Abstract
Cell-cell signaling is an important component of the stem cell microenvironment, affecting both differentiation and self-renewal. However, traditional cell-culture techniques do not provide precise control over cell-cell interactions, while existing cell patterning technologies are limited when used with proliferating or motile cells. To address these limitations, we created the Bio Flip Chip (BFC), a microfabricated polymer chip containing thousands of microwells, each sized to trap down to a single stem cell. We have demonstrated the functionality of the BFC by patterning a 50×50 grid of murine embryonic stem cells (mESCs), with patterning efficiencies > 75%, onto a variety of substrates – a cell-culture dish patterned with gelatin, a 3-D substrate, and even another layer of cells. We also used the BFC to pattern small groups of cells, with and without cell-cell contact, allowing incremental and independent control of contact-mediated signaling. We present quantitative evidence that cell-cell contact plays an important role in depressing mESC colony formation, and show that E-cadherin is involved in this negative regulatory pathway. Thus, by allowing exquisite control of the cellular microenvironment, we provide a technology that enables new applications in tissue engineering and regenerative medicine.
Introduction
Cell-cell interactions – consisting of diffusible signaling and cell-cell contact (juxtacrine signaling) – are important in numerous biological processes, including tumor growth [1], atherosclerotic plaque formation [2,3], and stem cell differentiation and self-renewal [4-7]. In addition, cell-cell interactions have proven to be crucial for generating functional tissues in vitro [8,9].
Cell signaling can be modulated in vitro in several ways, using molecular inhibitors [7,10,11] or genetic approaches [12-14], but both are limited to manipulating single or known molecules. Alternatively, one can manipulate the general class of cell-cell interactions, diffusible or cell-cell contact, by modulating the cells' relative positions. However, traditional cell-culture techniques can only modulate cell position by varying the cell seeding density, providing control of cell signaling only at the macroscopic level of the cell-culture dish. In order to precisely control the cellular microenvironment, we need to manipulate a cell's position at the single-cell level. To accomplish this, we require cell patterning.
There are numerous ways to pattern cells – including microcontact printing (μCP) [15,16], switchable substrates [17,18], elastomeric stencils [19,20], microwells [21,22], optical tweezers [23,24], electrophoresis [25], and dielectrophoresis [9,26]. When choosing a cell patterning technology, there are several capabilities to consider: patterning cells with single-cell resolution, patterning large numbers of cells, allowing the patterned cells room to grow and move, being gentle on the cells, and being easy to use. While the above cell patterning tools are sufficient for their developed applications, each technique necessarily has some limitations. In particular, we are interested in creating patterns of single (or small numbers of) cells onto a variety of substrates and monitoring their proliferation over time. Existing techniques are limited in their ability to provide this functionality. μCP, for instance, restricts motility and proliferation by chemically patterning the substrate, leading researchers to develop switchable substrates that in turn require chemical synthesis [18] or uncommon materials [17] (e.g., ultrapure gold). In addition, existing cell patterning techniques each impose requirements on the substrate, such as requiring specific surface chemistry [15-18], electrodes [9,25,26], or optical transparency [23,24]. This can impose serious restrictions on cells whose behavior is substrate-dependent (e.g., glass vs. tissue-culture polystyrene (TCPS)) or prevent researchers from patterning cells onto novel substrates (e.g., combinatorial surfaces [27]). Instead, we have developed a new cell patterning technology, called the Bio Flip Chip (BFC), which circumvents these limitations while remaining exceedingly simple to use.
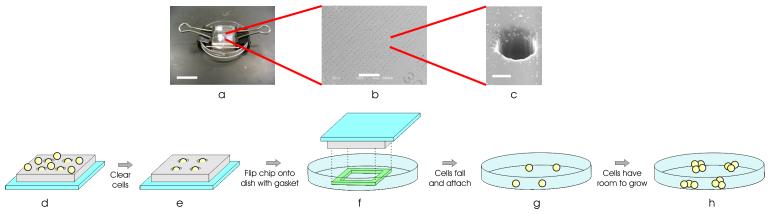
The BFC is a microfabricated polymer chip containing thousands of microwells (Fig. 1a-b), each sized to trap down to a single cell (Fig. 1c). To use the BFC, cells are pipetted onto the surface of the chip, allowing cells to fall into the microwells (Fig. 1d). After the cells are trapped in the microwells and the other cells are rinsed away (Fig. 1e), we flip the BFC upside down onto the desired substrate (Fig. 1f). The cells then fall out of the microwells onto the substrate, where they attach after a few hours (Fig. 1g). We can then remove the BFC from the substrate or leave it on, forming a closed chamber over the cells. In both modes of operation, the cells now have room to grow and move (Fig. 1h).
Figure 1.
BFC device and operation. (a) BFC packaging, consisting of a BFC, spacer gasket, and cell-culture dish, all held together using binder clips. Scale bar, 15 mm. (b) SEM image of the BFC microwells, spaced 200 μm apart. Scale bar, 1 mm. (c) SEM image of a single microwell, with diameter and height of 30 μm. Scale bar, 15 μm. (d) The cells are pipetted onto the surface of the chip, allowing cells to fall into the microwells. (e) The cells are trapped in the microwells and the other cells are rinsed away. (f) The BFC is flipped upside down onto the dish with a spacer gasket. (g) The cells then fall out of the microwells onto the substrate, where they attach after a few hours. (h) The cells now have room to grow and move.
This approach to cell patterning positions cells with single-cell resolution anywhere on a substrate and onto any substrate. In addition, using the BFC requires no external equipment or chemicals to pattern cells, making this technology easily adoptable in any lab. Here we describe the fabrication and operation of BFCs along with results showing the specific features of the technology: large-scale patterning, substrate-independence, multi-day tracking, and pattern control. Finally, we present the use of BFCs to control stem cell signaling, demonstrating that murine embryonic stem cells are sensitive to single-cell perturbations in cell signaling, and showing that increasing cell-cell contact contributes to a depression in colony-forming efficiency. These results therefore provide the first quantitative evidence that seeding mESCs as single cells maximizes colony formation, supporting the long-established protocols of single-cell seeding to maximize self-renewal. We thus provide a platform technology that can be used to manipulate cells in unique ways, facilitating the creation of novel biomaterials with increased functionality.
Materials and Methods
Fabrication of the BFC
We made the BFCs by molding polydimethylsiloxane (PDMS) over a 4″ Si master wafer. We dehydrated the Si wafers for 30 min at 130 °C. SU8-2050 (Microchem) was poured onto the wafer, ramped at 300 rpm/s to 3000-4000 rpm, and spun for 30 s to yield feature heights of 40-30 μm, respectively. After spinning, we placed the wafer on a 65 °C hotplate, immediately ramped up the temperature to 95 °C for 5-6 min, and then ramped it down to 65 °C. We exposed the wafers to a UV dose of 200 mJ/cm2 on a contact aligner (Karl Suss, MJB-3 Mask Aligner) using a dark-field mask printed at 20,000 dpi (CAD/Art Services). We placed the wafers on a 65 °C hotplate, immediately ramped up the temperature to 95 °C for 4-5 min, and then ramped it down to 65 °C. Next, we immersed the wafers in PM acetate (Doe and Ingalls) and Isopropanol while we spun them at 500 rpm for 3 min. Actual heights of the SU8 features were measured with a profilometer (Sloan, Dektak II). We then silanized the wafers for 30 min using hexamethyldisiloxane (Shin-Etsu MicroSi) to prevent PDMS from adhering to the Si master wafer. We mixed PDMS (Dow Corning, Sylgard 184) in a 10:1 ratio, poured it over the master Si wafer (∼10 g per wafer), and let it cure overnight at room temperature. The cured PDMS was then slowly peeled off the Si wafer and each chip was cut out using a razor blade and bonded to a 1” ×1” cut microscope slide for easy handling.
BFC Operation
We soaked the BFC overnight in phosphate-buffered saline (PBS) in order to prevent absorption of media into the PDMS during the experiment. After soaking, we sprayed the chips with ethanol and dried them with a Kimwipe (Kimtech Science). In the tissue-culture hood, we coated the BFC surface with 200 μL of 7.5% Bovine Serum Albumin (BSA) (Invitrogen, 15260-037) and scraped it with a pipette tip to disperse the BSA and remove any bubbles from the wells. The BSA helped to prevent cells from sticking in the microwells after flipping. We left the BSA on the BFC surface for ∼1 hr and agitated with a pipette tip every 20 min to prevent crusting.
To fit the BFC inside a 35×10 mm tissue-culture polystyrene (TCPS) dish (Falcon, 35-3001), we cut the rims of the TCPS dish half way down so that the ¾” binder clips (Office Depot) would fit over the dish rim to clamp the chip and dish together (Fig. 1a). We cut the PDMS spacer gasket (frame-shaped with 20×20 mm outer edge, 15×15 mm inner edge, and 250 μm thickness) from a PDMS sheet (Silicone Specialty Products) and then applied it to the dish using tweezers. This spacer gasket was required in order to provide the cells with enough media to prevent starvation (data not shown). We were able to pattern cells with a range of gasket thicknesses, from 100-600 μm, without any significant differences in cell patterning precision.
We sterilized the dish, gasket, BSA-coated chip, and 2 binder clips under UV light for 1 hr. Afterward, we aspirated the BSA and rinsed the BFC with 200 μL PBS. After aspirating the PBS, we applied 200 μL of cell solution (∼1×106 cells/mL ) to the BFC surface. We let the cells settle for 5-10 min. We then tilted the BFC to one corner at a 15° angle and slowly pipetted the cell solution off with a 200 μL pipette. Next, we placed the BFC flat and added 100 μL of PBS or blank media to the opposite BFC corner. We observed the BFC under the microscope to make sure cells were still loaded in the wells and rinsed the BFC an additional 2-4 times if necessary. We added 100 μL of media to both the BFC surface and the dish inside the PDMS gasket. We spread the media around to wet the entire dish surface in order to prevent bubbles from forming in the chamber after flipping. We then inverted the pre-wetted dish and slowly pushed up the BFC into the dish. We applied a binder clip to each side and removed the metal prongs so that dish remained level when flipped over. Lastly, we quickly flipped over the setup onto the incubator shelf while avoiding any unnecessary movement.
The cells fell out of the wells within minutes and attached to the dish after a few hours. The BFC can then be removed or left on the substrate. To remove the chip without disturbing the cells, we reinserted the metal prongs into the binder clips and removed them slowly at equal speeds. We then added 0.75 mL media to the dish, around the BFC, and returned the setup to the incubator. This allows the media to slowly release the BFC from the substrate, preventing disruption of the cell pattern (< 5% of the cells are disturbed). After 4 hrs, the BFC can be carefully removed. We aspirated and replaced the old media with 1.0 mL of fresh media. We then maintained the dishes by feeding daily with 1.0 mL media.
Preparation of the Substrates
To pattern the cells onto the surface with patterned gelatin, the 3-D substrate, and another layer of cells – we used the same protocol described above, with prior modification to the substrate before flipping the BFC. The surface with patterned gelatin was made by placing an elastomeric stencil onto a 60×15 mm TCPS dish (Falcon, 35-3002) and immersing the dish with fluorescent gelatin at 1 μg/mL (Molecular Probes, G13186). We incubated the dish for 15 min at 37 °C and then washed it twice with PBS before removing the stencil. The elastomeric stencils had 100 μm circular holes that formed the pattern (Fig. 2c).
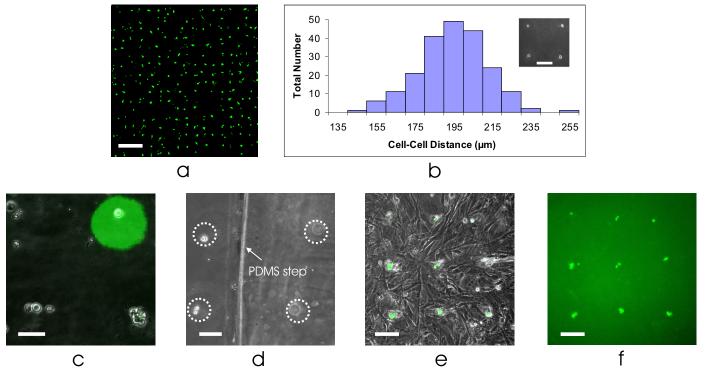
Figure 2.
BFC functionality. (a) Large-scale patterning of mESCs, stained with a vital dye on day 1 after seeding. Scale bar, 500 μm. (b) Histogram showing the distribution of cell-cell distances, with mean spacing of 195 ± 18 μm. Inset: high precision patterning, seeding cells within 1 cell diameter of their target. Inset scale bar, 100 μm. (c) Cell patterning of mESCs onto a TCPS dish with patterned fluorescent gelatin (top right), showing the compatibility of these two approaches. Scale bar, 50 μm. (d) Cell patterning of half the cells onto a TCPS dish (left, in-focus) and half the cells on a 100 μm step of polydimethylsiloxane (PDMS) (right, out-of-focus). Scale bar, 50 μm. (e) Cell patterning of mESCs onto a layer of murine embryonic fibroblasts. Scale bar, 100 μm. (f) Fluorescent picture of (e), showing only the mESCs. Scale bar, 100 μm.
To create the 3-D substrate, we placed a 100 μm thick layer of PDMS (Silicone Specialty Products) onto half of a 35×10 mm TCPS dish, creating a stepped surface. The cells were then flipped onto the dish, with half of the cells falling directly onto the dish and the other half falling onto the PDMS step (Fig. 2d).
To create the layer of cells, we seeded murine embryonic fibroblasts (MEFs) at a density of 3×106 cells/mL onto a 35×10 mm TCPS dish one day prior to patterning the murine embryonic stem cells. We incubated this dish with MEFs overnight and removed it from the incubator several minutes before flipping. We aspirated the old media from the dish, placed a PDMS gasket on the dish over the MEFs, and added 100 μL of media to the dish inside the gasket. We then inverted the dish with cells and slowly pushed up the BFC into dish, as described above.
Calculating Cell Patterning Efficiency and Precision
To calculate the cell patterning efficiency, a 50×50 grid of cells was patterned onto a 35×10 mm TCPS dish and the entire chip was scanned and imaged at 5× magnification (Zeiss, Axioplan 2 imaging microscope). We then used Matlab-based imaging software to stitch together the individual pictures into one stitched image of the entire chip. From the final stitched image, we calculated the cell patterning efficiency by dividing the number of spots with cells within one well diameter from the well edge, by the total number of available spots (n = 4 chips).
To calculate the cell patterning precision, we took pictures of the 50×50 grid of patterned cells across the chip at 10× magnification (Zeiss, Axiovert 200 microscope). From the pictures, we measured the number of pixels between adjacent cells (n = 211 measurements over 4 chips). We converted the number of pixels to a distance using a conversion factor, determined using a cell-counting chamber with a known grid spacing (VWR, 15170-208).
Cell Culture
We cultured ABJ1 mouse embryonic stem cells (mESCs) with a stably integrated green fluorescent protein (GFP) reporter for Oct-4 without feeders in mESC media: Dulbecco's Modified Eagle's Medium (DMEM) (Invitrogen, 11960-044) supplemented with 15% ES-qualified fetal bovine serum (Invitrogen, 16141-079), 4 mM L-glutamine (Invitrogen, 25030-081), 1 mM non-essential amino acids (Invitrogen, 11140-050), 50 U/mL penicillin, 50 μg/mL streptomycin (Invitrogen, 15140-122), 100 μM β-mercaptoethanol (Sigma-Aldrich, M7522), and 500 pM leukemia inhibitory factor (Chemicon, ESGRO ESG1107). We cultured cells directly on 100×20 mm TCPS dishes (Nunc, 172958) in a 37 °C humidified environment with 7.5% CO2. For maintenance of mESCs, we fed cells daily and passaged every other day using 0.25% trypsin-Ethylene-Diamine-Tetra-Acetic acid (trypsin-EDTA) (Invitrogen, 25200-056) at a density of 5×105 cells/cm2. The mESC passage number was always below P30. To demonstrate large-scale cell patterning, we stained the mESCs with a live green-fluorescent stain (Molecular Probes, Calcein AM). To inhibit E-cadherin signaling, we incubated mESCs with 40 μg/mL anti-E-cadherin antibody (clone DECMA-1, Sigma, U3254) in mESC media for 2 hrs; control experiments were run with cells incubated with mESC media for the same length of time.
We cultured the murine embryonic fibroblasts (MEFs) in media identical to mESC media, except without the leukemia inhibitory factor and β-mercaptoethanol. The MEFs were thawed, previously being mitotically inactivated with mitomycin C (Sigma-Aldrich, M4287), and seeded at a density of ∼3×106 cells/mL. The MEF passage number was P5.
Results
Large-Scale and Precise Cell Patterning
Scalability, efficiency, and precision are important capabilities of any new approach to cell patterning. We demonstrate the scalability of BFCs by creating large-scale (50×50) cell patterns of murine embryonic stem cells (mESCs) onto a tissue-culture polystyrene (TCPS) dish (Fig. 2a). We consistently achieved high cell patterning efficiencies, with an overall efficiency of 75.9% (n = 4 chips), calculated by dividing the number of spots with cells within one well diameter from the well edge, by the total number of available spots. In addition, we have achieved stem cell patterning with high precision (Fig. 2b inset). Using a microwell-to-microwell spacing of 200 μm, we patterned the cells onto a TCPS dish with a cell-cell spacing of 195 ± 18 μm (n = 211 measurements over 4 chips), thus seeding a cell within one cell diameter away from its target location (Fig. 2b). We have further demonstrated cell patterning down to single-cell resolution, with 50% of the patterned spots resulting in single cells (n = 4 chips).
Cell Patterning onto Any Substrate
Cell behavior can vary significantly depending on the substrate that the cells are seeded on (e.g., glass vs. TCPS). Since ESCs are very sensitive to cell-substrate interactions [27], we wanted to be able to pattern them onto TCPS, their standard in vitro substrate. Most existing cell patterning tools require modification of the substrate – adhesive patterning (e.g., μCP) requires patterning different ECMs [16,18] while techniques that use electromagnetic fields require metal electrodes to be deposited onto glass [28] or silicon [29]. Because the process of patterning with BFCs intrinsically transfers the patterned cells onto another substrate, substrate modification is avoided, allowing patterning of mESCs onto TCPS (Fig. 2a).
We also patterned cells onto a TCPS dish patterned with gelatin (Fig. 2c), showing the compatibility of BFCs with cell patterning techniques that do modify the substrate. One can even use BFCs to pattern over 3-D topography, which we demonstrated by patterning mESCs onto a stepped surface (Fig. 2d). Lastly, because most cell patterning techniques trap and pattern cells with the same substrate, they are incapable of patterning cells on top of an already-existing layer of cells, limiting their utility to one cell layer. However, patterning onto another layer of cells is possible using BFCs, which we demonstrated by patterning mESCs on top of a layer of murine embryonic fibroblasts (Fig. 2e-f). We can thus use BFCs to pattern onto any substrate – including different materials (e.g., glass, polystyrene), different chemistries (e.g., fibronectin, gelatin, matrigel), different topographies, or different cells.
Multi-Day Cell Tracking: Proliferation and Migration
One of our goals in developing BFCs was to allow patterned cells to proliferate and migrate. mESCs, for instance, divide rapidly (∼14 hour doubling time [7]) and the resulting colonies also migrate. In order to study these phenomena, tracking cell growth and movement over multiple days, one needs a technique that patterns cells onto an unconfined area. Currently, there are several existing cell patterning techniques that are suitable for cell tracking over timescales where proliferation or migration is not significant (usually < 24 hours). However, these techniques trap the cells in a confined area, such as an extracellular matrix island [16] or a microwell [21,22]. Using BFCs, however, we can pattern cells onto substrates that are entirely unconfined, enabling both proliferation and migration.
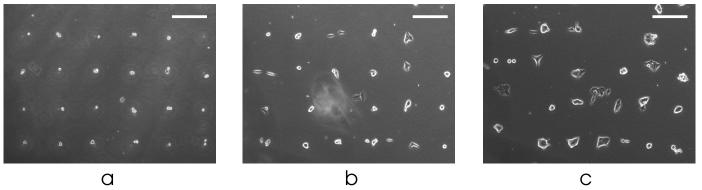
In addition, since each cell has a unique coordinate within the patterned grid, it is possible to track the resulting cells or colonies if desired. In Figure 3 we show tracking of one area of patterned mESCs over two days. By day 2, almost all of the initially patterned cells have proliferated into mESC colonies, suggesting that our technique is gentle on the cells and not grossly affecting cell health. The fast doubling time of mESCs causes some deterioration of the pattern by day 2 (Fig. 3c), which is strong evidence that the cells can indeed migrate freely and is a key feature of our approach. While this precludes the use of BFCs for creating long-term stationary patterns, we emphasize that stationary patterns are exactly what we are trying to avoid.
Figure 3.
Multi-day cell tracking. Proliferation and migration of mESCs on (a) day 0, (b) day 1, and (c) day 2. Scale bar, 200 μm.
Modulating the Cell Seeding Density
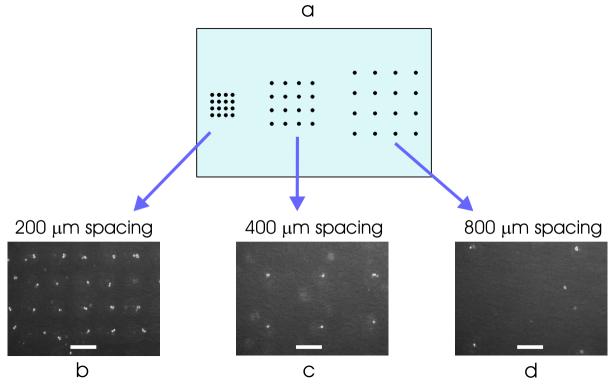
Diffusible signaling between cells has been shown to be an important component of the stem cell microenvironment. For instance, stem cell seeding density can affect both differentiation and self-renewal [5-7]. Traditional cell culture techniques control the cell density only at the macroscopic level of the cell-culture dish, creating poor uniformity in cell density and resulting in a cellular microenvironment that varies from cell to cell. Simply using BFCs to pattern the cells into a grid allows every cell to experience a more uniform microenvironment. Additionally, by varying the spacing of the microwells across the chip (Fig 4a), we can pattern cells at multiple seeding densities on the same substrate. We demonstrated this functionality by patterning mESCs on a 60×15 mm TCPS dish at three seeding densities – with a cell-cell spacing of 200, 400, and 800 μm (Fig. 4b-d).
Figure 4.
Modulating the cell seeding density. (a) By varying the microwell-to-microwell spacing across the chip, we can pattern cells at multiple seeding densities on the same 60×15 mm TCPS dish. Cell patterning with cell-cell spacing of: (b) 200 μm, (c) 400 μm, and (d) 800 μm. Scale bar, 200 μm.
Precise Control of the Stem Cell Microenvironment
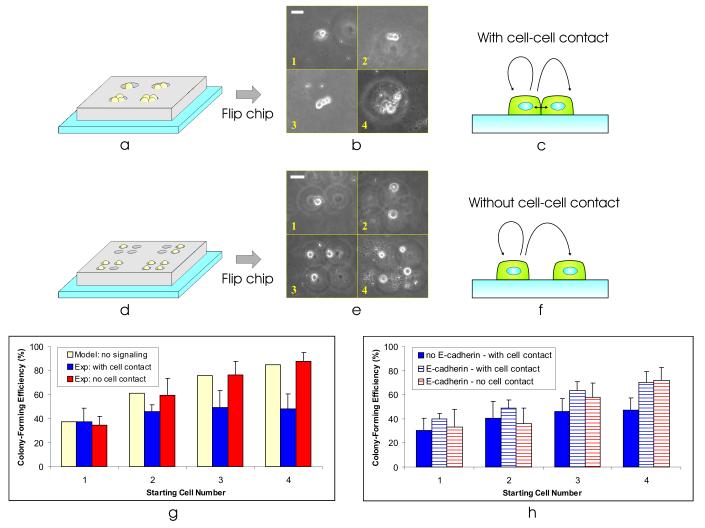
To locally control the mESC microenvironment, we would like to be able to incrementally and independently modulate cell-cell contact. Thus, we require the ability to seed a variable-sized group of cells, with and without cell-cell contact, spaced far enough away from other cell groups to prevent intercolony signaling. This type of precise cell seeding would not be possible using traditional cell-culture techniques. Instead, we used microwells that are larger than the average cell size to allow multiple numbers of cells to stochastically load into each well (Fig. 5a). Using this approach, we were able to pattern small clusters of 1-4 mESCs (Fig. 5b), allowing them to experience cell-cell contact (Fig. 5c).
Figure 5.
Precise control of the stem cell microenvironment. (a) By making the well dimensions large enough to trap multiple cells, we can vary the number of cells patterned at each spot. (b) Using a BFC with microwell-to-microwell spacing of 0.75 mm, well diameter of 40 μm, and well height of 50 μm, we patterned clusters of cells with 1-4 cells at each spot. Collage is from images taken randomly from different areas on the substrate. Scale bar, 40 μm. (c) Patterning the cells as clusters allows them to experience cell-cell contact. (d) Using multiple single-cell sized wells, we can pattern small groups of cells without any cell-cell contact. (e) Using a BFC with microwell-to-microwell spacing of 0.75 mm, well diameter of 30 μm, and well height of 30 μm, we patterned small groups of 1-4 single mESCs at each spot. Using microwells with edge-edge distance of ∼20 μm (n = 30), we patterned the cells with a membrane-membrane distance of 45 ± 15 μm (n = 33). Collage is from images taken randomly from different areas on the substrate. Scale bar, 40 μm. (f) Patterning mESCs as small groups of single cells allows us to turn off cell-cell contact. (g) Colony-forming efficiency (2 days after cell seeding) as a function of starting cell number for three cases – model of no cell-cell signaling (solid yellow bars), experiment with cell-cell contact (solid blue bars), experiment without cell-cell contact (solid red bars). Bars on experimental data indicate 1 standard deviation from the mean. (h) Colony-forming efficiency as a function of starting cell number for three cases – cells in contact without exposure to anti-E-cadherin (solid blue bars), cells in contact with exposure to anti-E-cadherin (striped blue bars), and cells without contact with exposure to anti-E-cadherin (striped red bars). Bars on experimental data indicate 1 standard deviation from the mean.
Patterning cells in this fashion, we found that the single-cell colony-forming efficiency was ∼37.5% (Fig. 5g), which is typical for mESCs [30]. Interestingly, as we increase the number of starting cells per cluster, we do not observe a proportional increase in colony-forming efficiency. Assuming there was no signaling between the cells in a group, the probability that at least one of n independent cells in a group would form a colony is 1 - (probability that all the cells in a cluster die). As the number of cells increases, the probability that a colony will form approaches 100%. For n = 4 cells, this translates into a 1−(1−0.375)4 = 85% colony-forming efficiency (Fig. 5g, solid yellow bars), whereas we observe a colony forming efficiency of 48 ± 13% with ABJ1s (n = 5 chips, 156 total colonies counted) (Fig. 5g, solid blue bars).
In order to investigate the cause of this decreased colony-forming efficiency, we used BFCs with different well configurations to selectively turn off cell-cell contact. Using small groups of single-cell sized microwells (Fig. 5d), we patterned groups of 1-4 single mESCs (Fig. 5e), without cell-cell contact (Fig. 5f). Turning off cell-cell contact, we find that the mESC colony-forming efficiency reverts to that expected for independently acting cells (e.g., no cell-cell signaling) (Fig. 5g, solid red bars). This suggests that cell-cell contact plays a primary role in depressing colony formation. We repeated the contact and no contact patterning experiments, both at least five times, with the same cell lines as well as with another cell line (D3) (data not shown) – resulting in the same cell contact-mediated depression in colony-forming efficiency – ruling out time, population, or cell line artifacts.
To investigate the molecules involved with this cell contact-mediated depression in colony formation, we used a blocking antibody known to inhibit E-cadherin signaling [31]. E-cadherin is a transmembrane protein that has been shown to regulate cell-cell adhesion signaling in mESCs [32]. We incubated a single-cell suspension of mESCs in anti-E-cadherin antibody before patterning and then repeated the patterning experiments (Fig. 5a-f). Since the antibody is not present in the media after patterning, E-cadherin signaling is primarily disrupted during seeding, allowing us to assay its initial effect on proliferation. Compared to the depressed colony-forming efficiency that we observed previously for cells in contact (Fig. 5h, solid blue bars), the cells exposed to anti-E-cadherin had colony-forming efficiencies that were similar with and without cell contact (Fig. 5h, striped blue and red bars), suggesting that E-cadherin is involved in this negative regulatory pathway.
Discussion
Implications of the Bio Flip Chip for Cell Patterning
One should be able to use BFCs, in principle, to pattern any cell type. We measured the unattached mESC diameter as 20 ± 3 μm (n = 30) and therefore tried a range of microwell diameters (30, 35, and 40 μm) and heights (30, 40, and 50 μm) to optimize single-cell trapping. The optimal well diameter and height was 30 μm. To use the BFC to pattern single cells of another cell type, we recommend using a microwell diameter and height equal to 10 μm greater than the unattached cell diameter. In general, a well diameter-to-height ratio of one maximized single-cell patterning efficiency. When the ratio was >1, the wells were too shallow and the cells were washed out of the wells during clearing, reducing overall efficiency. When the ratio was <1, the wells were too deep, and multiple cells were trapped in each well.
Our results demonstrate the key advantages of this new technology and suggest additional applications. First, the ability to pattern single motile cells (Fig. 3a-c) enables patterned chemotaxis assays. These are currently performed using switchable substrates [33], which trade off substrate-independence and ease of use for temporal control of motility. Second, BFCs can be used to pattern cells onto other cells (Fig. 2e). Many cell types or in vitro assays require support cells; including maintaining hESCs on feeders [34], differentiating stem cells in the presence of stroma [35], or co-culture of primary cells with non-parenchymal cells [36]. μCP is the predominant approach for patterning such interactions, but is limited to side-by-side patterning where the 2nd cell type fills the interstitial spaces of the first patterned cell type. This is a general feature of cell patterning methods that trap and pattern cells with the same substrate, whereas the BFC technique is completely substrate-independent. This substrate independence is what allows us to pattern one cell type onto another cell type.
A key factor for the impact of various patterning technologies is their ease-of-use and transferability. Many existing patterning technologies are either difficult to fabricate or use [9,17,18,23-26], a barrier that prevents their wide adoption. Indeed, the most common cell patterning technique is μCP [15,16], which is simple enough to be used by those with moderate skill in a typical biological laboratory. BFCs are poised to have similar adoptability, since they require no external equipment or chemicals. Using BFCs is complementary and orthogonal to use of μCP (Fig. 2c); μCP provides higher patterning efficiencies and a greater range of pattern shapes, while the BFCs provide substrate-independence with no restrictions on motility or proliferation.
Implications of the Bio Flip Chip for Biology
Mouse and human ESCs require very different cell seeding conditions in order to maximize self-renewal, defined as proliferation and maintenance of pluripotency. Convention has dictated that mESCs should be seeded as single cells [12,13,30,37-39], since clusters of thousands of cells can cause differentiation [40], while hESCs have < 1% single-cell colony-forming efficiencies [41] and prefer to be seeded as clusters of hundreds of cells [42]. These existing studies investigate ESC behavior at both extremes of cell seeding conditions, as single cells or hundreds to thousands of cells. However, there have been no reports studying the effects of seeding smaller cell clusters, allowing incremental changes in cell signaling. While traditional cell-culture techniques cannot reliably seed small groups of cells in a scalable fashion, with or without cell contact, we can use BFCs to make these types of experiments possible.
Our data provides the first quantitative evidence that seeding mESCs as single cells maximizes colony formation, supporting the long-established protocols of single-cell seeding to maximize self-renewal. Indeed, if each cell cluster has roughly the same colony-forming efficiency of a single cell (Fig. 5g, solid blue bars), then seeding cells in clusters effectively discards the majority of the cells in the culture. In addition, we show that E-cadherin is involved in this cell contact-mediated depression of colony formation. Previous studies have shown that cell agglomeration of mESCs in embryoid bodies negatively affects cell proliferation [43] and that E-cadherin mediates this process [31], possibly occurring in a similar fashion to what we observed here.
Thus, the BFC allows us to precisely modulate cell-cell contact to determine its effects on a specific cell behavior. Potential signaling molecules can then be investigated in the presence of various molecular inhibitors or genetic alterations. The BFC technology has allowed us to quickly determine the mechanisms and molecules involved in mESC colony-forming efficiency, quantitatively validating a protocol practiced for over two decades.
Conclusions
We have presented a novel cell patterning technology called the Bio Flip Chip. This technology offers a unique combination of capabilities – including patterning cells with single-cell resolution, patterning large numbers of cells, allowing the patterned cells room to grow and move, patterning onto any substrate, being gentle on the cells, and being easy to use. In addition, we have used the BFCs for precise control of the stem cell microenvironment, allowing us to investigate previously unanswered questions in stem cell biology. Thus, the BFC serves as a powerful tool, enabling us to create novel biomaterials for applications in tissue engineering and regenerative medicine.
Acknowledgements
We would like to thank Stephanie Flavin for her help with all the substrate experiments, Somponnat Sampattavanich for his help patterning the fluorescent gelatin, Dr. Sangeeta Bhatia for her informative discussions on patterning onto 3-D substrates, Dr. George Daley for his generous donation of ABJ1 cells, and the MIT Microsystems Technology Lab for their help with fabrication. This work was supported by: the National Institutes of Health (RR18878), a National Science Foundation Graduate Fellowship, a Harvard-MIT Health Sciences and Technology Medical Engineering and Medical Physics Fellowship, and an MIT Presidential Fellowship. The authors confirm that there are no known conflicts of interest associated with this publication and there has been no significant financial support for this work that could have influenced its outcome.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Heasley LE. Autocrine and paracrine signaling through neuropeptide receptors in human cancer. Oncogene. 2001;20(13):1563–1569. doi: 10.1038/sj.onc.1204183. [DOI] [PubMed] [Google Scholar]
- 2.Navab M, Imes SS, Hama SY, Hough GP, Ross LA, Bork RW, et al. Monocyte Transmigration Induced by Modification of Low-Density-Lipoprotein in Cocultures of Human Aortic-Wall Cells Is Due to Induction of Monocyte Chemotactic Protein-1 Synthesis and Is Abolished by High-Density-Lipoprotein. Journal of Clinical Investigation. 1991;88(6):2039–2046. doi: 10.1172/JCI115532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Quehenberger O. Molecular mechanisms regulating monocyte recruitment in atherosclerosis. Journal of Lipid Research. 2005;46(8):1582–1590. doi: 10.1194/jlr.R500008-JLR200. [DOI] [PubMed] [Google Scholar]
- 4.Tsai RYL, McKay RDG. Cell contact regulates fate choice by cortical stem cells. Journal of Neuroscience. 2000;20(10):3725–3735. doi: 10.1523/JNEUROSCI.20-10-03725.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Purpura KA, Aubin JE, Zandstra PW. Sustained in vitro expansion of bone progenitors is cell density dependent. Stem Cells. 2003;22(1):39–50. doi: 10.1634/stemcells.22-1-39. [DOI] [PubMed] [Google Scholar]
- 6.Javazon EH, Colter DC, Schwarz EJ, Prockop DJ. Rat marrow stromal cells are more sensitive to plating density and expand more rapidly from single-cell-derived colonies than human marrow stromal cells. Stem Cells. 2001;19(3):219–225. doi: 10.1634/stemcells.19-3-219. [DOI] [PubMed] [Google Scholar]
- 7.Zandstra PW, Le HV, Daley GQ, Griffith LG, Lauffenburger DA. Leukemia inhibitory factor (LIF) concentration modulates embryonic stem cell self-renewal and differentiation independently of proliferation. Biotechnology and Bioengineering. 2000;69(6):607–617. [PubMed] [Google Scholar]
- 8.Bhatia SN, Balis UJ, Yarmush ML, Toner M. Microfabrication of hepatocyte/fibroblast co-cultures: Role of homotypic cell interactions. Biotechnology Progress. 1998;14(3):378–387. doi: 10.1021/bp980036j. [DOI] [PubMed] [Google Scholar]
- 9.Albrecht DR, Underhill GH, Wassermann TB, Sah RL, Bhatia SN. Probing the role of multicellular organization in three-dimensional microenvironments. Nature Methods. 2006;3(5):369–375. doi: 10.1038/nmeth873. [DOI] [PubMed] [Google Scholar]
- 10.Gratsch TE, O'Shea KS. Noggin and chordin have distinct activities in promoting lineage commitment of mouse embryonic stem (ES) cells. Developmental Biology. 2002;245(1):83–94. doi: 10.1006/dbio.2002.0629. [DOI] [PubMed] [Google Scholar]
- 11.Sato N, Meijer L, Skaltsounis L, Greengard P, Brivanlou AH. Maintenance of pluripotency in human and mouse embryonic stem cells through activation of Wnt signaling by a pharmacological GSK-3-specific inhibitor. Nature Medicine. 2004;10(1):55–63. doi: 10.1038/nm979. [DOI] [PubMed] [Google Scholar]
- 12.Chambers I, Colby D, Robertson M, Nichols J, Lee S, Tweedie S, et al. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 2003;113(5):643–655. doi: 10.1016/s0092-8674(03)00392-1. [DOI] [PubMed] [Google Scholar]
- 13.Niwa H, Miyazaki J, Smith AG. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nature Genetics. 2000;24(4):372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- 14.Raz R, Lee CK, Cannizzaro LA, D'Eustachio P, Levy DE. Essential role of STAT3 for embryonic stem cell pluripotency. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(6):2846–2851. doi: 10.1073/pnas.96.6.2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Singhvi R, Kumar A, Lopez GP, Stephanopoulos GN, Wang DIC, Whitesides GM, et al. Engineering Cell-Shape and Function. Science. 1994;264(5159):696–698. doi: 10.1126/science.8171320. [DOI] [PubMed] [Google Scholar]
- 16.Chen CS, Mrksich M, Huang S, Whitesides GM, Ingber DE. Geometric control of cell life and death. Science. 1997;276(5317):1425–1428. doi: 10.1126/science.276.5317.1425. [DOI] [PubMed] [Google Scholar]
- 17.Jiang XY, Ferrigno R, Mrksich M, Whitesides GM. Electrochemical desorption of self-assembled monolayers noninvasively releases patterned cells from geometrical confinements. Journal of the American Chemical Society. 2003;125(9):2366–2367. doi: 10.1021/ja029485c. [DOI] [PubMed] [Google Scholar]
- 18.Lahann J, Mitragotri S, Tran TN, Kaido H, Sundaram J, Choi IS, et al. A reversibly switching surface. Science. 2003;299(5605):371–374. doi: 10.1126/science.1078933. [DOI] [PubMed] [Google Scholar]
- 19.Ostuni E, Kane R, Chen CS, Ingber DE, Whitesides GM. Patterning mammalian cells using elastomeric membranes. Langmuir. 2000;16(20):7811–7819. [Google Scholar]
- 20.Folch A, Jo B-H, Hurtado O, Beebe DJ, Tonor M. Microfabricated elastomeric stencils for micropatterning cell cultures. J Biomed Mater Res. 2000;52:346–353. doi: 10.1002/1097-4636(200011)52:2<346::aid-jbm14>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 21.Revzin A, Tompkins RG, Toner M. Surface engineering with poly(ethylene glycol) photolithography to create high-density cell arrays on glass. Langmuir. 2003;19(23):9855–9862. [Google Scholar]
- 22.Rettig JR, Folch A. Large-scale single-cell trapping and imaging using microwell arrays. Analytical Chemistry. 2005;77(17):5628–5634. doi: 10.1021/ac0505977. [DOI] [PubMed] [Google Scholar]
- 23.Birkbeck AL, Flynn RA, Ozkan M, Song DQ, Gross M, Esener SC. VCSEL Arrays as micromanipulators in chip-based biosystems. Biomedical Microdevices. 2003;5(1):47–54. [Google Scholar]
- 24.Dufresne ER, Grier DG. Optical tweezer arrays and optical substrates created with diffractive optics. Review of Scientific Instruments. 1998;69(5):1974–1977. [Google Scholar]
- 25.Ozkan M, Pisanic T, Scheel J, Barlow C, Esener S, Bhatia SN. Electro-optical platform for the manipulation of live cells. Langmuir. 2003;19(5):1532–1538. [Google Scholar]
- 26.Rosenthal A, Voldman J. Dielectrophoretic traps for single-particle patterning. Biophysical Journal. 2005;88(3):2193–2205. doi: 10.1529/biophysj.104.049684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anderson DG, Levenberg S, Langer R. Nanoliter-scale synthesis of arrayed biomaterials and application to human embryonic stem cells. Nature Biotechnology. 2004;22(7):863–866. doi: 10.1038/nbt981. [DOI] [PubMed] [Google Scholar]
- 28.Gray DS, Tan JL, Voldman J, Chen CS. Dielectrophoretic registration of living cells to a microelectrode array. Biosensors & Bioelectronics. 2004;19(7):771–780. doi: 10.1016/j.bios.2003.08.013. [DOI] [PubMed] [Google Scholar]
- 29.Frenea M, Faure SP, Le Pioufle B, Coquet P, Fujita H. Positioning living cells on a high-density electrode array by negative dielectrophoresis. Materials Science & Engineering C-Biomimetic and Supramolecular Systems. 2003;23(5):597–603. [Google Scholar]
- 30.Schratt G, Weinhold B, Lundberg AS, Schuck S, Berger J, Schwarz H, et al. Serum response factor is required for immediate-early gene activation yet is dispensable for proliferation of embryonic stem cells. Molecular and Cellular Biology. 2001;21(8):2933–2943. doi: 10.1128/MCB.21.8.2933-2943.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dang SM, Gerecht-Nir S, Chen J, Itskovitz-Eldor J, Zandstra PW. Controlled, scalable embryonic stem cell differentiation culture. Stem Cells. 2004;22(3):275–282. doi: 10.1634/stemcells.22-3-275. [DOI] [PubMed] [Google Scholar]
- 32.Nagano K, Taoka M, Yamauchi Y, Itagaki C, Shinkawa T, Nunomura K, et al. Large-scale identification of proteins expressed in mouse embryonic stem cells. Proteomics. 2005;5(5):1346–1361. doi: 10.1002/pmic.200400990. [DOI] [PubMed] [Google Scholar]
- 33.Jiang X, Bruzewicz DA, Wong AP, Piel M, Whitesides GM. Directing cell migration with asymmetric micropatterns. PNAS. 2005;102(4):975–978. doi: 10.1073/pnas.0408954102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Richards M, Fong CY, Chan WK, Wong PC, Bongso A. Human feeders support prolonged undifferentiated growth of human inner cell masses and embryonic stem cells. Nature Biotechnology. 2002;20(9):933–936. doi: 10.1038/nbt726. [DOI] [PubMed] [Google Scholar]
- 35.Kawasaki H, Mizuseki K, Nishikawa S, Kaneko S, Kuwana Y, Nakanishi S, et al. Induction of midbrain dopaminergic neurons from ES cells by stromal cell-derived inducing activity. Neuron. 2000;28(1):31–40. doi: 10.1016/s0896-6273(00)00083-0. [DOI] [PubMed] [Google Scholar]
- 36.Bhatia SN, Yarmush ML, Toner M. Controlling cell interactions by micropatterning in co-cultures: Hepatocytes and 3T3 fibroblasts. Journal of Biomedical Materials Research. 1997;34(2):189–199. doi: 10.1002/(sici)1097-4636(199702)34:2<189::aid-jbm8>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 37.Berrill A, Tan HL, Wuang SC, Fong WJ, Choo ABH, Oh SKW. Assessment of stem cell markers during long-term culture of mouse embryonic stem cells. Cytotechnology. 2004;44(12):77–91. doi: 10.1023/B:CYTO.0000043414.90681.c2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matsuda T, Nakamura T, Nakao K, Arai T, Katsuki M, Heike T, et al. STAT3 activation is sufficient to maintain an undifferentiated state of mouse embryonic stem cells. Embo Journal. 1999;18(15):4261–4269. doi: 10.1093/emboj/18.15.4261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ying QL, Nichols J, Chambers I, Smith A. BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell. 2003;115(3):281–292. doi: 10.1016/s0092-8674(03)00847-x. [DOI] [PubMed] [Google Scholar]
- 40.Hamazaki T, Oka M, Yamanaka S, Terada N. Aggregation of embryonic stem cells induces Nanog repression and primitive endoderm differentiation. Journal of Cell Science. 2004;117(23):5681–5686. doi: 10.1242/jcs.01489. [DOI] [PubMed] [Google Scholar]
- 41.Amit M, Carpenter MK, Inokuma MS, Chiu CP, Harris CP, Waknitz MA, et al. Clonally derived human embryonic stem cell lines maintain pluripotency and proliferative potential for prolonged periods of culture. Developmental Biology. 2000;227(2):271–278. doi: 10.1006/dbio.2000.9912. [DOI] [PubMed] [Google Scholar]
- 42.Suemori H, Yasuchika K, Hasegawa K, Fujioka T, Tsuneyoshi N, Nakatsuji N. Efficient establishment of human embryonic stem cell lines and long-term maintenance with stable karyotype by enzymatic bulk passage. Biochemical and Biophysical Research Communications. 2006;345(3):926–932. doi: 10.1016/j.bbrc.2006.04.135. [DOI] [PubMed] [Google Scholar]
- 43.Dang SM, Kyba M, Perlingeiro R, Daley GQ, Zandstra PW. Efficiency of embryoid body formation and hematopoietic development from embryonic stem cells in different culture systems. Biotechnology and Bioengineering. 2002;78(4):442–453. doi: 10.1002/bit.10220. [DOI] [PubMed] [Google Scholar]