the promoter of the cyclin-dependent kinase inhibitor p57Kip2. J Mol Endocrinol. 2003;30:359–368. doi: 10.1677/jme.0.0300359. [DOI] [PubMed] [Google Scholar]
- Presul E, Schmidt S, Kofler R, Helmberg A. Identification, tissue expression, and glucocorticoid responsiveness of alternative first exons of the human glucocorticoid receptor. J Mol Endocrinol. 2007;38:79–90. doi: 10.1677/jme.1.02183. [DOI] [PubMed] [Google Scholar]
- Milo M, Fazeli A, Niranjan M, Lawrence ND. A probabilistic model for the extraction of expression levels from oligonucleotide arrays. Biochem Soc Trans. 2003;31:1510–1512. doi: 10.1042/bst0311510. [DOI] [PubMed] [Google Scholar]
- Dong Y, Poellinger L, Okret S, Hoog JO, von Bahr-Lindstrom H, Jornvall H, Gustafsson JA. Regulation of gene expression of class I alcohol dehydrogenase by glucocorticoids. Proc Natl Acad Sci USA. 1988;85:767–771. doi: 10.1073/pnas.85.3.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bookout AL, Jeong Y, Downes M, Yu RT, Evans RM, Mangelsdorf DJ. Anatomical profiling of nuclear receptor expression reveals a hierarchical transcriptional network. Cell. 2006;126:789–799. doi: 10.1016/j.cell.2006.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye L, Mishina Y, Chen D, Huang H, Dallas SL, Dallas MR, Sivakumar P, Kunieda T, Tsutsui TW, Boskey A, et al. Dmp1-deficient mice display severe defects in cartilage formation responsible for a chondrodysplasia-like phenotype. J Biol Chem. 2005;280:6197–6203. doi: 10.1074/jbc.M412911200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein GS, van Wijnen AJ, Stein JL, Lian JB. Nuclear structure–skeletal gene expression interrelationships. Front Biosci. 1998;3:d849–864. doi: 10.2741/a328. [DOI] [PubMed] [Google Scholar]
- Ogata Y, Yamauchi M, Kim RH, Li JJ, Freedman LP, Sodek J. Glucocorticoid regulation of bone sialoprotein (BSP) gene expression. Identification of a glucocorticoid response element in the bone sialoprotein gene promoter. Eur J Biochem. 1995;230:183–192. doi: 10.1111/j.1432-1033.1995.0183i.x. [DOI] [PubMed] [Google Scholar]
- Sodek J, Kim RH, Ogata Y, Li J, Yamauchi M, Zhang Q, Freedman LP. Regulation of bone sialoprotein gene transcription by steroid hormones. Connect Tissue Res. 1995;32:209–217. doi: 10.3109/03008209509013725. [DOI] [PubMed] [Google Scholar]
- Fisher LW, Fedarko NS. Six genes expressed in bones and teeth encode the current members of the SIBLING family of proteins. Connect Tissue Res. 2003;44:33–40. doi: 10.1080/713713644. [DOI] [PubMed] [Google Scholar]
- Ramstad VE, Franzen A, Heinegard D, Wendel M, Reinholt FP. Ultrastructural distribution of osteoadherin in rat bone shows a pattern similar to that of bone sialoprotein. Calcif Tissue Int. 2003;72:57–64. doi: 10.1007/s00223-002-2047-9. [DOI] [PubMed] [Google Scholar]
- Olivotto E, Vitellozzi R, Fernandez P, Falcieri E, Battistelli M, Burattini S, Facchini A, Flamigni F, Santi S, Facchini A, et al. Chondrocyte hypertrophy and apoptosis induced by GROalpha require three-dimensional interaction with the extracellular matrix and a co-receptor role of chondroitin sulfate and are associated with the mitochondrial splicing variant of cathepsin B. J Cell Physiol. 2007;210:417–427. doi: 10.1002/jcp.20864. [DOI] [PubMed] [Google Scholar]
- Merz D, Liu R, Johnson K, Terkeltaub R. IL-8/CXCL8 and growth-related oncogene alpha/CXCL1 induce chondrocyte hypertrophic differentiation. J Immunol. 2003;171:4406–4415. doi: 10.4049/jimmunol.171.8.4406. [DOI] [PubMed] [Google Scholar]
- MacRae VE, Farquharson C, Ahmed SF. The restricted potential for recovery of growth plate chondrogenesis and longitudinal bone growth following exposure to pro-inflammatory cytokines. J Endocrinol. 2006;189:319–328. doi: 10.1677/joe.1.06609. [DOI] [PubMed] [Google Scholar]
- Grimaud E, Blanchard F, Charrier C, Gouin F, Redini F, Heymann D. Leukaemia inhibitory factor (lif) is expressed in hypertrophic chondrocytes and vascular sprouts during osteogenesis. Cytokine. 2002;20:224–230. doi: 10.1006/cyto.2002.2002. [DOI] [PubMed] [Google Scholar]
- De Benedetti F, Rucci N, Del Fattore A, Peruzzi B, Paro R, Longo M, Vivarelli M, Muratori F, Berni S, Ballanti P, et al. Impaired skeletal development in interleukin-6-transgenic mice: a model for the impact of chronic inflammation on the growing skeletal system. Arthritis Rheum. 2006;54:3551–3563. doi: 10.1002/art.22175. [DOI] [PubMed] [Google Scholar]
- Carda C, Silvestrini G, Gomez de Ferraris ME, Peydro A, Bonucci E. Osteoprotegerin (OPG) and RANKL expression and distribution in developing human craniomandibular joint. Tissue Cell. 2005;37:247–255. doi: 10.1016/j.tice.2005.03.002. [DOI] [PubMed] [Google Scholar]
- Sakakura Y, Tsuruga E, Irie K, Hosokawa Y, Nakamura H, Yajima T. Immunolocalization of receptor activator of nuclear factor-kappaB ligand (RANKL) and osteoprotegerin (OPG) in Meckel's cartilage compared with developing endochondral bones in mice. J Anat. 2005;207:325–337. doi: 10.1111/j.1469-7580.2005.00466.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvestrini G, Ballanti P, Patacchioli F, Leopizzi M, Gualtieri N, Monnazzi P, Tremante E, Sardella D, Bonucci E. Detection of osteoprotegerin (OPG) and its ligand (RANKL) mRNA and protein in femur and tibia of the rat. J Mol Histol. 2005;36:59–67. doi: 10.1007/s10735-004-3839-1. [DOI] [PubMed] [Google Scholar]
- Zheng ZH, Zhu P, Wang YH, Fan CM, Ding J, Shang P. [In vitro induction of directional differentiation of bone marrow mesenchymal stem cells towards chondrocytes] Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi. 2005;21:79–82. [PubMed] [Google Scholar]
- Engelbrecht Y, de Wet H, Horsch K, Langeveldt CR, Hough FS, Hulley PA. Glucocorticoids induce rapid up-regulation of mitogen-activated protein kinase phosphatase-1 and dephosphorylation of extracellular signal-regulated kinase and impair proliferation in human and mouse osteoblast cell lines. Endocrinology. 2003;144:412–422. doi: 10.1210/en.2002-220769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien CA, Jia D, Plotkin LI, Bellido T, Powers CC, Stewart SA, Manolagas SC, Weinstein RS. Glucocorticoids act directly on osteoblasts and osteocytes to induce their apoptosis and reduce bone formation and strength. Endocrinology. 2004;145:1835–1841. doi: 10.1210/en.2003-0990. [DOI] [PubMed] [Google Scholar]
- Stanton LA, Sabari S, Sampaio AV, Underhill TM, Beier F. p38 MAP kinase signalling is required for hypertrophic chondrocyte differentiation. Biochem J. 2004;378:53–62. doi: 10.1042/BJ20030874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigoriadis A, Heersche J, Aubin J. Differentiation of muscle, fat, cartilage, and bone from progenitor cells present in a bone-derived clonal cell population: effect of dexamethasone. J Cell Biol. 1988;106:2139–2151. doi: 10.1083/jcb.106.6.2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Zhou J, Bondy CA. Igf1 promotes longitudinal bone growth by insulin-like actions augmenting chondrocyte hypertrophy. Faseb J. 1999;13:1985–1990. doi: 10.1096/fasebj.13.14.1985. [DOI] [PubMed] [Google Scholar]
- Zelzer E, Mamluk R, Ferrara N, Johnson RS, Schipani E, Olsen BR. VEGFA is necessary for chondrocyte survival during bone development. Development. 2004;131:2161–2171. doi: 10.1242/dev.01053. [DOI] [PubMed] [Google Scholar]
- Abraham SM, Lawrence T, Kleiman A, Warden P, Medghalchi M, Tuckermann J, Saklatvala J, Clark AR. Antiinflammatory effects of dexamethasone are partly dependent on induction of dual specificity phosphatase 1. J Exp Med. 2006;203:1883–1889. doi: 10.1084/jem.20060336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai A, Han J, Cato AC, Akira S, Li JD. Glucocorticoids synergize with IL-1beta to induce TLR2 expression via MAP Kinase Phosphatase-1-dependent dual Inhibition of MAPK JNK and p38 in epithelial cells. BMC Mol Biol. 2004;5:2. doi: 10.1186/1471-2199-5-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanton LA, Underhill TM, Beier F. MAP kinases in chondrocyte differentiation. Dev Biol. 2003;263:165–175. doi: 10.1016/S0012-1606(03)00321-X. [DOI] [PubMed] [Google Scholar]
- Hulley PA, Gordon F, Hough FS. Inhibition of mitogen-activated protein kinase activity and proliferation of an early osteoblast cell line (MBA 15.4) by dexamethasone: role of protein phosphatases. Endocrinology. 1998;139:2423–2431. doi: 10.1210/en.139.5.2423. [DOI] [PubMed] [Google Scholar]
- Xu H, Yang Q, Shen M, Huang X, Dembski M, Gimeno R, Tartaglia LA, Kapeller R, Wu Z. Dual specificity MAPK phosphatase 3 activates PEPCK gene transcription and increases gluconeogenesis in rat hepatoma cells. J Biol Chem. 2005;280:36013–36018. doi: 10.1074/jbc.M508027200. [DOI] [PubMed] [Google Scholar]
- Rozsa FW, Reed DM, Scott KM, Pawar H, Moroi SE, Kijek TG, Krafchak CM, Othman MI, Vollrath D, Elner VM, et al. Gene expression profile of human trabecular meshwork cells in response to long-term dexamethasone exposure. Mol Vis. 2006;12:125–141. [PubMed] [Google Scholar]
- Grenier J, Tomkiewicz C, Trousson A, Rajkowski KM, Schumacher M, Massaad C. Identification by microarray analysis of aspartate aminotransferase and glutamine synthetase as glucocorticoid target genes in a mouse Schwann cell line. J Steroid Biochem Mol Biol. 2005;97:342–352. doi: 10.1016/j.jsbmb.2005.06.034. [DOI] [PubMed] [Google Scholar]
- Kolbus A, Blazquez-Domingo M, Carotta S, Bakker W, Luedemann S, von Lindern M, Steinlein P, Beug H. Cooperative signaling between cytokine receptors and the glucocorticoid receptor in the expansion of erythroid progenitors: molecular analysis by expression profiling. Blood. 2003;102:3136–3146. doi: 10.1182/blood-2003-03-0923. [DOI] [PubMed] [Google Scholar]
- Wang JC, Derynck MK, Nonaka DF, Khodabakhsh DB, Haqq C, Yamamoto KR. Chromatin immunoprecipitation (ChIP) scanning identifies primary glucocorticoid receptor target genes. Proc Natl Acad Sci USA. 2004;101:15603–15608. doi: 10.1073/pnas.0407008101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W, Chaudhuri S, Brickley DR, Pang D, Karrison T, Conzen SD. Microarray analysis reveals glucocorticoid-regulated survival genes that are associated with inhibition of apoptosis in breast epithelial cells. Cancer Res. 2004;64:1757–1764. doi: 10.1158/0008-5472.CAN-03-2546. [DOI] [PubMed] [Google Scholar]
- Yoshida NL, Miyashita T, U M, Yamada M, Reed JC, Sugita Y, Oshida T. Analysis of gene expression patterns during glucocorticoid-induced apoptosis using oligonucleotide arrays. Biochem Biophys Res Commun. 2002;293:1254–1261. doi: 10.1016/S0006-291X(02)00361-3. [DOI] [PubMed] [Google Scholar]
- Phuc Le P, Friedman JR, Schug J, Brestelli JE, Parker JB, Bochkis IM, Kaestner KH. Glucocorticoid receptor-dependent gene regulatory networks. PLoS Genet. 2005;1:e16. doi: 10.1371/journal.pgen.0010016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogatsky I, Hittelman AB, Pearce D, Garabedian MJ. Distinct glucocorticoid receptor transcriptional regulatory surfaces mediate the cytotoxic and cytostatic effects of glucocorticoids. Mol Cell Biol. 1999;19:5036–5049. doi: 10.1128/mcb.19.7.5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agbemafle BM, Oesterreicher TJ, Shaw CA, Henning SJ. Immediate early genes of glucocorticoid action on the developing intestine. Am J Physiol Gastrointest Liver Physiol. 2005;288:G897–906. doi: 10.1152/ajpgi.00454.2004. [DOI] [PubMed] [Google Scholar]
- Weber PS, Madsen-Bouterse SA, Rosa GJ, Sipkovsky S, Ren X, Almeida PE, Kruska R, Halgren RG, Barrick JL, Burton JL. Analysis of the Bovine Neutrophil Transcriptome During Glucocorticoid Treatment. Physiol Genomics. 2006 doi: 10.1152/physiolgenomics.00094.2006. [DOI] [PubMed] [Google Scholar]
- Gupta V, Galante A, Soteropoulos P, Guo S, Wagner BJ. Global gene profiling reveals novel glucocorticoid induced changes in gene expression of human lens epithelial cells. Mol Vis. 2005;11:1018–1040. [PubMed] [Google Scholar]
- Wang Y, Middleton F, Horton JA, Reichel L, Farnum CE, Damron TA. Microarray analysis of proliferative and hypertrophic growth plate zones identifies differentiation markers and signal pathways. Bone. 2004;35:1273–1293. doi: 10.1016/j.bone.2004.09.009. [DOI] [PubMed] [Google Scholar]
- Schoneveld OJ, Gaemers IC, Lamers WH. Mechanisms of glucocorticoid signalling. Biochim Biophys Acta. 2004;1680:114–128. doi: 10.1016/j.bbaexp.2004.09.004. [DOI] [PubMed] [Google Scholar]
- Saatcioglu F, Claret FX, Karin M. Negative transcriptional regulation by nuclear receptors. Semin Cancer Biol. 1994;5:347–359. [PubMed] [Google Scholar]
- Barnes PJ. Corticosteroid effects on cell signalling. Eur Respir J. 2006;27:413–426. doi: 10.1183/09031936.06.00125404. [DOI] [PubMed] [Google Scholar]
- Almawi WY, Beyhum HN, Rahme AA, Rieder MJ. Regulation of cytokine and cytokine receptor expression by glucocorticoids. J Leukoc Biol. 1996;60:563–572. doi: 10.1002/jlb.60.5.563. [DOI] [PubMed] [Google Scholar]
- U M, Shen L, Oshida T, Miyauchi J, Yamada M, Miyashita T. Identification of novel direct transcriptional targets of glucocorticoid receptor. Leukemia. 2004;18:1850–1856. doi: 10.1038/sj.leu.2403516. [DOI] [PubMed] [Google Scholar]
- Nuber UA, Kriaucionis S, Roloff TC, Guy J, Selfridge J, Steinhoff C, Schulz R, Lipkowitz B, Ropers HH, Holmes MC, et al. Up-regulation of glucocorticoid-regulated genes in a mouse model of Rett syndrome. Hum Mol Genet. 2005;14:2247–2256. doi: 10.1093/hmg/ddi229. [DOI] [PubMed] [Google Scholar]
- Croxtall JD, Choudhury Q, Flower RJ. Glucocorticoids act within minutes to inhibit recruitment of signalling factors to activated EGF receptors through a receptor-dependent, transcription-independent mechanism. Br J Pharmacol. 2000;130:289–298. doi: 10.1038/sj.bjp.0703272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasker JG. Rapid glucocorticoid actions in the hypothalamus as a mechanism of homeostatic integration. Obesity (Silver Spring) 2006;14:259S–265S. doi: 10.1038/oby.2006.320. [DOI] [PubMed] [Google Scholar]
- Buttgereit F, Scheffold A. Rapid glucocorticoid effects on immune cells. Steroids. 2002;67:529–534. doi: 10.1016/S0039-128X(01)00171-4. [DOI] [PubMed] [Google Scholar]
- Bartholome B, Spies CM, Gaber T, Schuchmann S, Berki T, Kunkel D, Bienert M, Radbruch A, Burmester GR, Lauster R, et al. Membrane glucocorticoid receptors (mGCR) are expressed in normal human peripheral blood mononuclear cells and up-regulated after in vitro stimulation and in patients with rheumatoid arthritis. Faseb J. 2004;18:70–80. doi: 10.1096/fj.03-0328com. [DOI] [PubMed] [Google Scholar]
- Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- Beato M, Chalepakis G, Schauer M, Slater EP. DNA regulatory elements for steroid hormones. J Steroid Biochem. 1989;32:737–747. doi: 10.1016/0022-4731(89)90521-9. [DOI] [PubMed] [Google Scholar]


