Short abstract
: The use of cis-Decoder, a new tool for discovery of conserved sequence elements that are shared between similarly regulating enhancers, suggests that enhancers use overlapping repertoires of highly conserved core elements.
Abstract
A systematic approach is described for analysis of evolutionarily conserved cis-regulatory DNA using cis-Decoder, a tool for discovery of conserved sequence elements that are shared between similarly regulated enhancers. Analysis of 2,086 conserved sequence blocks (CSBs), identified from 135 characterized enhancers, reveals most CSBs consist of shorter overlapping/adjacent elements that are either enhancer type-specific or common to enhancers with divergent regulatory behaviors. Our findings suggest that enhancers employ overlapping repertoires of highly conserved core elements.
Background
Tissue-specific coordinate gene expression requires multiple inputs that involve dynamic interactions between sequence specific DNA-binding transcription factors and their target DNAs. The enhancer or cis-regulatory module is the focal point of integration for many of these regulatory events. Enhancers, which usually span 0.5 to 1.0 kb, contain clusters of transcription factor DNA-binding sites (reviewed by [1-3]). DNA sequence comparisons of different co-regulating enhancers suggest that many may rely on different combinations of transcription factors to achieve coordinate gene regulation. For example, the Drosophila pan-neural genes deadpan, scratch and snail all have distinct central nervous system (CNS) enhancers that drive expression in the same embryonic neuroblasts, yet comparisons of these enhancers reveal that they have few sequences in common [4,5].
Comparative genomic analysis of orthologous cis-regulatory regions reveals that many contain multi-species conserved sequences (MCSs; reviewed by [6-8]). Close inspection of enhancer MCSs reveals that these sequences are made up of smaller blocks of conserved sequences, designated here as 'conserved sequence blocks' (CSBs). EvoPrint analysis of enhancer CSBs reveals that many have remained unchanged for over 160 million years (My) of collective divergence [9] (and see below). CSBs that are over 10 base-pairs (bp) long are likely to be made up of adjacent or overlapping sequence-specific transcription factor DNA-binding sites. For example, DNA-binding sites for transcription factors that play essential roles in the regulation of the previously characterized Drosophila Krüppel central domain enhancer [10-12] are found adjacent to or overlapping one another within enhancer CSBs [9]. Although transcription factor consensus DNA-binding sites are detected within CSBs, searches of 2,086 CSBs (27,996 total bp) curated from 35 mammalian and 99 Drosophila characterized enhancers reveal that well over half of the sequences do not correspond to known DNA-binding sites and, as yet, have no assigned function(s) (this paper).
In order to initiate the functional dissection of novel CSBs and to gain a better understanding of their substructure, we have developed a multi-step protocol and accompanying computer algorithms (collectively known as cis-Decoder; see Figure 1) that allow for the rapid identification of short 6 to 14 bp DNA sequence elements, called cis-Decoder tags (cDTs), within enhancer CSBs that are also present in CSBs from other enhancers with either related or divergent functions. There is no limit to the number of enhancer CSBs examined by this approach, which allows one to build large cDT-libraries. Due to their different copy numbers, positions and/or orientations within the different enhancers, the conserved short sequence elements may otherwise go unnoticed by more conventional DNA alignment programs. Because this approach does not rely on any previously described transcription factor consensus DNA-binding site information or any other predicted motif or the presence of overrepresented sequences, cis-Decoder analysis affords an unbiased 'evo-centric' view of shared single or multiple sequence homologies between different enhancers. The cDT-libraries and cis-Decoder alignment tools enable one to differentiate between functionally different enhancers before any experimental expression data have been collected. cis-Decoder analysis reveals that most CSBs have a modular structure made up of two classes of interlocking sequence elements: those that are conserved only in other enhancers that regulate overlapping expression patterns; and more common conserved sequence elements that are part of divergently regulated enhancers.
Figure 1.
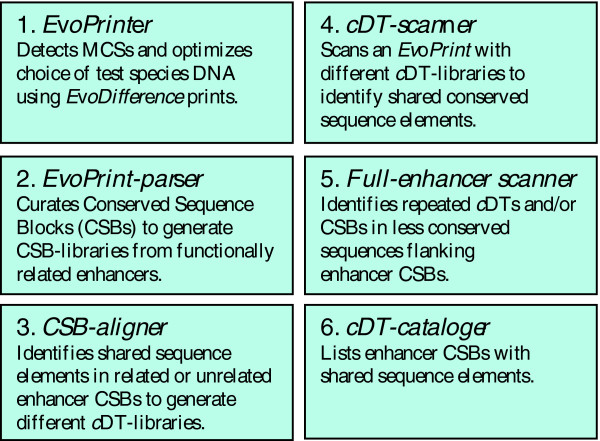
cis-Decoder methodology for identification of conserved sequence elements shared among different enhancers. The cis-Decoder methodology allows one to discover short 6 to 14 bp sequence elements within conserved enhancer sequences that are shared by other functionally related enhancers or are common to many enhancers with divergent regulatory behaviors. These shared sequence elements or cDTs can be used to identify and differentiate between cis-regulatory enhancer regions that regulate different tissue-specific expression patterns. cis-Decoder analysis involves the sequential use of the following web-accessed computer algorithms: EvoPrinter → EvoPrint-parser → CSB-aligner → cDT-scanner → Full-enhancer scanner → cDT-cataloger.
To demonstrate the efficacy of cis-Decoder analysis in identifying shared enhancer sequence elements, we show how cDT-library scans of different EvoPrinted mammalian and Drosophila enhancers accurately identify shared sequences within enhancers involved in similar regulatory behaviors. The cis-regulatory regions of the mammalian Delta-like 1 (Dll1) and Drosophila snail genes, which contain closely associated neural and mesodermal enhancers, were selected to highlight cis-Decoder's ability to differentiate between enhancers with different regulatory functions. We show how a cDT-library generated from both mammalian and Drosophila enhancer CSBs can be used to identify enhancer type-specific elements that have been conserved during the evolutionary diversification of metazoans. Finally, we show how cis-Decoder analysis can be used to examine novel putative enhancer regions.
Results and discussion
Generation of EvoPrints and CSB-libraries
Our analysis of mammalian cis-regulatory sequences included 14 neural and 21 mesodermal enhancers whose regulatory behaviors have been characterized in developing mouse embryos. A full list of enhancers used in this study and the references describing their embryonic expression patterns is given in Table 1. In most cases, their EvoPrints included orthologs from placental mammals (human, chimp, rhesus monkey, cow, dog, mouse, rat) or also included the opossum; these species afford enough additive divergence (≥200 My) to resolve most enhancer MCSs [13]. When possible, chicken and frog orthologs were also included in the EvoPrints. Except when EvoDifference profiles [9] revealed sequencing gaps or genomic rearrangements in one or more species that were not present in the majority of the different orthologous DNAs, pair-wise reference species versus test species readouts from all of the above BLAT formatted genomes [14] were used to generate the EvoPrints.
Table 1.
Enhancers analyzed
| Enhancer | Class | Reference |
| Drosophila | ||
| anterior open/yan | neur | [61] |
| atonal F:2.6 PNS | neur | [62] |
| bagpipe DS3.5 | meso | [63] |
| bearded PNS | neur | [57] |
| biparous/tap CNS | neur | [64] |
| charlatan PNS | neur | [65] |
| deadpan CNS | neur | [5] |
| deadpan PNS | neur | [5] |
| dpp 813 | meso | [28] |
| eve neuronal CNS | neur | [66] |
| eve EL CNS | neur | [18] |
| eve MES | meso | [67] |
| eve stripe 1 | seg | [18] |
| eve stripe 2 | seg | [68] |
| eve stripe 4+6 | seg | [18] |
| eve stripe 5 | seg | [18] |
| eve stripe 3+7 | seg | [69] |
| eve ftz-like | seg | [18] |
| eyeless 12 PNS | neur | [16] |
| ftz distal | meso | [70] |
| ftz proxA | meso | [70] |
| ftz CE8024 | seg | [71] |
| ftz neuro CNS | neur | [72] |
| ftz PS4* | seg | [70] |
| giant 1 | seg | [24] |
| giant 3 | seg | [24] |
| giant 6 | seg | [24] |
| giant 10 | seg | [24] |
| gooseberry-n CNS | neur | [73] |
| gooseberry GLE | neur | [74] |
| gooseberry fragIV | seg | [74] |
| hairy h7 | seg | [75] |
| hairy stripe 0 | seg | [44] |
| hairy stripe 1 | seg | [17] |
| hairy stripe 5 | seg | [76] |
| hairy stripe 3+4 | seg | [77] |
| hairy stripe 6+2 | seg | [77] |
| heartless early | meso | [78] |
| huckebein ventral | seg | [79] |
| hunchback CNS | neur | [19] |
| hunchback ant | seg | [80] |
| hunchback upstr | seg | [20] |
| knirps 5 | seg | [24] |
| Krüppel CD1 | seg | [10] |
| mir-1 | meso | [81] |
| Mef2 I-D | meso | [82] |
| Mef2 II-E | meso | [79] |
| nerfin-1 CNS | neur | AK (pers. com.) |
| odd skipped-3 | seg | [24] |
| odd skipped-5 | seg | [24] |
| paired cc | seg | [80] |
| paired O-E | seg | [44] |
| paired stripe P | seg | [83] |
| paired stripe 1 | seg | [84] |
| paired stripe 2P | seg | [83] |
| paired zebra | seg | [79] |
| pdm-1 Gap+CNS | seg/n | [84] |
| pdm-2 CE8012 | neur | [71] |
| pdp1 intron 1 | meso | [85] |
| pdp1 intron 2 | meso | [85] |
| runt stripe 1E+6 | seg | [86] |
| runt stripe 1+7 | seg | [86] |
| runt stripe 3+7 | seg | [86] |
| runt stripe 5 | seg | [86] |
| runt 15G CNS | neur | [86] |
| Schizo/loner PNS | neur | [65] |
| scratch sA | neur | [5] |
| scratch PNS | neur | [5] |
| Scr 3.0RR | meso | [23] |
| Scr 7.0RR | meso | [23] |
| Scr 8.2XX | meso | [23] |
| scute SCM | neur | [87] |
| serpent-A7.1EB | meso | [22] |
| snail CNS | neur | [4] |
| snail PNS | neur | [4] |
| snail MES | meso | [4] |
| string b-5.8 CNS | neur | [88] |
| teashirt del-1-5 | meso | [89] |
| tinman B | meso | [21] |
| tinman C | meso | [21] |
| tinman D | meso | [21] |
| toll-6.5RL | meso | [90] |
| β-tub 56DAS1 | meso | [91] |
| Tropomyosin1-M | meso | [92] |
| Tropomyosin1-P | meso | [92] |
| twist-del | meso | [48] |
| vnd CNS | neur | [93] |
| vnd A CNS | neur | [93] |
| wor CNS | neur | [94] |
| Mammalian | ||
| bagpipe Hox 1 | meso | [95] |
| Cbfa1 non-coding | meso | [96] |
| Dll1 HI CNS | neur | [35] |
| Dll1 HII CNS | neur | [35] |
| Dll1 msd | meso | [35] |
| Dll1 msd II | meso | [35] |
| forkhead box f1 | meso | [97] |
| Gata6 | meso | [38] |
| dHAND | meso | [98] |
| Hes 7 | meso | [99] |
| HoxA-5 | meso | [100] |
| H. domain only | neur | [101] |
| IA-1 CNS | neur | [102] |
| α7 integrin | meso | [103] |
| Mef2c | meso | [104] |
| Mash1 CNS | neur | [105] |
| Math1 CNS | neur | [27] |
| Myogenic factor-5 | meso | [106] |
| Nestin CNS | neur | [107] |
| Nfatc1 | meso | [108] |
| Neurogenin 2:5' | neur | [109] |
| Neurogenin 2:3' | neur | [109] |
| Nkx-2.5 | meso | [110] |
| Otx 2 CNS | neur | [111] |
| Pax 3 | meso | [112] |
| Phox2b CNS | neur | [25] |
| Serum response f | meso | [113] |
| Six2 | meso | [114] |
| Sox-2 CNS | neur | [115] |
| Sox-2 #2 CNS | neur | [116] |
| Sox 9p CNS | neur | [37] |
| Stem cell leukemia | meso | [117] |
| Tbx1 | meso | [118] |
| Wnt-1 | neur | [36] |
Meso, mesodermal; neur, neural; seg, segmental.
Using the EvoPrint-Parser program, both forward and reverse-complement sequences of each enhancer CSB of 6 bp or greater were extracted, named and consecutively numbered. Based on their enhancer regulatory expression pattern, CSBs were grouped into two different CSB-libraries, neural and mesodermal (Tables 1 and 2). Although there exists a distinction between expression in either neural or mesodermal tissues, each of the CSB-libraries represent a heterogeneous population of enhancers that drive gene expression in different cells and/or different developmental times in these tissues. For this study, CSBs of 5 bp or less were not included in the analysis. Although these shorter CSBs, particularly the 5 and 4 bp CSBs, are most likely important for enhancer function, the use of CSBs of 6 bp or larger (representing greater than 80% of the conserved MCS sequences) is sufficient to resolve sequence element differences between enhancers that regulate divergent expression patterns (see below). A total of 286 neural CSBs and 289 mesodermal CSBs were extracted from the mammalian enhancers (Table 2).
Table 2.
cis-Decoder libraries
| cis-Decoder tag libraries | cDTs | Enhancers | CSBs/Total bp |
| Mammalian/vertebrate | |||
| Neural specific | 336 | 14 | 286/4,162 |
| Mesodermal specific | 258 | 21 | 289/3,749 |
| Common | 137 | 35 | 575/7,911 |
| Neural enriched* | 60 | 35 | 575/7,911 |
| Mesodermal enriched* | 55 | 35 | 575/7,911 |
| Drosophila | |||
| Neural specific | 444 | 36 | 601/8,002 |
| Segmental specific | 284 | 38 | 513/6,608 |
| Mesodermal specific | 169 | 25 | 398/5,469 |
| Neural and segmental | 451 | 75 | 1,114/14,610 |
| Neural/segmental enriched* | 277 | 100 | 1,511/20,085 |
| Mesodermal enriched* | 104 | 63 | 1,511/20,085 |
| Common | 993 | 100 | 1,511/20,085 |
| Drosophila /mammalian/vertebrate | |||
| Neural specific | 873 | 50 | 887/12,164 |
| Mesodermal specific | 445 | 46 | 687/9,218 |
* cDTs have a ≥75% correspondence to a specific library but are also present at a low frequency in unrelated enhancers.
For Drosophila, three CSB-libraries, neural, segmental and mesodermal, were generated from CSBs identified by EvoPrinting (Tables 1 and 2): neural enhancers included those regulating both CNS and peripheral nervous system (PNS) determinants; segmental enhancers included those regulating both pair-rule and gap gene expression; and mesodermal enhancers included those regulating both presumptive and late expression. Many of the D. melanogaster reference sequences used to initiate the EvoPrints were curated from the regulatory element database REDfly [15], while others were identified from their primary reference (Table 1). The collection of neural enhancers includes both those that direct expression during early development, such as the snail [4], scratch, and deadpan CNS and PNS enhancers [5], and late nervous system regulators, such as the eyeless enhancer ey12 [16], which confers expression in the adult brain. The early embryonic segmental enhancers represent pair-rule regulators such as the hairy stripe 1 [17] and even-skipped stripe 1 [18] enhancers, and gap expression regulators, such as the hunchback enhancers [19,20]. The mesodermal enhancers include those directing mesodermal anlage expression of snail [4] and tinman [21], and late expressing enhancers, such as those directing serpent fat body expression [22] and mesodermal expression of Sex combs reduced [23]. The collective evolutionary divergence of all of the EvoPrints was greater than 100 My and in most cases EvoPrints represented over approximately 160 My of additive divergence. The average CSB length for both the Drosophila and mammalian CSBs is 13 bp; the longest identified CSBs were 99 bp from the giant (-10) segmental enhancer [15,24] and 95 bp from the Paired-like homeobox-2b mammalian neural enhancer [25]. Complete lists of all CSBs identified in this study are given at the cis-Decoder website [26].
Identification and use of cis-Decoder tags
As an initial step toward understanding the nature of the CSB substructure, we have developed a set of DNA sequence alignment tools, known collectively as cis-Decoder, that allow identification of 6 bp or greater perfect match identities, called cDTs, within two or more CSBs from either similar or divergent enhancers. The cDTs, which range in size from 6 to 14 bp with an average of 7 or 8 bp, are organized into cDT-libraries that identify sequence elements within CSBs of the same CSB-library. In addition, common cDT-libraries that represent sequence elements aligning to CSBs of two or more different CSB-libraries were also organized.
Mammalian CSB alignments, using the CSB-aligner program, yielded 336 neural specific and 60 neural-enriched cDTs and analysis of the mammalian mesodermal CSBs yielded 258 mesodermal specific and 55 mesodermal enriched cDTs (Table 2). The CSB alignments also produced 137 cDTs that are common to both neural and mesodermal CSBs. Alignments of the Drosophila enhancer CSBs yielded 444 neural specific cDTs (showing no hits on mesodermal or segmental enhancer CSBs), 284 segmental enhancer specific cDTs and an additional 451 cDTs found in neural and segmental enhancers but not part of mesodermal CSBs (Table 2). We also identified 451 cDTs that were enriched in neural and/or segmental CSBs but were also found at a lower frequency in mesodermal enhancer CSBs. From the mesodermal CSBs analyzed, 169 mesodermal specific cDTs (not in neural or segmental enhancer CSBs) were identified along with 104 additional cDTs enriched in mesodermal enhancers but also found at a lower frequency among neural and/or segmental enhancer CSBs. A common cDT-library was also generated that contains 993 cDTs that represent common sequence elements found in CSBs of both neural and mesodermal enhancers.
To search for enhancer sequence element conservation between taxa, we generated neural and mesodermal cDT-libraries from the combined alignments of mammalian and fly CSBs (Table 2) and many of the cDTs in these libraries align to both mammalian and fly CSBs. For example, the 11 bp neural specific cDT (CAGCTGACAGC) aligns with CSBs in the vertebrate Math-1 [27] and Drosophila deadpan [5] early CNS enhancers. All CSB-, cDT-libraries and alignment tools are available at the cis-Decoder website.
The constituent sequence elements of the different cDT-libraries are dependent on the enhancers used to identify them. As additional CSBs are included in the cDT-library construction, certain cDTs may be re-designated. For example, some that are currently considered neural specific will be discovered to be neural enriched, and others that are part of enriched libraries may be reassigned to common cDT-libraries.
Although each mammalian and fly cDT is present in at least two or more enhancers, most are not found as repeated sequences in any of the enhancers. In addition, one of the principle observations of our analysis is that enhancers of similarly regulated genes share different combinatorial sets of elements that are enhancer-type specific (see below).
Cross-library CSB alignments revealed that nearly all CSBs contain cDTs that are either shared by CSBs from divergent enhancer types or found only in CSBs from enhancers with related regulatory functions. For example, the 37 bp neural mastermind #10 CSB (TATTATTACTATATACAATATGGCATATTATTATTAC) contains a 9 bp sequence (first underlined sequence) also found in the 20 bp #8 CSB from the dpp mesodermal enhancer [15,28] and it also contains a 14 bp sequence (second underlined sequence) that constitutes the entire 14 bp #33 CSB from the neural enhancer region of nerfin-1 ([29] and unpublished results).
The analysis of both the mammalian and fly common cDT-libraries reveals that many cDTs contain core recognition sequences for known transcription factors. However, when additional flanking CSB sequences are considered, many common transcription factor binding sites become tissue specific cDTs. For example, the DNA-binding site for basic helix-loop-helix (bHLH) transcription factors, the E-box motif CAGCTG (reviewed by [30]) is present 22 times in different neural CSBs, and 2 and 4 times within the CSBs of segmental and mesodermal enhancers, respectively. However, when flanking sequences are included in the analysis, such as the sequences CAGCTGG, CAGCTGAT, CAGCTGTG, CAGCTGCA, CAGCTGCT and ACAGCTGCC, all are neural specific cDTs (E-box underlined). It has been previously shown that different E-boxes bind different bHLH transcription factors to regulate different neural target genes [31]. Although transcription factor consensus DNA-binding sites are well represented in the cDT-libraries, greater than 50% of the cDTs in all of the libraries, both mammalian and fly, represent novel sequences whose function(s) are currently unknown. The fact that there exists such a high percentage of novel sequences within these highly conserved sequences indicates that the identity, function and/or the combinatorial events that regulate enhancer behavior are as yet unknown.
cis-Decoder analysis of the murine Delta-like 1 enhancers identifies multiple shared elements with other related vertebrate embryonic enhancers
Although the resolution of cis-Decoder analysis increases as more enhancers and/or enhancer types are included in the CSB and cDT alignments, our analysis of mammalian enhancers found that many shared sequence elements can be identified among related enhancers when as few as two different enhancer groups are used to generate specific cDT-libraries. This is a particularly useful feature of cis-Decoder, especially when studying a biological process or developmental event where relatively little is known about the participating genes and their controlling enhancers. To demonstrate the ability of cis-Decoder to analyze relatively small subsets of enhancers, we show how cDT-libraries generated from 14 neural and 21 mesodermal mammalian enhancers can be used to distinguish between the neural and mesodermal enhancers that regulate embryonic expression of Dll1.
Dll1 encodes a Notch ligand that is essential for cell-cell signaling events that regulate multiple developmental events (reviewed by [32]). Studies in the mouse reveal that Dll1 is dynamically expressed in specific regions of the developing brain, spinal cord and also in a complex pattern within the embryonic mesoderm [33,34]. The 1.6 kb Dll1 cis-regulatory region, located 5' to its transcribed sequence, has been shown to contain distinct enhancers that direct gene expression in these different tissues [35]. These studies have identified two highly conserved neural enhancers, designated Homology I (H-I) and Homology II (H-II), and two mesodermal enhancers termed msd and msd-II. The H-I enhancer directs expression to the ventral neural tube, while the H-II enhancer primarily drives Dll1 expression in the marginal zone of the dorsal region of the neural tube [34]. The msd enhancer drives expression in paraxial mesoderm, and msd-II directs Dll1 expression to the presomitic and somitic mesoderm.
An EvoPrint of the Dll1 cis-regulatory region reveals clustered CSBs in each of the enhancer regions (Figure 2). Here, EvoPrint analysis used mouse (reference DNA), human, rhesus monkey, cow, rat, opossum and Xenopus tropicalis orthologs, representing over approximately 240 My of collective evolutionary divergence. EvoPrint-parser CSB extraction of the EvoPrint generated a total of 35 CSBs of 6 bp or longer, representing 83% of the total MCS. A cDT-scan of the four Dll1 enhancer regions using the mammalian neural and mesodermal specific cDT-libraries accurately differentiates between the neural and mesodermal enhancers (Figure 3; note intra-CSB sequences are not shown). The cDT-library scan identified 77 type-specific sequence elements within the Dll1 CSBs and over half (52%) align with three or more CSBs from different enhancers, indicating that, even if Dll1 had been excluded from the analysis that generated the specific cDT-libraries, there would still be extensive coverage of the Dll1 CSBs by type-specific cDTs. All but eight of the CSBs contain elements that align with one or more neural or mesodermal specific cDTs. The H-I and H-II early CNS enhancers exhibited 64% and 43% coverage, respectively, by neural specific cDTs. The CSBs of the two mesodermal enhancers, msd and msd-II, exhibited 48% and 56% coverage, respectively, by one or more mesodermal specific cDTs. When common cDTs, shared by mesodermal and neural enhancers, were taken into account, coverage of all four enhancers was 81% (data not shown).
Figure 2.

EvoPrint analysis of vertebrate Delta-like 1 enhancers. An EvoPrint of the vertebrate Dll1 cis-regulatory region generated from the following genomes: mouse (reference sequence), human, rhesus monkey, cow, rat, opossum and Xenopus tropicalis. Shown is the first codon (ATG) and 4,265 bp of upstream 5' flanking sequence of the mouse Dll1 gene containing, in 5' → 3' order, respectively, the Homology-I neural enhancer region (304 bp), the msd mesodermal enhancer (a 1,495 bp FokI restriction fragment), the Homology-II neural enhancer (207 bp fragment) and the msd-II mesodermal enhancer (1,615 bp HindIII restriction fragment) as described [35]. Multi-species conserved sequences within the murine DNA, shared by all orthologous DNAs that were used to generate the EvoPrint, are identified with uppercase black-colored letters and less or non-conserved DNA are denoted by lowercase gray-colored letters. Note that the chimpanzee, dog and chicken genomes were excluded from the analysis due either to sequence breaks and/or sequencing ambiguities as detected by EvoDifference profiles.
Figure 3.

cDT-scanner analysis of vertebrate Delta-like 1 enhancers. Alignment of vertebrate neural and mesodermal specific cDTs with the Dll1 upstream CSBs identifies its neural and mesodermal enhancers. Dll1 CSBs of 6 bp or greater were curated using the EvoPrint-parser from the EvoPrint shown in Figure 2 and aligned with cDTs from the vertebrate neural and mesodermal cDT-libraries described in Table 2. Designations adjacent to the aligned cDTs indicate the number of perfect matches to CSBs within neural (n) or mesodermal (m) enhancers analyzed in this study. Transcription factor DNA-binding site searches of the Delta-like 1 CSBs and their aligning cDTs revealed that many contained putative binding sites and, in several cases, the shared sequence elements correspond exactly to, or had significant sequence overlap with, the characterized binding sites. For example, several cDTs that align to H-I enhancer CSBs correspond to known binding sites: these include a YY1 binding site (GCCATTT), an E-box (CAGATG; reviewed by [30]), a variant Oct1 site (ATGAAAAT) and a predicted core Lef-1 binding site (underlined) within a cDT (GCAAAGA). Within H-II conserved sequences, one common and one neural specific cDT aligned with the E-boxes (CAGGTG and CAGCTG), respectively.
cDT-cataloger analysis of aligning cDTs with H-I and H-II early CNS enhancers revealed that the H-I enhancer shares a remarkable 9 different sequence elements with the Wnt-1 early CNS neural plate enhancer CSBs [36], representing 62 bp (32%) of the H-I CSB coverage, 7 elements with the Paired-like homeobox-2b (Phox2b) hindbrain-sensory ganglia enhancer CSBs (23% coverage) and 6 sequence elements (20% coverage) with the Sox9p hindbrain-spinal cord enhancer CSBs [37] as well as numerous other neural specific elements in common with CSBs of other neural enhancers (Figure 4; Additional data file 1). Comparisons of Dll1 H-I, Wnt-1, Phox2b and Sox9p enhancer CSBs reveal that the orientation and order of the shared cDTs are unique for each of the enhancers (data not shown). The H-I and H-II enhancer CSBs also share the 7 bp sequence element GCTCCCC, and H-I has a repeat sequence element (AGTTAAA) that is present in two of its CSBs (#11 and #13). The conserved AGTTAAA repeat is also part of a CSB in Phox2b enhancer [25]. cDT-cataloger analysis of the mesodermal enhancer cDT hits (Figure 4; Additional data file 1) reveal that, together, msd and msd-II share 7 elements in common with the mesodermal enhancer of Nkx2.5 [38] as well as numerous elements in common with CSBs of other mesodermal enhancers (Figure 2; Additional data file 1).
Figure 4.

cDT-cataloger analysis of vertebrate cDTs that align with the Delta-like 1 Homology I and msd enhancers. cDT-cataloger analysis identifies other neural and mesodermal enhancers with shared sequence elements. Homeodomain protein DNA binding sites (ATTA) and bHLH binding sites known as E-boxes (CAGATG) are underlined. Analysis of Dll1 Homology II and msd2 enhancers is given in Additional data file 1.
Previous cross-taxa comparative studies have demonstrated that, in many cases, the regulatory circuits controlling the spatial-temporal regulatory activities of certain enhancers have been conserved over large evolutionary distances (discussed in [1]). For example, the Deformed autoregulatory element from Drosophila functions in a conserved manner in mice [39] and its human ortholog, the Hox4B regulatory element, provides specific expression in Drosophila [40]. Given this degree of conservation, we reasoned that cDT-libraries built from the combined alignments of enhancer CSBs from both mammalian and Drosophila CSB-libraries would lead to the discovery of additional enhancer type-specific sequence elements and thereby enhance our understanding of the relationship between evolutionarily distant enhancers (Table 2). By including all of the neural enhancer CSBs (286 mammalian and 601 Drosophila) in the CSB alignments, the total number of neural specific cDTs increased to 873 compared to 336 mammalian and 322 Drosophila neural specific cDTs (Table 2). The combined mesodermal specific cDT-library (Table 2) also increased compared to the individual mammalian and fly libraries. The combined mammalian and fly neural and mesodermal specific cDT-libraries contain cDTs that align with both mammalian and fly CSBs and cDTs that align exclusively with only mammalian or fly CSBs. Whether the 'cross-taxa' cDTs indicate significant functional overlap remains to be tested. However, a cDT-scan of the EvoPrinted Dll1 cis-regulatory region, using the cross-taxa libraries, identifies multiple conserved sequence elements that are shared with CSBs from functionally related fly enhancers (Figure 5), suggesting that many of the core cis-regulatory elements that participate in enhancer function are conserved across taxonomic divisions.
Figure 5.
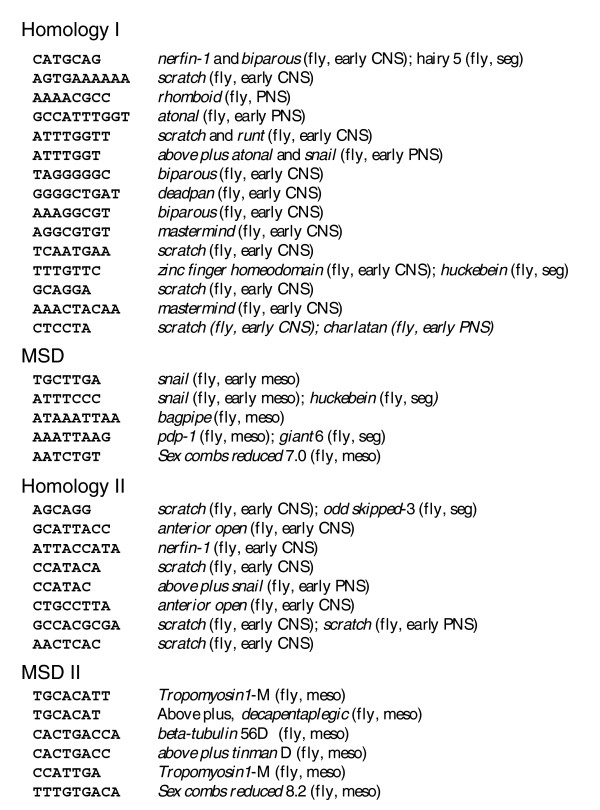
cDT-cataloger analysis of the Delta-like 1 upstream cDT hits using the combined mammalian and fly cDT-libraries. cDT-cataloger analysis using the combined mammalian and fly cDT-libraries (both neural and mesodermal specific libraries) identifies multiple Dll1 enhancer sequence elements (6 to 10 bp in length) that are shared among fly and mammalian enhancer CSBs. Note, only cDTs that align to Drosophila CSBs are shown.
cis-Decoder identifies sequence elements within the Drosophila snail and hairy stripe 1 enhancers that are also conserved in other functionally related tissue-specific enhancers
To demonstrate the ability of cis-Decoder to differentiate between Drosophila neural and mesodermal enhancers, we show an analysis of the snail upstream cis-regulatory region. The enhancers that regulate snail's dynamic embryonic expression have been mapped to a 2,974 bp upstream DNA fragment [4,41]. An EvoPrint of this sequence reveals that each of the restriction fragments that contain the different enhancer activities (CNS, mesodermal and PNS) harbor clusters of highly conserved CSBs (Figure 6). The combined evolutionary divergence of the snail upstream EvoPrint (generated from Drosophila melanogaster, D. sechellia, D. yakuba, D. erecta, D. ananassae, D. pseudoobscura, D. mojavensis, D. virilis and D. grimshawi orthologous sequences) is approximately 160 My, suggesting that many, if not all, of the identified CSBs are likely to be genus invariant and that each base-pair within a CSB has been evolutionarily challenged.
Figure 6.
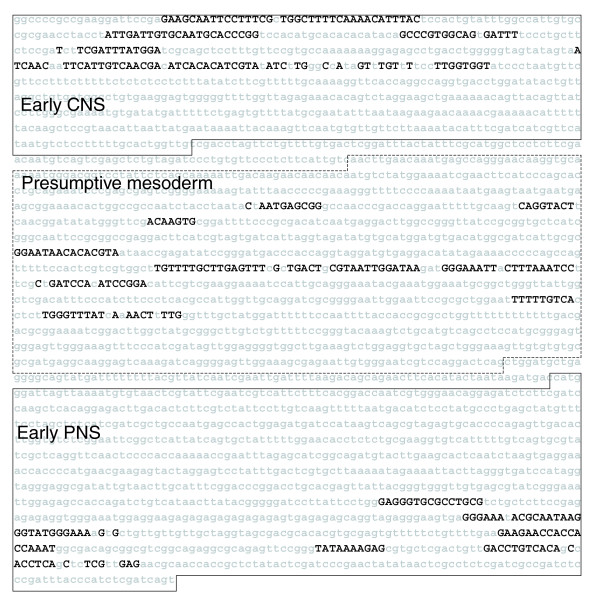
EvoPrint analysis of the Drosophila snail cis-regulatory region. An EvoPrint of the Drosophila snail upstream early CNS, presumptive mesodermal, and early PNS enhancer regulatory region (2,974 bp) [4,41] was generated using the following genomes: D. melanogaster (reference sequence), D. sechellia, D. yakuba, D. erecta, D. ananassae, D. pseudoobscura, D. virilis, D. mojavensis and D. grimshawi. Due to breaks in co-linearity, sequencing gaps and/or sequencing ambiguities, as detected by EvoDifference analysis, D. simulans and D. persimilis were not included in the analysis. Invariant MCSs, shared by all species, are identified with uppercase black-colored letters. The three previously identified genomic restriction fragments [4] containing the CNS, mesodermal and PNS enhancers are highlighted by solid lines for neural enhancers and dotted lines for the mesodermal enhancer.
To identify sequence elements within the snail upstream CSBs that are present in CSBs of other functionally related or unrelated enhancers, we carried out a cDT-scan of the snail EvoPrint using the neural, segmental and mesodermal specific cDTs and the enriched cDT-libraries (Figure 7). Within the snail early CNS neuroblast enhancer region, our cDT-library scan identified 22 different neural and neural/segmental cDT hits, distributed among all but one of the CSBs, covering 73% of the CSBs. Interestingly, 10 of the 22 cDTs that align with the early CNS enhancer CSBs are found in CSBs of both neural and segmentation enhancers. The high percentage of neural/segmental cDT hits most likely reflects the fact that this enhancer initially drives snail expression in the neuroectoderm in a pair-rule pattern and then in a segmental pattern corresponding to the first wave of delaminating neuroblasts [4]. cDT-cataloger analysis of the aligning cDTs reveals that many of the identified sequence elements are also part of other early neuroblast enhancer CSBs. For example, the 9 bp cDTs ATTCCTTTC, ATTGATTGT, ATTGTGCAA, TGCAATGCA and GATTTATGG are also present, respectively, in CSBs from the nerfin-1, biparous, string, scratch and worniu neuroblast enhancers (Figure 8; see Table 1 for references).
Figure 7.
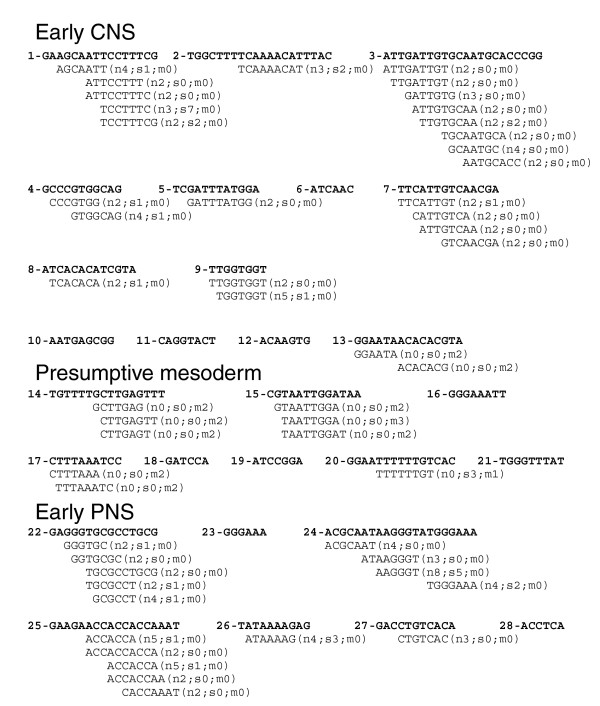
cDT-Scanner analysis of the Drosophila snail enhancer region. cDT-library scan of the snail enhancer region CSBs accurately differentiates between the neural, mesodermal and early PNS enhancers. Shown, in order of appearance within the EvoP, are 6 bp and greater CSBs aligned to cDTs from either the neural, segmentation or mesodermal cDT-libraries (described in Table 2). Designations adjacent to the aligned cDTs include number of perfect matches to neural (n), segmentation (s) and to mesodermal (m) enhancer CSBs analyzed in this study (enhancers used to generate cDT-libraries are listed in Table 1).
Figure 8.

cDT-cataloger analysis of the Drosophila snail enhancers. cDT-cataloger analysis reveals that the different enhancers share sequence elements with the snail CNS, presumptive mesoderm, and PNS enhancers. Shown are cDTs identified in the cDT-scan (Figure 7) followed by the different enhancers that also contain the sequence in one or more of their CSBs (see Table 1 for enhancer references).
Within the presumptive mesodermal enhancer CSBs, 11 cDTs mesodermal specific aligned with 5 of the 12 CSBs, covering 40% of the CSBs (Figure 7). Like the neural cDTs, some of the mesodermal cDTs contain putative DNA-binding sites for classes of known transcription factor families. For example, the seventh cDT (TAATTGGA) contains a consensus core DNA-binding sequence (underlined) for Antennapedia class homeodomain factors [42] (reviewed by [43]).
In the snail early PNS enhancer region, 5 of the 7 CSBs aligned with a total of 15 different cDTs that cover 69% of the total PNS CSB sequence (Figure 7). Similar to the CNS enhancer CSB cDT alignments, close to half of the PNS cDT hits represent sequence elements within both neural and segmental enhancer CSBs, again most likely a reflection of the segmental structure of the PNS. The significant overlap in cDTs found in both CNS and PNS enhancer CSBs may reflect the likelihood that many early neural specific transcriptional regulatory factors are pan-neural.
Many of the snail enhancer CSB-cDT hits represent sequences found only in two CSBs, snail itself and one other. In these instances it appears that these elements, although specific for neural or mesodermal CSBs, are relatively rare when compared to others. Only through analysis of additional enhancers will it be clear whether these rare elements are indeed type-specific or only enriched in the type-specific CSBs. Nevertheless, the fact that the sequence elements identified by these rare cDTs are conserved in two distinct enhancer CSBs that have both been under positive selection for over 160 My of collective divergence merits their inclusion in the analysis.
As part of our study of Drosophila enhancers, we carried out cis-Decoder analysis of 38 segmentation enhancers responsible for both gap and pair-rule gene expression during Drosophila embryogenesis. Although the segmentation enhancer specific library consisted of only 284 cDTs, these cDTs aligned with over 70% of bases of the CSBs of segmentation enhancers. As an example of alignment of these cDTs with a segmental enhancer, we present an alignment of segmentation specific cDTs with the hairy stripe 1 enhancer (Additional data file 2). cis-Decoder recognizes highly conserved Abdominal-B, HOX, Hunchback, Kruppel and Tramtrack binding sites, as well as additional uncharacterized sites, as being shared by hairy stripe 1 enhancer and other segmentation enhancers.
Full-enhancer scanner identifies less conserved repeated cDTs and CSBs
Previous studies have demonstrated that certain enhancers, particularly those controlling the dynamic expression of developmental genes, contain clusters of DNA-binding site motifs for specific transcription factors (for example, see [44,45]; reviewed by [46]). Comparative genomic studies of orthologous enhancers have also revealed that, within a binding site cluster, individual DNA-binding sites can undergo turnover (discussed in [47,48]). This loss of and/or gain of transcription factor docking sites during evolution suggests that the repeated motifs may be functionally redundant and that the stability of any one binding site is most likely due to selective pressure(s) to maintain: total number of binding sites for tight spatial/temporal regulation; functional interactions between a bound factor and adjacent factors and/or; competition between antagonistic regulatory factors for overlapping binding sites. For example, overlapping/linked binding sites have been identified in the 3' most CSB of the Krüppel central domain enhancer [9,10]. The 15 bp CSB (CTGAACTAAATCCG) contains overlapping sites for the transcriptional activator Bicoid and repressor Knirps proteins [11]. In vivo experiments reveal that these interlocking sites are functionally important [12]. Additional binding sites for both of these factors are also present in the Krüppel enhancer but not all are found in CSBs (data not shown).
The Full-enhancer scanner is used to identify less conserved repeated cDTs by rescanning the entire enhancer sequence with the aligning cDTs. For example, a Full-enhancer scan of the even-skipped stripe 1 enhancer with its aligning cDTs reveals that the #15 CSB (AATCCTTTCG) is present two additional times within the intra-CSB sequences (Figure 9). Interestingly, this CSB contains the consensus binding sequence for Tramtrack (underlined), a regulator of segmental gene expression [49]. EvoDifference analysis reveals that the 5' most inter-block (AATCCTTTCG) is conserved in all Drosophila species except D. ananassae and the 3' inter-block repeat is absent in six of the ten species used to generate the EvoPrint (data not shown).
Figure 9.

Full enhancer scanner analysis identifies less conserved sequences that are also part of conserved sequence blocks. The following Drosophila species were used to produce an EvoPrint of the Drosophila melanogaster 800 bp even-skipped stripe #1 enhancer [18]: D. melanogaster (reference sequence), D. simulans, D. sechellia, D. erecta, D. ananassae, D. persimilis, D. pseudoobscura, D. virilis, D. mojavensis and D. grimshawi. Drosophila yakuba was not included in the EvoPrint analysis due to lack of sequence co-linearity detected with EvoDifference prints. Invariant MCSs, shared by all species used to generate the EvoPrint, are identified with uppercase black-colored letters. A Full-enhancer scan of the enhancer with one of its 10 bp CSBs (blue highlight) revealed that it is repeated two additional times in the less conserved inter-block sequences (lowercase yellow highlighted sequences). Note that the underlined sequence in this CSB is the core DNA-binding sequence for the Tramtrack transcription factor.
Use of cis-Decoder to examine novel cis-regulatory sequences
One major use of the cis-Decoder methodology is the comparative analysis of different enhancer regions. To test cis-Decoder's efficacy in characterizing putative cis-regulatory regions that were not included in the preparation of the cDT-libraries, we have examined a number of genes both in Drosophila and vertebrates using EvoPrinter and cDT-library scans. Our analysis reveals that putative enhancer regions associated with CNS-expressed genes align with a higher proportion of neural-specific cDTs than with mesodermal-specific cDTs. For example, cis-Decoder analysis of the immediate upstream regions from Drosophila E(spl) region transcript mβ (HLHmβ) [50] and of the human gene encoding Tuberoinfundibular peptide of 39 residues (TIP39) [51-53] revealed that both of these neural expressed genes had significant coverage by neural-specific cDTs of their proximal cis-regulatory region CSBs. Figure 10 shows cis-Decoder analysis of HLHmβ, while our analysis of TIP39 is presented in Additional data file 3.
Figure 10.

cis-Decoder analysis of the Drosophila HLHmβ 5' upstream cis-regulatory region. cis-Decoder analysis of the Drosophila HLHmβ upstream region identifies neural enhancer sequences. (a) An EvoPrint of the 869 bp Drosophila HLHmβ cis-regulatory region [54] was generated using the following genomes: D. melanogaster (reference sequence), D. simulans, D. sechellia, D. yakuba, D. erecta, D. ananassae, D. persimilis, D. pseudoobscura, D. virilis, D. mojavensis and D. grimshawi. Uppercase nucleotide sequences are conserved in all of the above genomes. (b) cis-Decoder tag analysis of the HLHmβ enhancer CSBs. CSBs (6 bp or greater) were extracted from the EvoPrint shown in (a) and aligned with Drosophila cDTs from neural and mesodermal libraries. Designations adjacent to the aligned cDTs include number of perfect matches to neural (n) and mesodermal (m) enhancers analyzed in this study. (c) cDT-cataloger analysis of the aligning cDTs reveal that the HLHmβ enhancer contains elements shared with 26 other neural enhancer CSBs and one mesodermal CSB.
During embryonic development, HLHmβ expression is activated in the ventral neurogenic ectoderm immediately prior to neuroblast delamination [50,54] and enhancer-reporter constructs from the HLHmβ enhancer region [55] are expressed in proneural territories in the ventral ectoderm at the time of the first wave of neuroblast delamination(stages 9-10) and in neuroblasts (Figure 1 of [55]). Our EvoPrint analysis of the 883 bp enhancer region (Figure 10a) revealed that 338 bases were highly conserved, and over 90% of these were found in CSBs of 6 or more bases. Alignment of Drosophila neural-specific and mesodermal-specific cDTs revealed that 11 of the 15 HLHmβ CSBs aligned with a total of 28 neural specific cDTs, while only 1 of its CSBs aligned with a single mesodermal specific cDT (Figure 10b,c). Both proneural transcription factors and the Notch pathway, acting through the Su(H) transcription factor, are implicated in the regulation of E(spl) complex genes (reviewed by [56]). Among the cDTs aligning with the CSBs, one, GCATGTGC, contains an E-box (underlined), the focus of activity of proneural transcription factors, and two others, TTTCCCA and TCCCAC, align with the consensus Su(H) binding site.
Although higher specificity is obtained by alignment with cDTs of 7 bases or greater, we have found that it is not unusual for 80% of CSBs associated with neural expressed genes to align with neural-specific cDTs versus only 20% of the CSBs in the same putative enhancer regions aligning with mesodermal-specific cDTs even when 6 base long cDTs are included in the analysis (data not shown). As the size and specificity of these libraries grow, their use as predictors of enhancer function will most likely increase as well.
As an additional assessment of the specificity of cDT-library scans, we generated negative control CSB-libraries for alignment to cDTs. These datasets, both Drosophila and mammalian, consisted of conserved sequence blocks within exons of genes that are not predominantly expressed in the CNS (data not shown). For this analysis we use the percent coverage of CSBs by cDTs, as used above for the analysis of Dll1 enhancers in which we counted the percent of the bases in the CSBs that aligned with cDTs. Whereas Drosophila and mammalian neural-specific cDTs, including hexamers, cover approximately 56% and 70%, respectively, of CSBs from neural enhancers, alignment with control CSBs was 20% or less. Again, when the alignment was repeated with cDTs of 7 bp or greater the CSB coverage of neural sequence was 5-fold greater than that observed with the control datasets. Taken together, our cDT alignments demonstrate their utility in identifying enhancer type-specific conserved sequence elements.
Evaluation of the cis-Decoder method was also carried out by examining the contribution that each enhancer made to the cDT-libraries. As one adds new enhancer CSBs to a specific library, the number of cDTs increases, such that alignment coverage of enhancer type-specific CSBs also increases. We illustrate the contribution of each enhancer to the specific cDT-libraries in our study (Additional data file 4). Overall, for Drosophila enhancers, prior to their inclusion in a library, on average 41% of the conserved nucleotides of enhancers align with the tissue specific cDT-library appropriate for that enhancer, while after inclusion in a library, 65% of the conserved nucleotides align. For example, addition of the bearded proneural enhancer [57], consisting of 21 CSBs (a total of 303 bp), to the Drosophila neural-specific CSB library resulted in 26 new neural-specific cDTs that were shared with at least one other neural enhancer. Prior to its inclusion, coverage of the bearded CSBs by alignment of neural-specific cDTs was 43%, while after its inclusion in the cDT-library preparation the alignment coverage of its CSBs increased to 67%. Addition of new enhancers to the out-group, used to remove common cDTs from a specific library, also enhances the specificity of the type-specific library and frequently shifts cDTs from specific to enriched libraries. Taken together, increased specificity of an enhancer-type cDT-library can be achieved either by including new similarly regulated enhancers in the generation of the cDT-library or increasing the number of out-group CSBs used to remove non-specific cDTs. Ideally, both approaches should be pursued to increase the depth and resolution of a particular cDT-library.
Conclusion
This study describes a systematic approach for the identification and comparative analysis of highly conserved DNA sequences within enhancers. Because our approach focuses solely on conserved sequences, the probability that cis-Decoder analysis dissects functionally important DNA is greatly enhanced. Most of the 2,086 CSBs identified in this study have undergone negative selection during more than 160 My of collective evolutionary divergence. Alignment of hundreds of CSBs from both similarly regulating enhancers and functionally different enhancers assures that conserved cis-regulatory elements shared by as few as two enhancers are identified and included in the analyses. Our cDT-scans show that most CSBs have a modular organization made up of smaller overlapping/interlocking sequence elements that align with CSBs of other enhancers. A typical CSB is made up of both enhancer type-specific sequence elements and common elements that are found in enhancers with different regulatory functions and, surprisingly, more than half of all of the shared CSB sequence elements do not correspond to know transcription factor DNA-binding sites and, as of yet, are functionally novel.
cDT-library scans of EvoPrinted cis-regulatory DNA reveal that it is possible to differentiate between functionally different enhancer types before any experimental/expression data are known. For example, cDT-library scans of the mammalian Dll1 or Drosophila snail cis-regulatory DNA sequences accurately differentiate between neural and mesodermal enhancers (Figures 3 and 7). cDT-library scans of co-regulating enhancers, using multiple libraries, reveal the combinatorial complexity of the cis-regulatory sequence elements involved in coordinate gene expression. Our studies indicate that many co-regulating enhancers rely on different combinations of the tissue-specific cis-regulatory elements to achieve synchronous regulatory behaviors. Although not highlighted in this paper, information gleaned from the cDT-scans and subsequent cDT-cataloger analysis of multiple co-regulating enhancers can be used to construct 'higher resolution' cDT-libraries that harbor many, or most, of the sequence elements that direct coordinate gene expression.
For example, sub-libraries of the Drosophila neural specific library can be generated to identify neuroblast- and PNS-specific tags. Enhancer CSB analysis using cDT-libraries generated from the combined alignments of both mammalian and fly CSBs also suggests that many of the sequence elements represented by the different cDTs have been conserved across taxonomic divisions and may represent core elements used by many metazoans to direct tissue-specific gene expression patterns.
Although we have initially generated cDT-libraries from general classes of different enhancer types, this approach should be applicable to the analysis of gene co-regulation in any cell type involved in any biological event. As the variety and depth of the different cDT-libraries increase, we believe that cDT-library scans of EvoPrinted putative enhancer regions will have great utility for the identification and initial characterization of cis-regulatory sequences. Future efforts that address the role of individual enhancer CSBs and the dissection of their modular elements will undoubtedly yield new insights into the function of these 'evolutionarily hardened' sequences and ultimately produce a better understanding of the regulatory code underlying coordinate gene expression.
Materials and methods
cis-Decoder [26] is a six-step integrated series of protocols and web-based algorithms that can be used to identify evolutionarily conserved DNA sequences that are shared among different enhancers (Figure 1). The following sections provide a detailed description of each step of the cis-Decoder procedure: EvoPrint analysis [58], for the discovery of MCSs; EvoPrint-parser, for CSB extraction and annotation; CSB-aligner, for the identification of shared elements between CSBs; cDT-scanner, to reveal cDT positions and their relations to other cDTs within CSBs; Full-enhancer scanner, for the discovery of less-conserved repeated cDTs or CSBs within enhancers; and cDT-cataloger for the identification of enhancers with shared sequence elements. A more detailed description of these steps is given at the cis-Decoder website. The Java applets CSB-aligner, cDT-scanner, Full-enhancer scanner and cDT-cataloger are available on-line at the cis-Decoder website and can be downloaded to the users computer to avoid Java-web browser incompatibilities. In our experience, a current version of the Mozilla browser avoids many potential incompatibilities.
EvoPrinter
The first step in the cis-Decoder analysis of an enhancer is preparing CSB-libraries from enhancers with related and/or divergent expression patterns. Enhancer CSBs were identified by the phylogenetic footprinting algorithm EvoPrinter [9]. Unlike other multi-species alignment programs that identify CSBs by outputting multiple aligned sequences interrupted by sequence gaps to optimize alignments, EvoPrinter outputs a single uninterrupted sequence to reveal CSBs as they exist in a species of interest. In Drosophila, when 9 or more species are used to generate an EvoPrint, the combined mutagenic histories of all of the orthologous DNAs represent an excess of 160 My of collective evolutionary divergence, thus affording near base-pair resolution of the functionally important DNA within the species of interest (discussed in [9]). Likewise, EvoPrint analysis of orthologous DNAs that include placental mammals (human, chimpanzee, rhesus monkey, cow, dog, rat and mouse), and, optionally, the opossum, detects CSBs that have been maintained for over 200 My of collective divergence. The EvoPrinter and EvoDifference print analysis algorithms and companion protocols are described [9], and are found online at the EvoPrinter tutorial website.
EvoPrint-parser
The EvoPrint-parser is a JavaScript program that automatically extracts and generates reverse-complement sequence and then annotates and lists in their 5' to 3' order CSBs that are 6 bp or longer from a known or putative enhancer region. Tissue-specific enhancer CSB-libraries can then be generated by assembling CSBs from enhancers of known function (for example, neural or mesodermal enhancers).
CSB-aligner
CSB-aligner is a Java applet that allows one to identify short sequence elements shared between different CSBs. To generate a CSB-alignment, parsed CSBs from multiple enhancer regions are placed in the upper window of the CSB-aligner applet. Then, forward direction CSBs from one or more enhancers are placed in the lower window of the CSB-aligner. A box associated with the lower window of the CSB-aligner allows for the naming of the CSBs introduced into the lower box and selection of the minimum aligned length (6, 7 or 8 base windows have been routinely used). Output length of the alignments produced by CSB-aligner can be selected (default value 100 bases).
Output of the CSB-aligner consists of the CSBs that were input into the lower window aligned with the CSBs that were introduced into the upper window. The CSB-aligner does not record CSB self-alignments. A second output window, the results table, is a list of the aligned matches along with their positions. Each of the output columns of the results table can be sorted by selecting the column header of the column to be sorted. Contents of results tables can be copy-pasted into Microsoft Word.
The CSB-alignment can be saved as an HTML file. Saving the HTML file allows copy pasting from the saved file into Microsoft Word and, once in Word, the file can be reformatted and saved or printed as the original readout. The CSB-alignment program has functioned successfully with the introduction of thousands of CSBs in both windows. The following CSB-libraries were created from EvoPrints of enhancers listed in Table 1: mammalian neural, mammalian mesodermal, Drosophila neural, Drosophila mesodermal and Drosophila segmental.
Interpreting the CSB-aligner readout and generation of cDT-libraries
A cDT is a short sequence element of 6 bp or greater that is a perfect match to sequences within CSBs that are present in two or more enhancers. A cDT-library represents a collection of cDTs that are shared by the various enhancers examined. Two types of cDT-libraries have been generated in this study. First, a 'tissue-specific library' contains cDTs that are shared by a group of enhancers that regulate similar expression patterns but are absent from a second set of enhancers that direct expression in tissues outside of the first group. Second, a 'common cDT-library' contains cDTs that were shared between sets of enhancers of divergently regulated genes. A subset of common libraries included 'enriched' libraries that had a three-fold greater representation from one enhancer type (for example, neural) than from a second type (for example, mesodermal).
All libraries were generated from readouts of the CSB-aligner. Making enhancer-type specific libraries requires two different CSB-libraries generated from functionally different enhancers, a library from the tissue of interest (for example, neural), and a second library that serves as an 'out-group' (for example, mesodermal). For the generation of a neural cDT-library, neural CSBs in both forward and reverse directions were copy-pasted into both upper and lower windows of CSB-aligner. The resulting cDTs from this alignment are listed in the 'Result of CSB alignment table' of the CSB-aligner output, in the column titled 'Motif.' Since this cDT list contains multiple copies of different cDTs, the extra copies are removed using the Java applet Puzzamatic 1.0 [59], a freeware created by Ron Surratt. The cDT list that contains all unique cDTs is then alphabetized and sorted by size also using Puzzamatic 1.0. The cDTs, constituting a raw neural cDT-library, were then copy/pasted into a Microsoft Word document. A second CSB-alignment is then performed with the neural CSBs in the top window of CSB-aligner, and mesodermal CSBs in the lower window. The cDTs from this alignment were freed of extra copies as above. These cDTs constituted an unedited common neural/mesodermal cDT-library. The unedited neural and common cDT-libraries are combined and cDTs common to the two libraries (present in the first and second alignments) are removed using the JavaScript program cDT-cleaner [60], thus leaving only the neural-specific sequences. Neural enriched and common cDTs were curated from the unedited shared cDT-library.
For Drosophila, segmental, neural (treating CNS and PNS specific enhancers together), and mesodermal specific cDT-libraries were generated. The out-group for neural and segmental cDT-libraries was the mesodermal CSB-library, and the out-group for the mesodermal cDT-library was neural CSBs. For mammals, neural and mesodermal cDT-libraries were generated. All cDT-libraries are listed in Table 2 and full libraries are available online [26].
Identification of shared elements within enhancers with the cDT-scanner
The function of cDT-scanner is to determine the relationship between any enhancer and any other group of MCSs used to generate the CSB libraries. cDT-scanner aligns the cDTs contained within various cDT-libraries with CSBs within an EvoPrint. cDT-scanner is a Java applet that uses a variant of the cis-Decoder aligner; it looks for only perfect matches between cDTs and CSB sequences. Alignment of cDTs using cDT-scanner is accomplished by first pasting a cDT-library in the upper window of cDT-scanner and then pasting the EvoPrint or CSBs to which they are to be aligned in the lower window. The output of cDT-scanner consists of perfect matches of cDTs aligned under the input CSBs. Since each library consists of cDTs shared by different enhancers, cDT-scanner portrays the shared elements within each CSB. A cDT-scanner alignment should be saved; information from saved files can be copy-pasted into Microsoft Word without loss of formatting features. For details on how to format cDT-alignments, see the website. A second output window for the cDT-scanner, a results table, is a list of the aligned matches along with their positions. Selecting the output column header sorts the results under that header. Contents of results tables can then be copy-pasted into Microsoft Word.
Finding less-conserved sequence elements
The 'Full-enhancer scanner' is a Java applet that identifies additional repeated cDT or CSB sequences within less conserved sequences flanking CSBs of enhancers. For this alignment, cDTs or CSBs present within an enhancer can be curated from the output of cDT-scanner termed 'Results from cDT-scan.' Curate both forward and reverse/complement sequences and paste into the upper window of Full-enhancer scanner. The EvoPrinted enhancer should be copy-pasted into the lower window. The program aligns to both conserved and non-conserved sequences of the EvoPrint.
Identification of enhancers that share conserved elements using cDT-cataloger
cDT-cataloger uses a variant of the CSB-aligner; it records only perfect matches between CSBs and cDTs of a specified size. The output lists those CSBs containing perfect sequence matches to the cDTs, and can be used to identify enhancers and count the number of times each cDT aligns with any CSB-library. Cataloguing is accomplished by copy-pasting the CSB-libraries (both forward and reverse directions) into the upper window of the cDT-cataloger and the selected cDTs of a single uniform size in the lower window. The size of the cDT(s) must be entered into the window provided.
Additional data files
The following additional data are available with the online version of this paper. Additional data file 1 contains the cDT-cataloger analysis of the murine Delta-like 1 Homology-II and msd-II enhancers supplemental to Figure 4. Additional data file 2 contains the cis-Decoder analysis of the Drosophila hairy stripe 1 enhancer. Additional data file 3 is a figure that contains cis-Decoder analysis of the human TIP39 5' proximal promoter. Additional data file 4 is a table that documents the contribution of each Drosophila and mammalian enhancer to the specific cDT-libraries generated in this study.
Supplementary Material
cDT-cataloger analysis of the murine Delta-like 1 Homology-II and msd-II enhancers supplemental to Figure 4
cis-Decoder analysis of the Drosophila hairy stripe 1 enhancer
cis-Decoder analysis of the human TIP39 5' proximal promoter
Contribution of each Drosophila and mammalian enhancer to the specific cDT-libraries generated in this study
Acknowledgments
Acknowledgements
We thank Laura Elnitski and Brian Mozer for critically reading the manuscript and Anthonois Ekatomatis for technical assistance. We are also indebted to Judy Brody for help with the cis-Decoder website construction and editorial assistance. This research was supported by the Intramural Research Program of the NIH, NINDS and NIMH.
Contributor Information
Thomas Brody, Email: brodyt@ninds.nih.gov.
Wayne Rasband, Email: wsr@nih.gov.
Kevin Baler, Email: kb263@cornell.edu.
Alexander Kuzin, Email: kuzina@mail.nih.gov.
Mukta Kundu, Email: muktakundu@mail.nih.gov.
Ward F Odenwald, Email: ward@codon.nih.gov.
References
- Wray GA, Hahn MW, Abouheif E, Balhoff JP, Pizer M, Rockman MV, Romano LA. The evolution of transcriptional regulation in eukaryotes. Mol Biol Evol. 2003;20:1377–1419. doi: 10.1093/molbev/msg140. [DOI] [PubMed] [Google Scholar]
- Levine M, Davidson EH. Gene regulatory networks for development. Proc Natl Acad Sci USA. 2005;102:4936–4942. doi: 10.1073/pnas.0408031102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Istrail S, Davidson EH. Logic functions of the genomic cis-regulatory code. Proc Natl Acad Sci USA. 2005;102:4954–4959. doi: 10.1073/pnas.0409624102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ip YT, Levine M, Bier E. Neurogenic expression of snail is controlled by separable CNS and PNS promoter elements. Development. 1994;120:199–207. doi: 10.1242/dev.120.1.199. [DOI] [PubMed] [Google Scholar]
- Emery JF, Bier E. Specificity of CNS and PNS regulatory subelements comprising pan-neural enhancers of the deadpan and scratch genes is achieved by repression. Development. 1995;121:3549–3560. doi: 10.1242/dev.121.11.3549. [DOI] [PubMed] [Google Scholar]
- Wasserman WW, Palumbo M, Thompson W, Fickett JW, Lawrence CE. Human-mouse genome comparisons to locate regulatory sites. Nat Genet. 2000;26:225–228. doi: 10.1038/79965. [DOI] [PubMed] [Google Scholar]
- Yuh CH, Brown CT, Livi CB, Rowen L, Clarke PJ, Davidson EH. Patchy interspecific sequence similarities efficiently identify positive cis-regulatory elements in the sea urchin. Dev Biol. 2002;246:148–161. doi: 10.1006/dbio.2002.0618. [DOI] [PubMed] [Google Scholar]
- Berezikov E, Guryev V, Plasterk RH, Cuppen E. CONREAL: conserved regulatory elements anchored alignment algorithm for identification of transcription factor binding sites by phylogenetic footprinting. Genome Res. 2004;14:170–178. doi: 10.1101/gr.1642804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odenwald WF, Rasband W, Kuzin A, Brody T. EVOPRINTER, a multigenomic comparative tool for rapid identification of functionally important DNA. Proc Natl Acad Sci USA. 2005;102:14700–14705. doi: 10.1073/pnas.0506915102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoch M, Schröder C, Seifert E, Jäckle H. cis-acting control elements for Krüppel expression in the Drosophila embryo. EMBO J. 1990;9:2587–2595. doi: 10.1002/j.1460-2075.1990.tb07440.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoch M, Seifert E, Jackle H. Gene expression mediated by cis-acting sequences of the Kruppel gene in response to the Drosophila morphogens bicoid and hunchback. EMBO J. 1991;10:2267–2278. doi: 10.1002/j.1460-2075.1991.tb07763.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoch M, Gerwin N, Taubert H, Jackle H. Competition for overlapping sites in the regulatory region of the Drosophila gene Kruppel. Science. 1992;256:94–97. doi: 10.1126/science.1348871. [DOI] [PubMed] [Google Scholar]
- Prabhakar S, Poulin F, Shoukry M, Afzal V, Rubin EM, Couronne O, Pennacchio LA. Close sequence comparisons are sufficient to identify human cis-regulatory elements. Genome Res. 2006;16:855–863. doi: 10.1101/gr.4717506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ. BLAT - the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. 10.1101/gr.229202. Article published online before March 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo SM, Li L, Hu Z, Halfon MS. REDfly: a regulatory element database for Drosophila. Bioinformatics. 2006;22:381–383. doi: 10.1093/bioinformatics/bti794. [DOI] [PubMed] [Google Scholar]
- Adachi Y, Hauck B, Clements J, Kawauchi H, Kurusu M, Totani Y, Kang YY, Eggert T, Walldorf U, Furukubo-Tokunaga K, Callaerts P. Conserved cis-regulatory modules mediate complex neural expression patterns of the eyeless gene in the Drosophila brain. Mech Dev. 2003;120:1113–1126. doi: 10.1016/j.mod.2003.08.007. [DOI] [PubMed] [Google Scholar]
- Riddihough G, Ish-Horowicz D. Individual stripe regulatory elements in the Drosophila hairy promoter respond to maternal, gap, and pair-rule genes. Genes Dev. 1991;5:840–854. doi: 10.1101/gad.5.5.840. [DOI] [PubMed] [Google Scholar]
- Fujioka M, Emi-Sarker Y, Yusibova GL, Goto T, Jaynes JB. Analysis of an even-skipped rescue transgene reveals both composite and discrete neuronal and early blastoderm enhancers, and multi-stripe positioning by gap gene repressor gradients. Development. 1999;126:2527–2538. doi: 10.1242/dev.126.11.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröder C, Tautz D, Seifert E, Jäckle H. Differential regulation of the two transcripts from the Drosophila gap segmentation gene hunchback. EMBO J. 1988;7:2881–2887. doi: 10.1002/j.1460-2075.1988.tb03145.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolis JS, Borowsky ML, Steingrimsson E, Shim CW, Lengyel JA, Posakony JW. Posterior stripe expression of hunchback is driven from two promoters by a common enhancer element. Development. 1995;121:3067–3077. doi: 10.1242/dev.121.9.3067. [DOI] [PubMed] [Google Scholar]
- Yin Z, Xu XL, Frasch M. Regulation of the twist target gene tinman by modular cis-regulatory elements during early mesoderm. Development. 1997;124:4971–4982. doi: 10.1242/dev.124.24.4971. [DOI] [PubMed] [Google Scholar]
- Miller JM, Oligino T, Pazdera M, Lopez AJ, Hoshizaki DK. Identification of fat-cell enhancer regions in Drosophila melanogaster. Insect Mol Biol. 2002;11:67–77. doi: 10.1046/j.0962-1075.2001.00310.x. [DOI] [PubMed] [Google Scholar]
- Gindhart JG, Jr, King AN, Kaufman TC. Characterization of the cis-regulatory region of the Drosophila homeotic gene Sex combs reduced. Genetics. 1995;139:781–795. doi: 10.1093/genetics/139.2.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder MD, Pearce M, Fak J, Fan H, Unnerstall U, Emberly E, Rajewsky N, Siggia ED, Gaul U. Transcriptional control in the segmentation gene network of Drosophila. PLoS Biol. 2004;2:E271. doi: 10.1371/journal.pbio.0020271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samad OA, Geisen MJ, Caronia G, Varlet I, Zappavigna V, Ericson J, Goridis C, Rijli FM. Integration of anteroposterior and dorsoventral regulation of Phox2b transcription in cranial motoneuron progenitors by homeodomain proteins. Development. 2004;131:4071–4083. doi: 10.1242/dev.01282. [DOI] [PubMed] [Google Scholar]
- cis-Decoder http://evoprinter.ninds.nih.gov/cisdecoder/index.htm
- Helms AW, Abney AL, Ben-Arie N, Zoghbi HY, Johnson JE. Autoregulation and multiple enhancers control Math1 expression in the developing nervous system. Development. 2000;127:1185–1196. doi: 10.1242/dev.127.6.1185. [DOI] [PubMed] [Google Scholar]
- Manak JR, Mathies LD, Scott MP. Regulation of a decapentaplegic midgut enhancer by homeotic proteins. Development. 1994;120:3605–3619. doi: 10.1242/dev.120.12.3605. [DOI] [PubMed] [Google Scholar]
- Kuzin A, Brody T, Moore AW, Odenwald WF. Nerfin-1 is required for early axon guidance decisions in the developing Drosophila CNS. Dev Biol. 2005;277:347–365. doi: 10.1016/j.ydbio.2004.09.027. [DOI] [PubMed] [Google Scholar]
- Murre C, McCaw PS, Baltimore D. A new DNA binding and dimerization motif in immunoglobulin enhancer daughterless, MyoD and myc proteins. Cell. 1989;56:777–783. doi: 10.1016/0092-8674(89)90682-X. [DOI] [PubMed] [Google Scholar]
- Powell LM, zur Lage PI, Prentice DRA, Senthinathan B, Jarman AP. The proneural proteins Atonal and Scute regulate neural target genes through different E-Box binding sites. Mol Cell Biol. 2004;24:9517–9526. doi: 10.1128/MCB.24.21.9517-9526.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- Bettenhausen B, Hrabe de Angelis M, Simon D, Guenet JL, Gossler A. Transient and restricted expression during mouse embryogenesis of Dll1, a murine gene closely related to Drosophila Delta. Development. 1995;121:2407–2418. doi: 10.1242/dev.121.8.2407. [DOI] [PubMed] [Google Scholar]
- Beckers J, Clark A, Wünsch K, De Angelis MH, Gossler A. Expression of the mouse Delta-1 gene during organogenesis and fetal development. Mech Dev. 1999;84:165–168. doi: 10.1016/S0925-4773(99)00065-9. [DOI] [PubMed] [Google Scholar]
- Beckers J, Caron A, Hrabe de Angelis M, Hans S, Campos-Ortega JA, Gossler A. Distinct regulatory elements direct Delta-1 expression in the nervous system and paraxial mesoderm of transgenic mice. Mech Dev. 2000;95:23–34. doi: 10.1016/S0925-4773(00)00322-1. [DOI] [PubMed] [Google Scholar]
- Rowitch DH, Echelard Y, Danielian PS, Gellner K, Brenner S, McMahon AP. Identification of an evolutionarily conserved 110 base-pair cis-acting regulatory sequence that governs Wnt-1 expression in the murine neural plate. Development. 1998;125:2735–2746. doi: 10.1242/dev.125.14.2735. [DOI] [PubMed] [Google Scholar]
- Bagheri-Fam S, Barrionuevo F, Dohrmann U, Gunther T, Schule R, Kemler R, Mallo M, Kanzler B, Scherer G. Long-range upstream and downstream enhancers control distinct subsets of the complex spatiotemporal Sox9 expression pattern. Dev Biol. 2006;291:382–397. doi: 10.1016/j.ydbio.2005.11.013. [DOI] [PubMed] [Google Scholar]
- Molkentin JD, Antos C, Mercer B, Taigen T, Miano JM, Olson EN. Direct activation of a GATA6 cardiac enhancer by Nkx2.5: evidence for a reinforcing regulatory network of Nkx2.5 and GATA transcription factors in the developing heart. Dev Biol. 2000;217:301–309. doi: 10.1006/dbio.1999.9544. [DOI] [PubMed] [Google Scholar]
- Awgulewitsch A, Jacobs D. Deformed autoregulatory element from Drosophila functions in a conserved manner in transgenic mice. Nature. 1992;358:341–344. doi: 10.1038/358341a0. [DOI] [PubMed] [Google Scholar]
- Malicki J, Cianetti LC, Peschie C, McGinnis W. A human HOX4B regulatory element provides head-specific expression in Drosophila. Nature. 1992;358:345–347. doi: 10.1038/358345a0. [DOI] [PubMed] [Google Scholar]
- Ip YT, Park RE, Kosman D, Yazdanbakhsh K, Levine M. dorsal-twist interactions establish snail expression in the presumptive mesoderm of the Drosophila embryo. Genes Dev. 1992;6:1518–1530. doi: 10.1101/gad.6.8.1518. [DOI] [PubMed] [Google Scholar]
- Odenwald WF, Garbern J, Arnheiter H, Tournier-Lasserve E, Lazzarini RA. The Hox-1.3 homeo box protein is a sequence-specific DNA-binding phosphoprotein. Genes Dev. 1989;3:158–172. doi: 10.1101/gad.3.2.158. [DOI] [PubMed] [Google Scholar]
- Gehring WJ. On the homeobox and its significance. Bioessays. 1986;5:3–4. doi: 10.1002/bies.950050102. [DOI] [PubMed] [Google Scholar]
- Markstein M, Zinzen R, Markstein P, Yee KP, Erives A, Stathopoulos A, Levine MA. A regulatory code for neurogenic gene expression in the Drosophila embryo. Development. 2004;131:2387–2394. doi: 10.1242/dev.01124. [DOI] [PubMed] [Google Scholar]
- Ochoa-Espinosa A, Yucel G, Kaplan L, Pare A, Pura N, Oberstein A, Papatsenko D, Small S. The role of binding site cluster strength in Bicoid-dependent patterning in Drosophila. Proc Natl Acad Sci USA. 2005;102:4960–4965. doi: 10.1073/pnas.0500373102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markstein M, Markstein P, Markstein V, Levine MS. Genome-wide analysis of clustered Dorsal binding sites identifies putative target genes in the Drosophila embryo. Proc Natl Acad Sci USA. 2002;99:763–768. doi: 10.1073/pnas.012591199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig M, Bergman C, Patel N, Kreitman M. Evidence for stabilizing selection in a eukaryotic enhancer element. Nature. 2000;403:564–567. doi: 10.1038/35000615. [DOI] [PubMed] [Google Scholar]
- Papatsenko D, Levine M. Quantitative analysis of binding motifs mediating diverse spatial readouts of the Dorsal gradient in the Drosophila embryo. Proc Natl Acad Sci USA. 2005;102:4966–4971. doi: 10.1073/pnas.0409414102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairall L, Schwabe JW, Chapman L, Finch JT, Rhodes D. The crystal structure of a two zinc-finger peptide reveals an extension to the rules for zinc-finger/DNA recognition. Nature. 1993;366:483–487. doi: 10.1038/366483a0. [DOI] [PubMed] [Google Scholar]
- Schrons H, Knust E, Campos-Ortega JA. The Enhancer of split complex and adjacent genes in the 96F region of Drosophila melanogaster are required for segregation of neural and epidermal progenitor cells. Genetics. 1992;32:481–503. doi: 10.1093/genetics/132.2.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usdin TB, Hoare SR, Wang T, Mezey E, Kowalak JA. TIP39: a new neuropeptide and PTH2-receptor agonist from hypothalamus. Nat Neurosci. 1999;2:941–943. doi: 10.1038/14724. [DOI] [PubMed] [Google Scholar]
- Dobolyi A, Palkovits M, Usdin TB. Expression and distribution of tuberoinfundibular peptide of 39 residues in the rat central nervous system. J Comp Neurol. 2003;455:547–566. doi: 10.1002/cne.10515. [DOI] [PubMed] [Google Scholar]
- Usdin TB, Dobolyi A, Ueda H, Palkovits M. Emerging functions for tuberoinfundibular peptide of 39 residues. Trends Endocrinol Metab. 2003;14:14–19. doi: 10.1016/S1043-2760(02)00002-4. [DOI] [PubMed] [Google Scholar]
- de Celis JF, de Celis J, Ligoxygakis P, Preiss A, Delidakis C, Bray S. Functional relationships between Notch, Su(H) and the bHLH genes of the E(spl) complex: the E(spl) genes mediate only a subset of Notch activities during imaginal development. Development. 1996;122:2719–2728. doi: 10.1242/dev.122.9.2719. [DOI] [PubMed] [Google Scholar]
- Nellesen DT, Lai EC, Posakony JW. Discrete enhancer elements mediate selective responsiveness of enhancer of split complex genes to common transcriptional activators. Dev Biol. 1999;213:33–53. doi: 10.1006/dbio.1999.9324. [DOI] [PubMed] [Google Scholar]
- Bray SJ. Expression and function of Enhancer of split bHLH proteins during Drosophila neurogenesis. Perspect Dev Neurobiol. 1997;4:313–323. [PubMed] [Google Scholar]
- Lai EC, Posakony JW. The Bearded box, a novel 3' UTR sequence motif, mediates negative post-transcriptional regulation of Bearded and Enhancer of split Complex gene expression. Development. 1997;124:4847–4856. doi: 10.1242/dev.124.23.4847. [DOI] [PubMed] [Google Scholar]
- EvoPrinter http://evoprinter.ninds.nih.gov/
- Puzzamatic http://www.mcn.org/c/rsurratt/Puzzamatic/WordSearchJar.html
- cDT-cleaner http://evoprinter.ninds.nih.gov/cisdecoder/Cdt_cleaner.htm
- Ramos E, Price M, Rohrbaugh M, Lai ZC. Identifying functional cis-acting regulatory modules of the yan gene in Drosophila melanogaster. Dev Genes Evol. 2003;213:83–89. doi: 10.1007/s00427-003-0297-9. [DOI] [PubMed] [Google Scholar]
- Sun Y, Jan LY, Jan YN. Transcriptional regulation of atonal during development of the Drosophila peripheral nervous system. Development. 1998;125:3731–3740. doi: 10.1242/dev.125.18.3731. [DOI] [PubMed] [Google Scholar]
- Lee HH, Frasch M. Nuclear integration of positive Dpp signals antagonistic Wg inputs and mesodermal competence factors during Drosophila visceral mesoderm induction. Development. 2005;132:1429–1442. doi: 10.1242/dev.01687. [DOI] [PubMed] [Google Scholar]
- Bush A, Hiromi Y, Cole M. Biparous: a novel bHLH gene expressed in neuronal and glial precursors in Drosophila. Dev Biol. 1996;180:759–772. doi: 10.1006/dbio.1996.0344. [DOI] [PubMed] [Google Scholar]
- Reeves N, Posakony JW. Genetic programs activated by proneural proteins in the developing Drosophila PNS. Dev Cell. 2005;8:413–425. doi: 10.1016/j.devcel.2005.01.020. [DOI] [PubMed] [Google Scholar]
- McDonald JA, Fujioka M, Odden JP, Jaynes JB, Doe CQ. Specification of motoneuron fate in Drosophila: integration of positive and negative transcription factor inputs by a minimal eve enhancer. J Neurobiol. 2003;57:193–203. doi: 10.1002/neu.10264. [DOI] [PubMed] [Google Scholar]
- Jiang J, Hoey T, Levine M. Autoregulation of a segmentation gene in Drosophila: combinatorial interaction of the even-skipped homeo box protein with a distal enhancer element. Genes Dev. 1991;5:265–277. doi: 10.1101/gad.5.2.265. [DOI] [PubMed] [Google Scholar]
- Small S, Blair A, Levine M. Regulation of even-skipped stripe 2 in the Drosophila embryo. EMBO J. 1992;11:4047–4057. doi: 10.1002/j.1460-2075.1992.tb05498.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small S, Blair A, Levine M. Regulation of two pair-rule stripes by a single enhancer in the Drosophila embryo. Dev Biol. 1996;175:314–324. doi: 10.1006/dbio.1996.0117. [DOI] [PubMed] [Google Scholar]
- Pick L, Schier A, Affolter M, Schmidt-Glenewinkel T, Gehring WJ. Analysis of the ftz upstream element: germ layer-specific enhancers are independently autoregulated. Genes Dev. 1990;4:1224–1239. doi: 10.1101/gad.4.7.1224. [DOI] [PubMed] [Google Scholar]
- Berman BP, Pfeiffer BD, Laverty TR, Salzberg SL, Rubin GM, Eisen MB, Celniker SE. Computational identification of developmental enhancers: conservation and function of transcription factor binding-site clusters in Drosophila melanogaster and Drosophila pseudoobscura. Genome Biol. 2004;5:R61. doi: 10.1186/gb-2004-5-9-r61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiromi Y, Kuroiwa A, Gehring WJ. Control elements of the Drosophila segmentation gene fushi tarazu. Cell. 1985;43:603–613. doi: 10.1016/0092-8674(85)90232-6. [DOI] [PubMed] [Google Scholar]
- Li X, Gutjahr T, Noll M. Separable regulatory elements mediate the establishment and maintenance of cell states by the Drosophila segment-polarity gene gooseberry. EMBO J. 1993;12:1427–1436. doi: 10.1002/j.1460-2075.1993.tb05786.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchard M, St-Amand J, Cote S. Combinatorial activity of pair-rule proteins on the Drosophila gooseberry early enhancer. Dev Biol. 2000;222:135–146. doi: 10.1006/dbio.2000.9702. [DOI] [PubMed] [Google Scholar]
- La Rosée A, Häder T, Taubert H, Rivera-Pomar R, Jäckle H. Mechanism and Bicoid dependent control of hairy stripe 7 expression in the posterior region of the Drosophila embryo. EMBO J. 1997;16:4403–4411. doi: 10.1093/emboj/16.14.4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langeland JA, Carroll SB. Conservation of regulatory elements controlling hairy pair-rule stripe formation. Development. 1993;117:585–596. doi: 10.1242/dev.117.2.585. [DOI] [PubMed] [Google Scholar]
- Howard KR, Struhl G. Decoding positional information: regulation of the pair-rule gene hairy. Development. 1990;110:1223–1231. doi: 10.1242/dev.110.4.1223. [DOI] [PubMed] [Google Scholar]
- Stathopoulos A, Tam B, Ronshaugen M, Frasch M, Levine M. pyramus and thisbe: FGF genes that pattern the mesoderm of Drosophila embryos. Genes Dev. 2004;18:687–699. doi: 10.1101/gad.1166404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hader T, Wainwright D, Shandala T, Saint R, Taubert H, Bronner G, Jäckle H. Receptor tyrosine kinase signaling regulates different modes of Groucho-dependent control of Dorsal. Curr Biol. 2000;10:51–54. doi: 10.1016/S0960-9822(99)00265-1. [DOI] [PubMed] [Google Scholar]
- Driever W, Thoma G, Nusslein-Volhard C. Determination of spatial domains of zygotic gene expression in the Drosophila embryo by the affinity of binding sites for the bicoid morphogen. Nature. 1989;340:363–367. doi: 10.1038/340363a0. [DOI] [PubMed] [Google Scholar]
- Biemar F, Zinzen R, Ronshaugen M, Sementchenko V, Manak JR, Levine MS. Spatial regulation of microRNA gene expression in the Drosophila embryo. Proc Natl Acad Sci USA. 2005;102:15907–15911. doi: 10.1073/pnas.0507817102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen HT, Xu X. Drosophila mef2 expression during mesoderm development is controlled by a complex array of cis-acting regulatory modules. Dev Biol. 1998;204:550–566. doi: 10.1006/dbio.1998.9081. [DOI] [PubMed] [Google Scholar]
- Gutjahr T, Vanario-Alonso CE, Pick L, Noll M. Multiple regulatory elements direct the complex expression pattern of the Drosophila segmentation gene paired. Mech Dev. 1994;48:119–128. doi: 10.1016/0925-4773(94)90021-3. [DOI] [PubMed] [Google Scholar]
- Kambadur R, Koizumi K, Stivers C, Nagle J, Poole SJ, Odenwald WF. Regulation of POU genes by castor and hunchback establishes layered compartments in the Drosophila CNS. Genes Dev. 1998;12:246–260. doi: 10.1101/gad.12.2.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy KL, Wohlwill A, Dzitoeva S, Lin MH, Holbrook S, Storti RV. The Drosophila PAR domain protein 1 (Pdp1) gene encodes multiple differentially expressed mRNAs and proteins through the use of multiple enhancers and promoters. Dev Biol. 2000;224:401–414. doi: 10.1006/dbio.2000.9797. [DOI] [PubMed] [Google Scholar]
- Klingler M, Soong J, Butler B, Gergen JP. Disperse versus compact elements for the regulation of runt stripes in Drosophila. Dev Biol. 1996;177:73–84. doi: 10.1006/dbio.1996.0146. [DOI] [PubMed] [Google Scholar]
- Culi J, Modolell J. Proneural gene self-stimulation in neural precursors: an essential mechanism for sense organ development that is regulated by Notch signaling. Genes Dev. 1998;12:2036–2047. doi: 10.1101/gad.12.13.2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehman DA, Patterson B, Johnston LA, Balzer T, Britton JS, Saint R, Edgar BA. Cis-regulatory elements of the mitotic regulator, string/Cdc25. Development. 1999;126:1793–1803. doi: 10.1242/dev.126.9.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick A, Core N, Kerridge S, Scott MP. Homeotic response elements are tightly linked to tissue-specific elements in a transcriptional enhancer of the teashirt gene. Development. 1995;121:2799–2812. doi: 10.1242/dev.121.9.2799. [DOI] [PubMed] [Google Scholar]
- Wharton KA, Jr, Crews ST. CNS midline enhancers of the Drosophila slit and Toll genes. Mech Dev. 1993;40:141–154. doi: 10.1016/0925-4773(93)90072-6. [DOI] [PubMed] [Google Scholar]
- Buttgereit D. Redundant enhancer elements guide beta 1 tubulin gene expression in apodemes during Drosophila embryogenesis. J Cell Sci. 1993;105:721–727. doi: 10.1242/jcs.105.3.721. [DOI] [PubMed] [Google Scholar]
- Lin SC, Lin MH, Horvath P, Reddy KL, Storti RV. PDP1, a novel Drosophila PAR domain bZIP transcription factor expressed in developing mesoderm, endoderm and ectoderm, is a transcriptional regulator of somatic muscle genes. Development. 1997;124:4685–4696. doi: 10.1242/dev.124.22.4685. [DOI] [PubMed] [Google Scholar]
- Shao X, Koizumi K, Nosworthy N, Tan DP, Odenwald WF, Nirenberg M. Regulatory DNA required for vnd/NK-2 homeobox gene expression pattern in neuroblasts. Proc Natl Acad Sci USA. 2002;99:113–117. doi: 10.1073/pnas.012584599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashraf SI, Hu X, Roote J, Ip YT. The mesoderm determinant Snail collaborates with related zinc-finger proteins to control Drosophila neurogenesis. EMBO J. 1999;18:6426–6438. doi: 10.1093/emboj/18.22.6426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigo I, Bovolenta P, Mankoo BS, Imai K. Meox homeodomain proteins are required for bapx1 expression in the sclerotome and activate its transcription by direct binding to its promoter. Mol Cell Biol. 2004;24:2757–2766. doi: 10.1128/MCB.24.7.2757-2766.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tou L, Quibria N, Alexander JM. Transcriptional regulation of the human Runx2/Cbfa1 gene promoter by bone morphogenetic protein-7. Mol Cell Endocrinol. 2003;205:121–129. doi: 10.1016/S0303-7207(03)00151-5. [DOI] [PubMed] [Google Scholar]
- Kim IM, Zhou Y, Ramakrishna S, Hughes DE, Solway J, Costa RH, Kalinichenko VV. Functional characterization of evolutionarily conserved DNA regions in forkhead box f1 gene locus. J Biol Chem. 2005;280:37908–37916. doi: 10.1074/jbc.M506531200. [DOI] [PubMed] [Google Scholar]
- McFadden DG, Charite J, Richardson JA, Srivastava D, Firulli AB, Olson EN. A GATA-dependent right ventricular enhancer controls dHAND transcription in the developing heart. Development. 2000;127:5331–5341. doi: 10.1242/dev.127.24.5331. [DOI] [PubMed] [Google Scholar]
- Bessho Y, Sakata R, Komatsu S, Shiota K, Yamada S, Kageyama R. Dynamic expression and essential functions of Hes7 in somite segmentation. Genes Dev. 2001;15:2642–2647. doi: 10.1101/gad.930601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabaries S, Lapointe J, Besch T, Carter M, Woollard J, Tuggle CK, Jeannotte L. Cdx protein interaction with Hoxa5 regulatory sequences contributes to Hoxa5 regional expression along the axial skeleton. Mol Cell Biol. 2005;25:1389–1401. doi: 10.1128/MCB.25.4.1389-1401.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mühlfriedel S, Kirsch F, Gruss P, Stoykova A, Chowdhury K. A roof plate-dependent enhancer controls the expression of Homeodomain only protein in the developing cerebral cortex. Dev Biol. 2005;283:522–534. doi: 10.1016/j.ydbio.2005.04.033. [DOI] [PubMed] [Google Scholar]
- Breslin MB, Zhu M, Lan MS. Neurod1/E47 regulates the E-box element of a novel zinc finger transcription factor, IA-1, in developing nervous system. J Biol Chem. 2003;278:38991–3899. doi: 10.1074/jbc.M306795200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jethanandani P, Kramer RH. α7 integrin expression is negatively regulated by δEF1 during skeletal myogenesis. J Biol Chem. 2005;280:36037–36046. doi: 10.1074/jbc.M508698200. [DOI] [PubMed] [Google Scholar]
- Wang DZ, Valdez MR, McAnally J, Richardson J, Olson EN. The Mef2c gene is a direct transcriptional target of myogenic bHLH and MEF2 proteins during skeletal muscle development. Development. 2001;128:4623–4633. doi: 10.1242/dev.128.22.4623. [DOI] [PubMed] [Google Scholar]
- Verma-Kurvari S, Savage T, Gowan K, Johnson JE. Lineage-specific regulation of the neural differentiation gene MASH1. Dev Biol. 1996;180:605–617. doi: 10.1006/dbio.1996.0332. [DOI] [PubMed] [Google Scholar]
- Buchberger A, Nomokonova N, Arnold HH. Myf5 expression in somites and limb buds of mouse embryos is controlled by two distinct distal enhancer activities. Development. 2003;130:3297–3307. doi: 10.1242/dev.00557. [DOI] [PubMed] [Google Scholar]
- Zimmerman L, Parr B, Lendahl U, Cunningham M, McKay R, Gavin B, Mann J, Vassileva G, McMahon A. Independent regulatory elements in the nestin gene direct transgene expression to neural stem cells or muscle precursors. Neuron. 1994;12:11–24. doi: 10.1016/0896-6273(94)90148-1. [DOI] [PubMed] [Google Scholar]
- Zhou B, Wu B, Tompkins KL, Boyer KL, Grindley JC, Baldwin HS. Characterization of Nfatc1 regulation identifies an enhancer required for gene expression that is specific to pro-valve endocardial cells in the developing heart. Development. 2005;132:1137–1146. doi: 10.1242/dev.01640. [DOI] [PubMed] [Google Scholar]
- Simmons AD, Horton S, Abney AL, Johnson JE. Neurogenin2 expression in ventral and dorsal spinal neural tube progenitor cells is regulated by distinct enhancers. Dev Biol. 2001;229:327–339. doi: 10.1006/dbio.2000.9984. [DOI] [PubMed] [Google Scholar]
- Lien CL, McAnally J, Richardson JA, Olson EN. Cardiac-specific activity of an Nkx2-5 enhancer requires an evolutionarily conserved Smad binding site. Dev Biol. 2002;244:257–266. doi: 10.1006/dbio.2002.0603. [DOI] [PubMed] [Google Scholar]
- Kurokawa D, Takasaki N, Kiyonari H, Nakayama R, Kimura-Yoshida C, Matsuo I, Aizawa S. Regulation of Otx2 expression and its functions in mouse epiblast and anterior neuroectoderm. Development. 2004;134:3307–3317. doi: 10.1242/dev.01219. [DOI] [PubMed] [Google Scholar]
- Brown CB, Engleka KA, Wenning J, Lu MM, Epstein JA. Identification of a hypaxial somite enhancer element regulating Pax3 expression in migrating myoblasts and characterization of hypaxial muscle Cre transgenic mice. Genesis. 2005;41:202–209. doi: 10.1002/gene.20116. [DOI] [PubMed] [Google Scholar]
- Barron MR, Belaguli NS, Zhang SX, Trinh M, Iyer D, Merlo X, Lough JW, Parmacek MS, Bruneau BG, Schwartz RJ. Serum response factor, an enriched cardiac mesoderm obligatory factor, is a downstream gene target for Tbx genes. J Biol Chem. 2005;280:11816–11828. doi: 10.1074/jbc.M412408200. [DOI] [PubMed] [Google Scholar]
- Kutejova E, Engist B, Mallo M, Kanzler B, Bobola N. Hoxa2 downregulates Six2 in the neural crest-derived mesenchyme. Development. 2005;132:469–478. doi: 10.1242/dev.01536. [DOI] [PubMed] [Google Scholar]
- Catena R, Tiveron C, Ronchi A, Porta S, Ferri A, Tatangelo L, Cavallaro M, Favaro R, Ottolenghi S, Reinbold R, et al. Conserved POU binding DNA sites in the Sox-2 upstream enhancer regulate gene expression in embryonic and neural stem cells. J Biol Chem. 2004;279:41846–41857. doi: 10.1074/jbc.M405514200. [DOI] [PubMed] [Google Scholar]
- Miyagi S, Nishimoto M, Saito T, Ninomiya M, Sawamoto K, Okano H, Muramatsu M, Oguro H, Iwama A, Okuda A. The Sox2 regulatory region 2 functions as a neural stem cell specific enhancer in the telencephalon. J Biol Chem. 2006;281:13374–13381. doi: 10.1074/jbc.M512669200. [DOI] [PubMed] [Google Scholar]
- Gottgens B, Nastos A, Kinston S, Piltz S, Delabesse EC, Stanley M, Sanchez MJ, Ciau-Uitz A, Patient R, Green AR. Establishing the transcriptional programme for blood: the SCL stem cell enhancer is regulated by a multiprotein complex containing Ets and GATA factors. EMBO J. 2002;21:3039–3050. doi: 10.1093/emboj/cdf286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagishi H, Maeda J, Hu T, McAnally J, Conway SJ, Kume T, Meyers N, Yamagishi C, Srivastava D. Tbx1 is regulated by tissue-specific forkhead proteins through a common Sonic hedgehog-responsive enhancer. Genes Dev. 2003;17:269–281. doi: 10.1101/gad.1048903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll SB, Laughon A, Thalley BS. Expression, function, and regulation of the hairy segmentation protein in the Drosophila embryo. Genes Dev. 1988;2:883–890. doi: 10.1101/gad.2.7.883. [DOI] [PubMed] [Google Scholar]
- Stanojevic D, Hoey T, Levine M. Sequence-specific DNA-binding activities of the gap proteins encoded by hunchback and Kruppel in Drosophila. Nature. 1989;341:331–335. doi: 10.1038/341331a0. [DOI] [PubMed] [Google Scholar]
- Treisman J, Desplan C. The products of the Drosophila gap genes hunchback and Kruppel bind to the hunchback promoters. Nature. 1989;341:335–337. doi: 10.1038/341335a0. [DOI] [PubMed] [Google Scholar]
- Ekker SC, Jackson DG, von Kessler DP, Sun BI, Young KE, Beachy PA. The degree of variation in DNA sequence recognition among four Drosophila homeotic proteins. EMBO J. 1994;13:3551–3560. doi: 10.1002/j.1460-2075.1994.tb06662.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsako S, Hyer J, Panganiban G, Oliver I, Caudy M. Hairy function as a DNA-binding helix-loop-helix repressor of Drosophila sensory organ formation. Genes Dev. 1994;8:2743–2755. doi: 10.1101/gad.8.22.2743. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
cDT-cataloger analysis of the murine Delta-like 1 Homology-II and msd-II enhancers supplemental to Figure 4
cis-Decoder analysis of the Drosophila hairy stripe 1 enhancer
cis-Decoder analysis of the human TIP39 5' proximal promoter
Contribution of each Drosophila and mammalian enhancer to the specific cDT-libraries generated in this study


