Abstract
Introduction
Single-nucleotide polymorphisms (SNPs) in codon 72 of the TP53 (also known as p53) gene (rs1042522) and in the promoter region of the MDM2 gene (SNP309; rs2279744) have been suggested to play roles in many cancers. We investigated whether these SNPs were associated with patient outcome and the effect of adjuvant systemic therapy.
Methods
The genotypes of TP53 codon 72 and MDM2 SNP309 were defined among 557 primary Japanese breast cancer patients (median follow-up, 61.7 months). The effects of several variables on survival were tested by Cox's proportional hazards regression analysis.
Results
We showed that the Pro/Pro genotype of TP53 codon 72 was associated with poorer disease-free survival (DFS) than other genotypes by Kaplan-Meier analysis (P = 0.049) and multivariate Cox's proportional hazards regression analysis (P = 0.047, risk ratio of recurrence = 1.67), whereas MDM2 SNP309 status was not associated with DFS. The association of the Pro/Pro TP53 genotype with poorer DFS was especially significant in patients who received adjuvant chemotherapy (P = 0.009). In contrast, among the patients who had received adjuvant hormonal therapy or no adjuvant systemic therapy, TP53 codon 72 genotype was not associated with DFS.
Conclusion
The Pro/Pro genotype of TP53 codon 72 appears to be an independent prognostic marker in breast cancer patients.
Introduction
The TP53 tumor suppressor pathway is well-known to be crucial for maintaining genomic integrity and preventing cells from undergoing oncogenic transformation [1,2]. MDM2 plays a key role in regulating the TP53 pathway by binding directly to the p53 protein, inhibiting its activity and mediating degradation via the ubiquitination system [3]. p53 also positively regulates MDM2 expression, thereby creating a negative feedback loop [3]. Overexpression of MDM2 is observed both in epithelial cells of transgenic mice with induced mammary carcinomas [4] and in multiple human tumors, including breast cancer [5-7].
The TP53 codon 72 Arg>Pro (CGC to CCC) polymorphism of exon 4 [8] (National Center for Biotechnology Information single-nucleotide polymorphism (SNP) identification number rs1042522) has been suggested to play a role in several different cancer types. These two variant protein forms may behave differently, as the Arg/Arg genotype has been reported to induce apoptosis more effectively than the Pro/Pro genotype [9,10], which may be due to enhanced mitochondrial localization of p53 protein in cells with the Arg/Arg genotype [9]. In contrast, the Pro/Pro genotype appears to induce a higher level of G1 arrest than the Arg/Arg genotype [11,12]. Patients with the Pro/Pro genotype of TP53 in breast cancers have poorer survival than those with other genotypes [13]. Furthermore, retention of the Arg allele of TP53 in tumor tissue of Arg/Pro heterozygous breast cancer patients has been associated with reduced disease-free and overall survival [14]. Taiwanese lung cancer patients and Israeli colorectal cancer patients with the Pro/Pro genotype of TP53 also showed poorer survival [15,16]. A recent study showed that breast cancer patients with the Pro/Pro genotype were less sensitive to chemotherapy than those with Arg/Arg or Arg/Pro genotypes [17]. Similar results were reported in head and neck carcinoma [11]. On the other hand, estrogen receptor (ER) positive patients possessing the Pro allele had better distant recurrence-free survival when randomized to tamoxifen compared to those who did not receive tamoxifen, while homozygous Arg/Arg patients did not [18]. After the initial report of a statistically significantly increased risk of breast cancer in women homozygous for the Pro allele [19], numerous studies examined a possible role of this TP53 polymorphism in breast cancer risk. Meta-analysis of nine studies has recently shown that this TP53 polymorphism is not associated with breast cancer risk [20].
A SNP in the promoter of the MDM2 gene, referred to as SNP309 (a T→G change) (rs2279744), has been implicated in earlier age of onset of Li-Fraumeni syndrome and sporadic cancers [21]. The MDM2 SNP309 G/G homozygous genotype elevates MDM2 protein expression [21]. A recent study showed that cells that harbor this genotype had a compromised TP53 response pathway and formed transcriptionally inactive p53-MDM2 complexes in response to stress [22]. The G/G genotype was also associated with increased incidence of esophageal squamous cell carcinoma [23]. Colorectal cancer patients who had both the SNP309 G allele and wild-type TP53 were diagnosed at a younger age than those with the T/T genotype and wild-type TP53 [24]. On the other hand, no association was found between SNP309 status and breast cancer incidence [25-27]. However, a recent study showed that, in women whose breast cancers expressed high levels of ER, those having the MDM2 SNP309 G/G genotype showed earlier age of onset than those with the T/T genotype [28]. Another study showed that among patients with the T/T genotype, mutant status of TP53 and aberrant p53 protein expression were associated with poor survival, suggesting an interaction between MDM2 SNP309 and tumor TP53 status [25].
Here, we have examined germ-line DNA samples from 557 consecutive primary breast cancer patients to investigate the association between these SNPs and breast cancer prognosis.
Materials and methods
Study subjects
Blood samples were obtained from a total of 557 consecutive primary breast cancer patients at Nagoya City University Hospital, Nagoya, Japan, between January 1983 and December 2003. Informed consent was obtained from all patients before surgery. The histological grade was estimated according to the Bloom and Richardson method proposed by Elston and Ellis [29]. Treatment decisions were based on consensus recommendations at that time in Japan. After surgery, 77 patients received no adjuvant therapy. A total of 137 patients received adjuvant chemotherapy alone (64 received oral 5-fluorouracil (5-FU), 41 were treated with a CMF (cyclophosphamide/methotrexate/5-FU)-based regimen, 26 with an anthracycline-based regimen, and 6 received other regimens). A total of 195 patients received adjuvant hormonal therapy alone (157 received tamoxifen, 28 tamoxifen plus luteinizing hormone-releasing hormone (LH-RH) agonist, 5 were treated with LH-RH alone, and 5 with aromatase inhibitor). A total of 144 patients received a combination of hormonal therapy and chemotherapy. The adjuvant therapy received by four patients could not be determined due to missing records. Patients were followed postoperatively every three months for the first five years, then annually. The median follow-up period was 61.7 months (range 3 to 258 months). Relapse data were available in 497 of 557 patients examined: 105 (21.1%) experienced disease recurrence; 95 (19.1%) showed distant relapse; and 58 (11.6%) had died. This protocol was approved by the Institutional Review Board of Nagoya City University Graduate School of Medical Sciences.
DNA extraction and genotyping
Genomic DNA was isolated from peripheral blood lymphocytes using a phenol-chloroform extraction method. Samples of genomic DNA were genotyped for TP53 codon 72 and MDM2 SNP309 polymorphisms. Genotyping was performed using TaqMan SNP genotyping assays. One TP53 probe (CTGCTCCCCGCGTGGCCC) was labeled at the 5'-end with VIC (Applied Biosystems' proprietary dye) and at the 3'-end with TAMRA. The other (CTGCTCCCCCCGTGGCCC) was labeled at the 5'-end with 6-carboxyfluorescein (FAM) and at the 3'-end with TAMRA. The TP53 primers were: forward, 5'-ATGAAGCTCCCAGAATGC-3', and reverse, 5'-GCCGGTGTAGGAGCT-3'. One MDM2 probe (CGCTGCGGCGCGGGA) was labeled at the 5'-end with VIC and labeled at the 3'-end with TAMRA. The other (CCGCTTCGGCGCGGGA) was labeled at the 5'-end with FAM and at the 3'-end with TAMRA. The MDM2 primers used for genotyping and sequencing were: forward, 5'-ATTTCGGACGGCTCTCGC-3', and reverse, 5'-GCGCAGCGTTCACACTAGTG-3'. Real-time TaqMan PCR and genotyping analyses were performed on an Applied Biosystems 7500 real-time PCR system (Applied Biosystems, Foster City, CA, USA) according to the manufacturer's standard PCR protocol. The results were analyzed on a 7500 real-time PCR system using the allelic discrimination assay program of Sequence Detection software version 1.3.1 (Applied Biosystems).
DNA sequencing
DNA sequencing was performed to confirm the results of TaqMan SNP genotyping assays. The primers used for sequencing of TP53 were: forward, 5'-CCCGGACGATATTGAACAATGG-3', and reverse, 5'-CAGAATGCAAGAAGCCCAGAC-3'. As mentioned above, the primers used for sequencing of MDM2 were the same as those utilized in the TaqMan genotyping assays. The PCR products were sequenced by an ABI PRISM 3100 Genetic Analyzer and analyzed by ABI SeqScape Software version 2.1.1 (Applied Biosystems).
Immunohistochemical analysis for p53
Serial sections (4 μm) for p53 staining were prepared from 289 consecutive breast cancer tissue blocks from 1983 to 1999 at Nagoya City University Hospital [30]. A monoclonal mouse antihuman p53 protein antibody (PAb1801; Novocastra, Newcastle, UK) was used at a 1:50 dilution. Bound antibody was detected with a streptavidin-biotin system (SAB-PO kit; Nichirei Co., Inc., Tokyo, Japan). Immunostaining was scored after the entire slide had been evaluated by light microscopy. The expression status of p53 was assessed according to the estimated proportion of tumor cells displaying positive nuclear staining. Scoring criteria for p53 protein expression were as follows (in the form, proportion of nuclear staining = score): none = 0, <1/10 = 1, 1/10 to 1/2 = 2, and >1/2 = 3. Tumors with a score of 1 or greater were considered to be positive for p53 protein accumulation.
Estrogen receptor measurement
Frozen tissue specimens were analyzed for ER by an enzyme immunoassay using ER EIA 'Abbott' (Dinabot, Tokyo, Japan). The cut-off ER level was less than 13 fmol/mg protein.
Statistical analyses
All molecular and immunohistochemical analyses were performed blinded to clinical data. Statistical calculations were done with StatView-J 5.0 software (SAS Institute Inc., Cary, NC, USA). The relationship between the genotype frequency of TP53 codon 72 or MDM2 SNP309 and clinicopathological factors were assessed by χ2 and Fisher's exact probability tests. Disease-free survival (DFS) curves were generated by the Kaplan-Meier method and verified by the log-rank test. Cox's proportional hazards regression analysis was used for univariate and multivariate analyses of prognostic values. Differences were considered significant when a P value < 0.05 was obtained.
Results
Distributions of TP53 and MDM2 polymorphisms
Each of the TP53 codon 72 genotypes (Arg/Arg, Arg/Pro, or Pro/Pro) and MDM2 SNP309 genotypes (T/T, T/G, or G/G) was clearly discriminated using TaqMan SNP genotyping assays. The allelic discrimination data from these assays were confirmed by direct sequencing of representative PCR products. We did not find an association between TP53 codon 72 or MDM2 SNP309 genotypes and clinicopathological features in our series (Tables 1 and 2).
Table 1.
Association between TP53 codon 72 status and clinicopathological characteristics
| Total, n (percent) | Arg/Arg, n (percent) | Arg/Pro, n (percent) | Arg/Arg +Arg/Pro, n (percent) | Pro/Pro, n (percent) | Pa | |
| Patients | 557 (100) | 63 (11) | 281 (51) | 344 (62) | 213 (38) | |
| Age (median, year) (n = 557) | 55 | 57 | 56 | 58 | ||
| Tumor size (n = 557) | 0.16 | |||||
| ≤ 2 cm | 202 (36) | 17 (27) | 99 (35) | 116 (33) | 86 (40) | |
| 2 cm to ≤ 5 cm | 286 (51) | 37 (59) | 151 (54) | 188 (55) | 98 (46) | |
| >5 cm | 57 (10) | 6 (9) | 28 (10) | 34 (10) | 23 (11) | |
| Unknown | 12 (2) | 3 (5) | 3 (1) | 6 (2) | 6 (3) | |
| Node status (n = 557) | >0.99 | |||||
| Negative | 321 (58) | 34 (54) | 167 (60) | 201 (58) | 120 (56) | |
| Positive | 198 (35) | 25 (40) | 99 (35) | 124 (36) | 74 (35) | |
| Unknown | 38 (7) | 4 (6) | 15 (5) | 19 (6) | 19 (9) | |
| Histology (n = 557) | 0.98 | |||||
| IDC | 486 (87) | 54 (86) | 246 (87) | 300 (87) | 186 (87) | |
| ILC | 20 (4) | 2 (3) | 10 (4) | 12 (4) | 8 (4) | |
| Others | 51 (9) | 7 (11) | 25 (9) | 32 (9) | 19 (9) | |
| ER status (n = 557) | 0.13 | |||||
| Positive | 344 (62) | 33 (52) | 171 (61) | 204 (59) | 140 (66) | |
| Negative | 181 (32) | 25 (40) | 95 (34) | 120 (35) | 61 (29) | |
| Unknown | 32 (6) | 5 (8) | 15 (5) | 20 (6) | 12 (5) | |
| Grade (n = 289) | 0.27 | |||||
| 1 | 71 (24) | 3 (9) | 48 (32) | 51 (28) | 20 (19) | |
| 2 | 147 (51) | 21 (62) | 67 (45) | 89 (48) | 59 (56) | |
| 3 | 54 (19) | 8 (23) | 28 (19) | 36 (19) | 18 (17) | |
| Unknown | 17 (6) | 2 (6) | 7 (4) | 9 (5) | 8 (8) | |
| p53 IHC (n = 289) | >0.99 | |||||
| Positive | 72 (25) | 15 (44) | 29 (19) | 44 (24) | 28 (27) | |
| Negative | 196 (68) | 19 (56) | 108 (72) | 127 (69) | 69 (66) | |
| Unknown | 21 (7) | 0 (0) | 13 (9) | 13 (7) | 8 (7) |
aArg/Arg and Arg/Pro genotypes compared with Pro/Pro genotype. ER, estrogen receptor; IDC, invasive ductal carcinoma; IHC, immunohistochemistry; ILC, invasive lobular carcinoma.
Table 2.
Association between MDM2 SNP309 status and clinicopathological characteristics
| Total, n (percent) | T/T, n (percent) | T/G, n (percent) | T/T + T/G, n (percent) | G/G, n (percent) | Pa | |
| Patients | 557 (100) | 111 (20) | 263 (47) | 374 (67) | 183 (33) | |
| Age (median, year) (n = 557) | 57 | 56 | 56 | 58 | ||
| Tumor size (n = 557) | 0.49 | |||||
| ≤ 2 cm | 202 (36) | 49 (44) | 88 (33) | 137 (37) | 65 (36) | |
| 2 cm to ≤ 5 cm | 286 (51) | 53 (48) | 139 (53) | 192 (51) | 94 (51) | |
| >5 cm | 57 (10) | 6 (5) | 28 (11) | 34 (9) | 23 (12) | |
| Unknown | 12 (2) | 3 (3) | 8 (3) | 11 (3) | 1 (1) | |
| Node status (n = 557) | 0.63 | |||||
| Negative | 321 (58) | 59 (53) | 159 (60) | 218 (58) | 103 (56) | |
| Positive | 198 (35) | 42 (38) | 88 (34) | 130 (35) | 68 (37) | |
| Unknown | 38 (7) | 10 (9) | 16 (6) | 26 (7) | 12 (7) | |
| Histology (n = 557) | 0.44 | |||||
| IDC | 486 (87) | 97 (87) | 233 (89) | 330 (88) | 156 (85) | |
| ILC | 20 (4) | 3 (3) | 8 (3) | 11 (3) | 9 (5) | |
| Others | 51 (9) | 11 (10) | 22 (8) | 33 (9) | 18 (10) | |
| ER status (n = 557) | 0.14 | |||||
| Positive | 344 (62) | 68 (61) | 153 (58) | 221 (59) | 123 (67) | |
| Negative | 181 (32) | 33 (30) | 95 (36) | 128 (34) | 53 (29) | |
| Unknown | 32 (6) | 10 (9) | 15 (6) | 25 (7) | 7 (4) | |
| Grade (n = 289) | 0.19 | |||||
| 1 | 71 (24) | 19 (31) | 36 (26) | 55 (27) | 16 (18) | |
| 2 | 147 (51) | 29 (47) | 66 (47) | 95 (47) | 52 (60) | |
| 3 | 54 (19) | 10 (16) | 29 (21) | 39 (19) | 15 (17) | |
| Unknown | 17 (6) | 4 (6) | 9 (6) | 13 (6) | 4 (5) | |
| p53 IHC (n = 289) | >0.99 | |||||
| Positive | 72 (25) | 19 (31) | 31 (22) | 50 (25) | 22 (25) | |
| Negative | 196 (68) | 41 (66) | 95 (68) | 136 (67) | 60 (69) | |
| Unknown | 21 (7) | 2 (3) | 14 (10) | 16 (8) | 5 (6) |
aT/T + T/G genotypes versus G/G genotype. ER, estrogen receptor; IDC, invasive ductal carcinoma; IHC, immunohistochemistry; ILC, invasive lobular carcinoma.
TP53 and MDM2 genotypes and breast cancer survival
We investigated whether these SNPs were associated with breast cancer survival. We found no association between MDM2 SNP309 status and DFS, as shown in Figure 1a. In contrast, the Pro/Pro genotype of TP53 codon 72 was significantly associated with poor DFS by Kaplan-Meier analysis, as shown in Figure 1b (P = 0.049), and multivariate Cox's proportional hazards regression analysis (P = 0.047, risk ratio of recurrence = 1.67, 95% confidence interval = 1.01 to 2.76) (Table 3). Our data indicates that the Pro/Pro genotype of TP53 codon 72 is an independent prognostic marker in breast cancer. In this analysis, we combined the Arg/Arg homozygous and the Arg/Pro heterozygous genotypes of TP53 codon 72, because the survival curve of the patients with the Arg/Arg genotype was similar to that of patients with the Arg/Pro genotype (data not shown). When the analysis was stratified by nodal status, no significant association between TP53 codon 72 genotype and DFS was seen for node-negative patients (P = 0.95; Figure 1c). In node-positive patients, however, the Pro/Pro genotype of TP53 codon 72 was significantly associated with poor DFS (P = 0.004; Figure 1d). Only 85 (26%) of 321 node-negative patients had received chemotherapy while 151 (76%) of 198 node-positive patients had. Therefore, we analyzed the relationship between breast cancer survival and this TP53 polymorphism in subgroups based on the type of adjuvant therapy.
Figure 1.
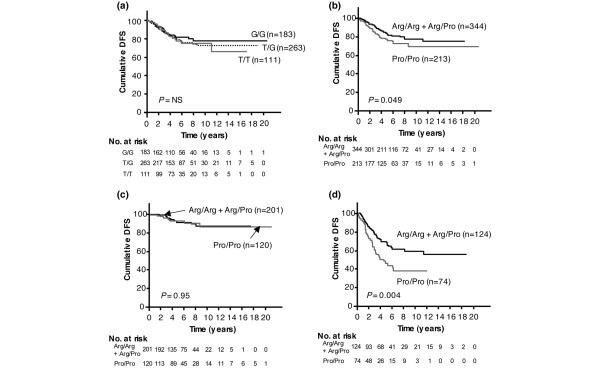
Kaplan-Meier survival curves of breast cancer patients. Disease-free survival (DFS) among patients categorized by (a) MDM2 SNP309 status and (b) TP53 codon 72 status. DFS in patients (c) without axillary lymph node involvement and (d) with lymph node metastasis according to their TP53 codon 72 status. NS, not significant.
Table 3.
Uni- and mutivariate Cox's model (disease-free survival) of prognostic factors
| Univariate | Multivariate | |||
| Variables | n | P value | P value | RR of recurrence (95 percent CI) |
| TP53 codon 72 status | 557 | |||
| Arg/Arg or Arg/Pro | 1.0 (referent) | |||
| Pro/Pro | 0.049 | 0.047 | 1.67 (1.01 to 2.76) | |
| MDM2 SNP309 status | 557 | |||
| T/T or T/G | 1.0 (referent) | |||
| G/G | 0.41 | 0.99 | 1.00 (0.57 to 1.75) | |
| Tumor size | 545 | |||
| <2 cm | 1.0 (referent) | |||
| ≥2 cm | 0.001 | 0.69 | 1.14 (0.61 to 2.12) | |
| Lymph node | 519 | |||
| Negative | 1.0 (referent) | |||
| Positive | <0.0001 | <0.0001 | 4.09 (2.35 to 7.12) | |
| ER status | 525 | |||
| Positive | 1.0 (referent) | |||
| Negative | <0.0001 | 0.002 | 2.48 (1.40 to 4.39) | |
| Grade | 272 | |||
| 1 | 1.0 (referent) | |||
| 2 or 3 | 0.009 | 0.29 | 1.50 (0.71 to 3.14) | |
| p53 IHC status | 268 | |||
| Negative | 1.0 (referent) | |||
| Positive | 0.18 | 0.14 | 0.64 (0.35 to 1.16) | |
CI, confidence interval; ER, estrogen receptor; IHC, immunohistochemistry; RR, risk ratio.
TP53 and MDM2 genotypes and adjuvant systemic therapy
Previous in vitro studies showed that cells with the TP53 codon 72 Arg/Arg genotype induced apoptosis markedly better than those with the Pro/Pro genotype [9,11]. A recent clinical study also suggested that breast cancer patients with the Pro/Pro genotype might be less sensitive to neoadjuvant chemotherapy than those with the Arg/Arg or Arg/Pro genotype [17]. Therefore, we hypothesized that this polymorphism might alter the sensitivity of tumors to adjuvant therapeutic agents. We analyzed the association between TP53 codon 72 polymorphism and DFS under different types of adjuvant systemic therapy. Our results show that, among patients who had received adjuvant chemotherapy alone (n = 137), those with the Pro/Pro genotype had poorer DFS than those with the Arg/Arg or Arg/Pro genotype, according to a Kaplan-Meier survival analysis (P = 0.009; Figure 2a). Similar results were observed when the analysis was expanded to include all patients receiving chemotherapy, with or without hormonal therapy (n = 281, P = 0.007; Figure 2b). In contrast, TP53 codon 72 genotype was not associated with DFS among patients who had received adjuvant hormonal therapy without chemotherapy (n = 195, P = 0.58; Figure 2c) nor among those who had received no adjuvant systemic therapy (n = 77, P = 0.24; Figure 2d).
Figure 2.
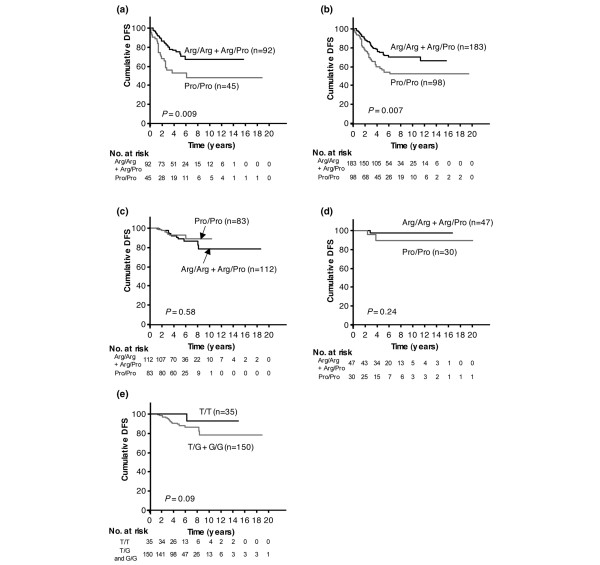
The effect of adjuvant systemic therapy on prognostic impact of polymorphisms. (a) TP53 codon 72 and adjuvant chemotherapy alone (n = 137); (b) TP53 codon 72 and adjuvant chemotherapy with or without hormonal therapy (n = 281); (c) TP53 codon 72 and adjuvant hormonal therapy alone (n = 195); (d) TP53 codon 72 and no adjuvant systemic therapy (n = 77); (e) MDM2 SNP309 and adjuvant tamoxifen with or without luteinizing hormone-releasing hormone analog (n = 185). DFS, disease-free survival.
A recent study proposed a model in which an estrogen-signaling pathway allows the G-allele of MDM2 SNP309 to accelerate breast cancer formation [28]. We hypothesized that this allele might also alter the sensitivity of tumors to adjuvant hormonal therapy. We therefore examined the correlation between MDM2 SNP309 and prognosis in patients who received adjuvant tamoxifen with or without LH-RH agonist. Although the T/T genotype tended to be associated with better DFS compared to other genotypes of SNP309 (Figure 2e), this association did not achieve significance (P = 0.09), and no statistically significant correlation was found between prognosis and MDM2 SNP309 genotype.
Discussion
We demonstrate here that SNPs of TP53 codon 72 may have an important role in breast cancer. We show that the Pro/Pro genotype at codon 72 of TP53 was associated with poor DFS, particularly in patients who received adjuvant chemotherapy while MDM2 SNP309 was not associated with prognosis.
TP53 codon 72 encodes two distinct functional allelic forms: arginine (Arg) or proline (Pro) [8]. Polymorphism at this codon has been suggested to modulate TP53-dependent apoptosis and modify sensitivity to chemotherapeutic agents [9,11,12,17]. Recent studies reported that the Pro/Pro genotype of TP53 codon 72 was associated with poorer survival in Finnish breast cancer patients [13], and suggested that MDM2 SNP309 status is associated with p53 protein function [21,22]. These findings inspired us to investigate the association of breast cancer prognosis with SNPs of TP53 codon 72 and MDM2 SNP309. We used the TaqMan SNP genotyping assay, which is amenable to high-throughput genotyping and avoids many problems of traditional genotyping assays, such as PCR-restriction fragment length polymorphism [31]. Although the TaqMan assay is convenient and reliable, it is less accurate than direct sequencing. Therefore, we analyzed our data carefully. Our results show that the Pro/Pro genotype of TP53 was associated with poorer DFS in Japanese breast cancers patients, thus supporting the Finnish study mentioned above [13]. However, the authors of that study did not address the question of why the Pro/Pro genotype of TP53 adversely affected the prognosis of breast cancer patients. Analysis of our entire set of 557 patients showed a relationship between the TP53 Pro/Pro genotype and DFS, but with a P value (0.047) at the borderline of significance, leaving doubt as to whether the Pro/Pro genotype is an independent risk factor for poor DFS.
We therefore attempted to identify subgroups in which the effect of the TP53 codon 72 was more significant. A recent study showed that breast cancer patients with the Pro/Pro genotype demonstrated less sensitivity to a neoadjuvant chemotherapy regimen that included 5-FU, cyclophosphamide, and the anthracycline derivative pirarubicin [17]. This suggested that TP53 codon 72 polymorphism might be a strong predictive marker for chemotherapy response in breast cancer patients. Our data show that the Pro/Pro genotype was associated with poor DFS in node-positive but not in node-negative patients, and 76% of node-positive patients in our series had received adjuvant chemotherapy while only 26% of node-negative patients had.
Although lymph node status is an important factor in classical staging of breast cancer based on histological/anatomical markers, there is little evidence that breast cancers with lymph node involvement are biologically different from those without it [32-34]. We therefore considered that the apparent effect of node status on the relationship between TP53 genotype and node status was due to a correlation between codon 72 polymorphism and effect of adjuvant chemotherapy, since 76% of node-positive patients in our series had received such therapy while only 26% of node-negative patients had. We thus analyzed breast cancer survival with respect to this TP53 polymorphism and the type of adjuvant therapy administered. Our data show that among patients who had received adjuvant chemotherapy, those with the Pro/Pro genotype of TP53 exhibited poorer DFS. Our finding is also consistent with a previous study of head and neck carcinoma showing that among patients who had received chemo-radiotherapy, those with the Pro/Pro genotype of TP53 showed poorer survival compared to patients with other genotypes [11].
Moreover, in ovarian cancer patients who received adjuvant cisplatinum/paclitaxel chemotherapy, the TP53 Pro allele was associated with a poorer prognosis [35]. An in vitro study showed that anticancer agents such as doxorubicin, 5-FU, and cisplatin induced a higher level of apoptosis in human H1299 cells expressing the Arg/Arg genotype of TP53 codon 72 than in those expressing the Pro/Pro genotype [11]. In addition, in a colony-survival assay, doxorubicin and cisplatin were more cytotoxic to cells expressing the Arg variant than to those expressing the Pro variant [11]. Our data are consistent with this in vitro study. The results among patients receiving both chemotherapy and hormonal therapy were similar to those among patients who had received adjuvant chemotherapy alone (data not shown), but no correlation was found between TP53 polymorphism and DFS among the patients receiving adjuvant hormonal therapy alone. Positive correlation between TP53 polymorphism and DFS was observed among patients receiving any chemotherapeutic agents. Since these compounds are cytotoxic while hormonal therapeutic agents are cytostatic, our results may reflect differential effects of TP53 polymorphism on different apoptotic pathways. Recently, ER positive patients possessing the Pro allele of TP53 codon 72 have been reported to show better distant recurrence-free survival when randomized to tamoxifen compared to those not receiving tamoxifen [18]. That report suggested that the Pro allele of TP53 codon 72 might be a predictor of tamoxifen response [18], although the authors did not present a Kaplan-Meier analysis of DFS by TP53 codon 72 genotype among patients receiving tamoxifen. We did not find any correlation between the genotype of TP53 codon 72 and prognosis in patients receiving tamoxifen alone (data not shown).
Neither did we find any statistically significant association between MDM2 SNP309 and survival in any subgroups or in the total population, although we did observe a non-significant tendency toward better DFS with the T/T genotype in patients who received adjuvant tamoxifen with or without LH-RH analog. Bond et al. [28] proposed a model in which an estrogen-signaling pathway allows the G-allele of MDM2 SNP309 to accelerate breast cancer formation. This allele might also alter the efficacy of tamoxifen, although the mechanism is unclear. A recent report showed that the G/G genotype of MDM2 SNP309 was associated with poor prognosis, as well as TP53 mutations and p53 protein immunopositivity, in gastric carcinoma [36]. TP53 alteration is correlated with shortened survival of patients with gastric carcinoma [37,38]. This suggests that the G/G genotype of MDM2 SNP309 might, therefore, be predictive of poor survival in gastric carcinoma patients. Many reports demonstrate that TP53 mutations confer a worse overall and disease-free survival in breast cancer patients [39,40], while the prognostic value of p53 protein accumulation has not been consistently demonstrated [41,42]. Although TP53 mutation status was not available in our series, we analyzed the correlation between MDM2 SNP309 and p53-immunopositivity, but did not find any association between them.
In previous reports, the frequency of the Pro/Pro genotype of TP53 codon 72 was about 7% to 30% [13,15,17,18,23,43,44], although 39.1% of patients with signet ring cell gastric carcinomas and 37.7% of female patients with lung carcinomas had the Pro/Pro genotype in subgroup analyses [15,44]. The reported frequency of the G/G genotype of MDM2 SNP309 is about 10% to 30% [23-28,36,43]. We found that 38% of patients had the Pro/Pro genotype of TP53 codon 72 and 33% had the G/G genotype of MDM2 SNP309. Therefore, the allelic discrimination data from TaqMan SNP genotyping assays were confirmed by direct sequencing of representative PCR products.
One of the goals of translational cancer research is to identify molecular predictors of chemotherapy treatment. Molecular genetic determinants of treatment outcome are important to facilitate identification of patients most likely to benefit from chemotherapy. The present results suggest that the SNP of TP53 codon 72 is a potentially useful marker. However, this is a retrospective pilot study. Further work will be needed to verify the effect of this SNP in breast cancer.
Conclusion
The genotypes of TP53 codon 72 and MDM2 SNP309 were defined among 557 primary Japanese breast cancer patients. Our data show that the Pro/Pro genotype of TP53 codon 72 appears to be an independent prognostic marker in breast cancer patients.
Abbreviations
DFS = disease-free survival; ER = estrogen receptor; FU = fluorouracil; LH-RH = luteinizing hormone-releasing hormone; SNP = single-nucleotide polymorphism.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
TT conceived the design, carried out genotyping assays and drafted the manuscript. ZZ carried out genotyping assays and DNA sequencing. MN carried out genotyping assays. MH performed the statistical analysis. NK collected the patients' data. H Iwase and ST provided study material. H Iwata participated in the study design. HY participated in the study design. YF supported the study financially. All authors read and approved the final manuscript.
Acknowledgments
Acknowledgements
This work was supported by Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan, grant #18390381.
Contributor Information
Tatsuya Toyama, Email: toyamat-ncu@umin.ac.jp.
Zhenhuan Zhang, Email: zhangz@kaiju.medic.kumamoto-u.ac.jp.
Mariko Nishio, Email: nishio_m@med.nagoya-cu.ac.jp.
Maho Hamaguchi, Email: mahoh@med.nagoya-cu.ac.jp.
Naoto Kondo, Email: nao0613@med.nagoya-cu.ac.jp.
Hirotaka Iwase, Email: hiwase@kaiju.medic.kumamoto-u.ac.jp.
Hiroji Iwata, Email: hiwata@aichi-cc.jp.
Satoru Takahashi, Email: sattak@med.nagoya-cu.ac.jp.
Hiroko Yamashita, Email: hirokoy@med.nagoya-cu.ac.jp.
Yoshitaka Fujii, Email: yosfujii@med.nagoya-cu.ac.jp.
References
- Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- Levine AJ. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/S0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- Michael D, Oren M. The p53-Mdm2 module and the ubiquitin system. Semin Cancer Biol. 2003;13:49–58. doi: 10.1016/S1044-579X(02)00099-8. [DOI] [PubMed] [Google Scholar]
- Lundgren K, Montes de Oca Luna R, McNeill YB, Emerick EP, Spencer B, Barfield CR, Lozano G, Rosenberg MP, Finlay CA. Targeted expression of MDM2 uncouples S phase from mitosis and inhibits mammary gland development independent of p53. Genes Dev. 1997;11:714–725. doi: 10.1101/gad.11.6.714. [DOI] [PubMed] [Google Scholar]
- Cordon-Cardo C, Latres E, Drobnjak M, Oliva MR, Pollack D, Woodruff JM, Marechal V, Chen J, Brennan MF, Levine AJ. Molecular abnormalities of mdm2 and p53 genes in adult soft tissue sarcomas. Cancer Res. 1994;54:794–799. [PubMed] [Google Scholar]
- Bueso-Ramos CE, Manshouri T, Haidar MA, Yang Y, McCown P, Ordonez N, Glassman A, Sneige N, Albitar M. Abnormal expression of MDM-2 in breast carcinomas. Breast Cancer Res Treat. 1996;37:179–188. doi: 10.1007/BF01806499. [DOI] [PubMed] [Google Scholar]
- Baccouche S, Daoud J, Frikha M, Mokdad-Gargouri R, Gargouri A, Jlidi R. Immunohistochemical status of p53, MDM2, bcl2, bax, and ER in invasive ductal breast carcinoma in Tunisian patients. Ann NY Acad Sci. 2003;1010:752–763. doi: 10.1196/annals.1299.136. [DOI] [PubMed] [Google Scholar]
- Matlashewski GJ, Tuck S, Pim D, Lamb P, Schneider J, Crawford LV. Primary structure polymorphism at amino acid residue 72 of human p53. Mol Cell Biol. 1987;7:961–963. doi: 10.1128/mcb.7.2.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont P, Leu JI, Della Pietra AC, 3rd, George DL, Murphy M. The codon 72 polymorphic variants of p53 have markedly different apoptotic potential. Nat Genet. 2003;33:357–365. doi: 10.1038/ng1093. [DOI] [PubMed] [Google Scholar]
- Thomas M, Kalita A, Labrecque S, Pim D, Banks L, Matlashewski G. Two polymorphic variants of wild-type p53 differ biochemically and biologically. Mol Cell Biol. 1999;19:1092–1100. doi: 10.1128/mcb.19.2.1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan A, Syed N, Gasco M, Bergamaschi D, Trigiante G, Attard M, Hiller L, Farrell PJ, Smith P, Lu X, Crook T. Polymorphism in wild-type p53 modulates response to chemotherapy in vitro and in vivo. Oncogene. 2004;23:3328–3337. doi: 10.1038/sj.onc.1207428. [DOI] [PubMed] [Google Scholar]
- Pim D, Banks L. p53 polymorphic variants at codon 72 exert different effects on cell cycle progression. Int J Cancer. 2004;108:196–199. doi: 10.1002/ijc.11548. [DOI] [PubMed] [Google Scholar]
- Tommiska J, Eerola H, Heinonen M, Salonen L, Kaare M, Tallila J, Ristimaki A, von Smitten K, Aittomaki K, Heikkila P, et al. Breast cancer patients with p53 Pro72 homozygous genotype have a poorer survival. Clin Cancer Res. 2005;11:5098–5103. doi: 10.1158/1078-0432.CCR-05-0173. [DOI] [PubMed] [Google Scholar]
- Bonafe M, Ceccarelli C, Farabegoli F, Santini D, Taffurelli M, Barbi C, Marzi E, Trapassi C, Storci G, Olivieri F, Franceschi C. Retention of the p53 codon 72 arginine allele is associated with a reduction of disease-free and overall survival in arginine/proline heterozygous breast cancer patients. Clin Cancer Res. 2003;9:4860–4864. [PubMed] [Google Scholar]
- Wang YC, Chen CY, Chen SK, Chang YY, Lin P. p53 codon 72 polymorphism in Taiwanese lung cancer patients: association with lung cancer susceptibility and prognosis. Clin Cancer Res. 1999;5:129–134. [PubMed] [Google Scholar]
- Starinsky S, Figer A, Ben-Asher E, Geva R, Flex D, Fidder HH, Zidan J, Lancet D, Friedman E. Genotype phenotype correlations in Israeli colorectal cancer patients. Int J Cancer. 2005;114:58–73. doi: 10.1002/ijc.20645. [DOI] [PubMed] [Google Scholar]
- Xu Y, Yao L, Ouyang T, Li J, Wang T, Fan Z, Lin B, Lu Y, Xie Y. p53 Codon 72 polymorphism predicts the pathologic response to neoadjuvant chemotherapy in patients with breast cancer. Clin Cancer Res. 2005;11:7328–7333. doi: 10.1158/1078-0432.CCR-05-0507. [DOI] [PubMed] [Google Scholar]
- Wegman P, Stal O, Askmalm MS, Nordenskjold B, Rutqvist LE, Wingren S. p53 polymorphic variants at codon 72 and the outcome of therapy in randomized breast cancer patients. Pharmacogenet Genomics. 2006;16:347–351. doi: 10.1097/01.fpc.0000204997.84182.69. [DOI] [PubMed] [Google Scholar]
- Sjalander A, Birgander R, Hallmans G, Cajander S, Lenner P, Athlin L, Beckman G, Beckman L. p53 polymorphisms and haplotypes in breast cancer. Carcinogenesis. 1996;17:1313–1316. doi: 10.1093/carcin/17.6.1313. [DOI] [PubMed] [Google Scholar]
- Breast Cancer Association Consortium Commonly studied single-nucleotide polymorphisms and breast cancer: results from the Breast Cancer Association Consortium. J Natl Cancer Inst. 2006;98:1382–1396. doi: 10.1093/jnci/djj374. [DOI] [PubMed] [Google Scholar]
- Bond GL, Hu W, Bond EE, Robins H, Lutzker SG, Arva NC, Bargonetti J, Bartel F, Taubert H, Wuerl P, et al. A single nucleotide polymorphism in the MDM2 promoter attenuates the p53 tumor suppressor pathway and accelerates tumor formation in humans. Cell. 2004;119:591–602. doi: 10.1016/j.cell.2004.11.022. [DOI] [PubMed] [Google Scholar]
- Arva NC, Gopen TR, Talbott KE, Campbell LE, Chicas A, White DE, Bond GL, Levine AJ, Bargonetti J. A chromatin-associated and transcriptionally inactive p53-Mdm2 complex occurs in mdm2 SNP309 homozygous cells. J Biol Chem. 2005;280:26776–26787. doi: 10.1074/jbc.M505203200. [DOI] [PubMed] [Google Scholar]
- Hong Y, Miao X, Zhang X, Ding F, Luo A, Guo Y, Tan W, Liu Z, Lin D. The role of P53 and MDM2 polymorphisms in the risk of esophageal squamous cell carcinoma. Cancer Res. 2005;65:9582–9587. doi: 10.1158/0008-5472.CAN-05-1460. [DOI] [PubMed] [Google Scholar]
- Menin C, Scaini MC, De Salvo GL, Biscuola M, Quaggio M, Esposito G, Belluco C, Montagna M, Agata S, D'Andrea E, et al. Association between MDM2-SNP309 and age at colorectal cancer diagnosis according to p53 mutation status. J Natl Cancer Inst. 2006;98:285–288. doi: 10.1093/jnci/djj054. [DOI] [PubMed] [Google Scholar]
- Boersma BJ, Howe TM, Goodman JE, Yfantis HG, Lee DH, Chanock SJ, Ambs S. Association of breast cancer outcome with status of p53 and MDM2 SNP309. J Natl Cancer Inst. 2006;98:911–919. doi: 10.1093/jnci/djj245. [DOI] [PubMed] [Google Scholar]
- Millikan RC, Heard K, Winkel S, Hill EJ, Massa B, Mayes L, Williams P, Holston R, Conway K, Edmiston S, de Cotret AR. No association between the MDM2-309 T/G promoter polymorphism and breast cancer in African-Americans or Whites. Cancer Epidemiol Biomarkers Prev. 2006;15:175–177. doi: 10.1158/1055-9965.EPI-05-0692. [DOI] [PubMed] [Google Scholar]
- Wilkening S, Bermejo JL, Burwinkel B, Klaes R, Bartram CR, Meindl A, Bugert P, Schmutzler RK, Wappenschmidt B, Untch M, et al. The single nucleotide polymorphism IVS1+309 in mouse double minute 2 does not affect risk of familial breast cancer. Cancer Res. 2006;66:646–648. doi: 10.1158/0008-5472.CAN-05-3168. [DOI] [PubMed] [Google Scholar]
- Bond GL, Hirshfield KM, Kirchhoff T, Alexe G, Bond EE, Robins H, Bartel F, Taubert H, Wuerl P, Hait W, et al. MDM2 SNP309 accelerates tumor formation in a gender-specific and hormone-dependent manner. Cancer Res. 2006;66:5104–5110. doi: 10.1158/0008-5472.CAN-06-0180. [DOI] [PubMed] [Google Scholar]
- Elston CW, Ellis IO. Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: experience from a large study with long-term follow-up. Histopathology. 1991;19:403–410. doi: 10.1111/j.1365-2559.1991.tb00229.x. [DOI] [PubMed] [Google Scholar]
- Yamashita H, Nishio M, Toyama T, Sugiura H, Zhang Z, Kobayashi S, Iwase H. Coexistence of HER2 over-expression and p53 protein accumulation is a strong prognostic molecular marker in breast cancer. Breast Cancer Res. 2004;6:R24–30. doi: 10.1186/bcr738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson VJ, Yucesoy B, Luster MI. Genotyping of single nucleotide polymorphisms in cytokine genes using real-time PCR allelic discrimination technology. Cytokine. 2004;27:135–141. doi: 10.1016/j.cyto.2004.05.002. [DOI] [PubMed] [Google Scholar]
- Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorlie T, Tibshirani R, Parker J, Hastie T, Marron JS, Nobel A, Deng S, Johnsen H, Pesich R, Geisler S, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci USA. 2003;100:8418–8423. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang HY, Nuyten DS, Sneddon JB, Hastie T, Tibshirani R, Sorlie T, Dai H, He YD, van't Veer LJ, Bartelink H, et al. Robustness, scalability, and integration of a wound-response gene expression signature in predicting breast cancer survival. Proc Natl Acad Sci USA. 2005;102:3738–3743. doi: 10.1073/pnas.0409462102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos AM, Sousa H, Portela C, Pereira D, Pinto D, Catarino R, Rodrigues C, Araujo AP, Lopes C, Medeiros R. TP53 and P21 polymorphisms: response to cisplatinum/paclitaxel-based chemotherapy in ovarian cancer. Biochem Biophys Res Commun. 2006;340:256–262. doi: 10.1016/j.bbrc.2005.11.176. [DOI] [PubMed] [Google Scholar]
- Ohmiya N, Taguchi A, Mabuchi N, Itoh A, Hirooka Y, Niwa Y, Goto H. MDM2 promoter polymorphism is associated with both an increased susceptibility to gastric carcinoma and poor prognosis. J Clin Oncol. 2006;24:4434–4440. doi: 10.1200/JCO.2005.04.1459. [DOI] [PubMed] [Google Scholar]
- Lim BH, Soong R, Grieu F, Robbins PD, House AK, Iacopetta BJ. p53 accumulation and mutation are prognostic indicators of poor survival in human gastric carcinoma. Int J Cancer. 1996;69:200–204. doi: 10.1002/(SICI)1097-0215(19960621)69:3<200::AID-IJC9>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Kubicka S, Claas C, Staab S, Kuhnel F, Zender L, Trautwein C, Wagner S, Rudolph KL, Manns M. p53 mutation pattern and expression of c-erbB2 and c-met in gastric cancer: relation to histological subtypes, Helicobacter pylori infection, and prognosis. Dig Dis Sci. 2002;47:114–121. doi: 10.1023/A:1013275706401. [DOI] [PubMed] [Google Scholar]
- Pharoah PD, Day NE, Caldas C. Somatic mutations in the p53 gene and prognosis in breast cancer: a meta-analysis. Br J Cancer. 1999;80:1968–1973. doi: 10.1038/sj.bjc.6690628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivier M, Langerod A, Carrieri P, Bergh J, Klaar S, Eyfjord J, Theillet C, Rodriguez C, Lidereau R, Bieche I, et al. The clinical value of somatic TP53 gene mutations in 1,794 patients with breast cancer. Clin Cancer Res. 2006;12:1157–1167. doi: 10.1158/1078-0432.CCR-05-1029. [DOI] [PubMed] [Google Scholar]
- Barbareschi M. Prognostic value of the immunohistochemical expression of p53 in breast carcinomas. Appl Immunohistochem. 1996;4:106–116. [Google Scholar]
- Norberg T, Lennerstrand J, Inganas M, Bergh J. Comparison between p53 protein measurements using the luminometric immunoassay and immunohistochemistry with detection of p53 gene mutations using cDNA sequencing in human breast tumors. Int J Cancer. 1998;79:376–383. doi: 10.1002/(SICI)1097-0215(19980821)79:4<376::AID-IJC12>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Sotamaa K, Liyanarachchi S, Mecklin JP, Jarvinen H, Aaltonen LA, Peltomaki P, de la Chapelle A. p53 codon 72 and MDM2 SNP309 polymorphisms and age of colorectal cancer onset in Lynch syndrome. Clin Cancer Res. 2005;11:6840–6844. doi: 10.1158/1078-0432.CCR-05-1139. [DOI] [PubMed] [Google Scholar]
- Yi SY, Lee WJ. A p53 genetic polymorphism of gastric cancer: difference between early gastric cancer and advanced gastric cancer. World J Gastroenterol. 2006;12:6536–6539. doi: 10.3748/wjg.v12.i40.6536. [DOI] [PMC free article] [PubMed] [Google Scholar]


