Abstract
Photosynthetic water oxidation occurs at the Mn-containing catalytic site of photosystem II (PSII). By the use of 14C-labeled amines and SDS-denaturing PAGE, covalent adducts derived from primary amines and the PSII subunits, CP47, D2/D1, and the Mn-stabilizing protein, can be observed. When PSII contains the 18- and 24-kDa extrinsic proteins, which restrict access to the active site, no 14C labeling is obtained. NaCl, but not Na2SO4, competes with 14C labeling in Mn-containing PSII preparations, and the concentration dependence of this competition parallels the activation of oxygen evolution. Formation of 14C-labeled adducts is observed in the presence or in the absence of a functional manganese cluster. However, no significant Cl− effect on 14C labeling is observed in the absence of the Mn cluster. Isolation and quantitation of the 14C-labeled aldehyde product, produced from [14C]benzylamine, gives yields of 1.8 ± 0.3 mol/mol PSII and 2.9 ± 0.2 mol/mol in Mn-containing and Mn-depleted PSII, respectively. The corresponding specific activities are 0.40 ± 0.07 μmol(μmol PSII-hr)−1 and 0.64 ± 0.04 μmol(μmol PSII-hr)−1. Cl− suppresses the production of [14C]benzaldehyde in Mn-containing PSII, but does not suppress the production in Mn-depleted preparations. Control experiments show that these oxidation reactions do not involve the redox-active tyrosines, D and Z. Our results suggest the presence of one or more activated carbonyl groups in protein subunits that form the active site of PSII.
Keywords: redox-active amino acid, amine oxidase, photosystem II, activated carbonyl
Photosystem II (PSII) is the photosynthetic reaction center that catalyzes the light-driven oxidation of water to O2 and reduction of plastoquinone to plastoquinol. PSII is a membrane protein that contains hydrophobic and extrinsic subunits. A Mn-containing catalytic site is located on the membrane’s luminal side. Four photo-induced charge separations are required to release O2; the sequentially oxidized forms of the catalytic site are called the Sn states. PSII contains two well characterized redox-active tyrosines, D and Z. Redox-active tyrosine D forms a stable radical with no known role in oxygen evolution; tyrosine Z acts as an intermediate electron carrier (reviewed in refs. 1 and 2). Recently, a new EPR signal has been observed in site-directed mutants of the cyanobacterium Synechocystis sp. PCC 6803 (3–6). It has been proposed that this signal arises from a third redox-active tyrosine, M, that is posttranslationally modified (5, 6).
Photosynthetic oxygen evolution requires Ca+2 and Cl− as essential cofactors, and primary amines are known to inhibit oxygen evolution (reviewed in ref. 7). Previous work with PSII has shown that some primary amines, such as methylamine, inhibit oxygen evolution in a Cl−-dependent fashion. Biochemical analysis has led to the conclusion that amine binding occurs at a Cl−-binding site on Mn (8–10).
Here, we report new evidence concerning the interaction of primary amines with PSII. The experiments described here can be distinguished from earlier work (8–10), because we have employed PSII preparations from which extrinsic polypeptides have been depleted. Removal of these subunits decreases the binding affinities for Ca+2 (11) and Cl− (12), so that an exogenous source is required to maintain the maximum rate of steady-state O2 evolution activity (13, 14). Removal of these subunits also increases the access of reductants into the catalytic site (15, 16). Our data show that removal of extrinsic subunits allows the incorporation of 14C from primary amines into PSII subunits and the oxidation of benzylamine to form benzaldehyde. Control experiments establish that these reactions occur independently of high valence manganese and tyrosyl radicals, D⋅ and Z⋅. To explain our results, we propose the presence of one or more activated carbonyl groups at the active site of PSII.
MATERIALS AND METHODS
Purified BSAO (bovine serum amine oxidase), at a protein concentration of 17.5 mg/ml and specific activity of 0.30 units/mg, was a generous gift from J. Klinman and S. Wang. The enzyme is a disulfide-linked homodimer; the monomeric molecular mass is 85,000 (17). The specific activity predicts a topaquinone content of 0.6 per monomeric subunit (18, 19). For binding experiments, the BSAO sample was diluted 50- to 100-fold with buffer A (400 mM sucrose/50 mM Hepes-NaOH, pH 7.5).
PSII-1 (PSII membranes) (20), PSII-2 (salt-washed PSII membranes that are depleted of the 24- and 18-kDa proteins and other extrinsic subunits) (21), PSII-3 (PSII membranes that are depleted of the manganese cluster, the manganese stabilizing protein, and other extrinsic subunits) (22), and core complexes (23) from spinach were isolated. Cl− depletion of PSII-1, PSII-2, and PSII-3 was performed by repeated homogenization into buffer A (remaining Cl− <1.6 mM). Spinach MSP (manganese stabilizing protein) (24), spinach LHC (light-harvesting complex) (23), and cyanobacterial PSI (photosystem I) and PSII (25) samples were isolated by procedures previously described. These samples were exchanged into buffer A, and Cl− was depleted by several rounds of concentration with a Centricon 10 or 100 (Amicon) and subsequent dilution with buffer A. O2 evolution and chlorophyll assays were performed by methods described previously (26).
Binding experiments were performed on 10-μl samples containing 40 pmol of PSII, 40 pmol of PSI, 30 pmol of LHC, and 80 pmol of the MSP (27). The number of moles of PSII reaction centers was determined as previously described (28). The number of moles of LHC and PSI was determined from assumed antenna sizes of 160 mol Chl/mol and 80 mol Chl/mol, respectively (see refs. 3 and 23 and references therein). The amount of amine oxidase homodimer was 20 pmol in the experiments described in Fig. 1 A–C and F and 40 pmol in Fig. 1E. The appropriate compound was added by mixing at room temperature and under room light (7–9 μE s−1m−2) or in the dark. In competition studies, the concentration of NaCl, Na2SO4, Tris, or dimethylamine was adjusted by the addition of a concentrated stock solution in buffer A. Where appropriate, the stock solution was pH adjusted by the addition of H2SO4. In both anion and amine competition studies, the anion or unlabeled amine was added, and samples were equilibrated for 30 min in the dark at 0°C before the addition of the [14C]amine. Denaturing/reducing buffer was added to give the final concentrations: 2.7 M urea, 2.1% SDS, 62.5 mM DTT, and 52 mM Na2CO3, and samples were incubated for 30 min at room temperature. Some samples also contained 0.04% bromophenol blue, which did not change the pattern of 14C labeling. Samples were subjected to PAGE according to the modified Neville method (29). In some experiments, all manipulations were performed in the dark.
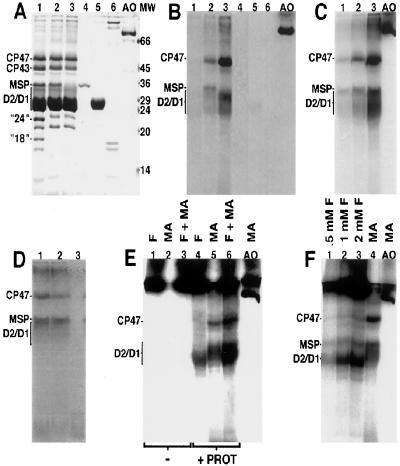
Figure 1.
SDS-denaturing PAGE of PSII and other proteins. In A and B, samples were incubated with 4 mM [14C]methylamine-hydrochloride for 30 min. In A, the gel was stained with Coomassie brilliant blue. Samples not treated with [14C]methylamine give similar staining patterns to those shown here. In B, fluorography was performed. Lanes: 1, PSII-1; 2, PSII-2; 3, PSII-3; 4, MSP; 5, LHC; 6, PSI; AO, BSAO. In C, electrophoresis and fluorography were performed on PSII and amine oxidase. PSII-2 was incubated with 4 mM [14C]methylamine-hydrochloride for 30 min (lane 1), 60 min (lane 2), and 120 min (lane 3). Lane AO contains amine oxidase incubated with 4 mM [14C]methylamine-hydrochloride for 30 min. In D, electrophoresis and fluorography were performed on PSII-2 (lane 1), PSII-2 plus 5 molar equivalents of plastoquinone (lane 2), and purified plastoquinone (lane 3), after incubation of samples with 4 mM [14C]methylamine-hydrochloride for 30 min. Note that, unlike gels in A–C and E and F, this gel was run so that the dye front can be visualized. In E, no protein (lanes 1–3) or PSII-3 (lanes 4–6) was incubated with 2 mM [14C]formaldehyde (lanes 1 and 4), 4 mM [14C]methylamine-hydrochloride (lanes 2 and 5), or [14C]formaldehyde (2 mM) and [14C]methylamine-hydrochloride (4 mM) (lanes 3 and 6). The incubation time was 30 min. Lane AO contains amine oxidase incubated with 4 mM [14C]methylamine-hydrochloride for 15 min. In F, PSII-2 was incubated with 0.5 mM (lane 1), 1.0 mM (lane 2), 2.0 mM [14C]formaldehyde (lane 3), or 4 mM [14C]methylamine-hydrochloride (lane 4). The incubation time was 60 min. Lane AO contains amine oxidase incubated with 4 mM [14C]methylamine-hydrochloride for 15 min.
Gels were either stained with Coomassie brilliant blue R (30) or processed for fluorography (31). We have not attempted to distinguish the D2 and D1 polypeptides in these experiments, because the bands overlap. The average length of exposure was 12–13 days at −80°C using Kodak Full Speed Blue film and an intensifying screen (DuPont Cronex Lightning Plus). Bands could be visualized after 4-day exposures. A Personal Densitometer SI (Molecular Dynamics) was used to scan the fluorogram or stained gel. Bands were analyzed separately. The number of pixels exposed in the background was subtracted from the number of pixels in each lane. To ease comparison of gels run on different days, the number of counts in each lane was divided by the total number of counts in all lanes. The first data point was then normalized to 100%.
Extraction and purification of [14C]benzaldehyde produced from PSII samples was performed with a Gilson HPLC system (28) and by a procedure previously described (18), except that column chromatography was performed at room temperature and the flow rate was 0.9 ml/min. PSII samples at 1 mg Chl/ml were mixed and incubated with 200 or 400 μM [14C]benzylamine-hydrochloride and [12C]benzylamine-hydrochloride to give a 1 mM total concentration in 248 μl. Acetone was added to a final concentration of 55%; samples were vortexed, sonicated for 30 min, and centrifuged at 13,000 × g, and the supernatant was filtered and injected. Buffer B, containing 50 mM Hepes-NaOH, pH 7.5, was employed. Buffers A and B gave similar results in the SDS/PAGE binding assay.
Room temperature EPR spectra were recorded, and illumination in the cavity was provided (26). Spectral conditions were: frequency, 9.5 GHz; modulation amplitude, 4 G; power, 15 mW; scan range, 100 G; time constant, 2 sec; scan time, 4 min; gain, 1.6 × 104; concentration, 3.0 mg Chl/ml; acceptor, 1 mM potassium ferricyanide. Red and heat-filtered light was employed.
RESULTS
PAGE, [14C]Methylamine, and BSAO.
Novel cofactors, containing activated carbonyl groups and arising from oxidation of tyrosine, have been identified in enzymes (32–37). For example, the tyrosine-derived “topa” (2,4,5-trihydroxyphenylalanine) in BSAO reacts with primary amines and catalyzes the oxidation of these compounds to form ammonia, hydrogen peroxide, and the corresponding aldehyde (32). Oxidation of amines is diagnostic of this type of functional group; model compounds catalyze this reaction in vitro (38, 39). We will use this enzyme as a positive control, because the protein is known to contain an activated carbonyl group. In the proposed mechanism of BSAO, the carbonyl carbon at position 5 of the topaquinone ring undergoes a nucleophilic attack by a primary amine forming a substrate–Schiff base complex (reviewed in refs. 33 and 36). It has been shown previously that it is possible to trap the substrate–Schiff base complex of primary amines and the topaquinone under reducing conditions (40).
Fig. 1 demonstrates that if BSAO is incubated with 14C-labeled methylamine-hydrochloride (specific activity: 54–57 mCi/mmol; 1 Ci = 37 GBq) and then treated with the reductant, DTT, and denaturants, labeling of a species migrating at an apparent molecular mass of 69 kDa (Fig. 1 A and B, lane AO), corresponding to monomeric BSAO, is observed. Although it is known that sulfhydryl reagents can act as reducing agents for Schiff bases (41), NaCNBH3 was previously employed (40) and would be expected to be a more effective reductant (41). However, NaCNBH3 is not a good reducing agent for disulfide linkages and, in our hands, results in aberrant electrophoretic migration of BSAO. Therefore, we compared the effects of DTT with the effects of a mixture of DTT and NaCNBH3. Use of both reductants resulted in an increase in the amount of 14C labeling of BSAO by a factor of not more than two, suggesting that this concentration of DTT is quite effective in the reduction of the substrate–Schiff base complex under these denaturing conditions.
PAGE, [14C]Methylamine, and PSII.
Extrinsic subunits of PSII can be removed from the reaction center by treatment with solutions of high ionic strength (Fig. 1A, lanes 1 and 2). Among these subunits are the so-called 18- and 24-kDa polypeptides (for example, see ref. 21). In our gel system, the 24-kDa protein comigrates with another subunit of similar molecular mass (Fig. 1A, compare lanes 1, 2, and 3).
As isolated, PSII membranes, which contain the 18- and 24-kDa proteins (PSII-1), give no detectable stable binding of radioactive compound (Fig. 1 A and B, lane 1). However, after removal of the 18- and 24-kDa subunits, generating PSII-2 and increasing access to the active site, treatment with [14C]methylamine and reduction with DTT result in covalent binding of 14C-labeled compounds to protein subunits (Fig. 1 A and B, lane 2). The labeled subunits with the lowest Rf factors have apparent molecular masses of 49 and 36 kDa and correspond to subunits called CP47 (chlorophyll-binding protein of approximate molecular mass of 47,000) and MSP (20, 42). The band migrating with an apparent molecular mass between 35 and 27 kDa corresponds to either the D2/D1 polypeptides (intrinsic subunits that bind most of the prosthetic groups in photosystem II) or to LHC (20, 42). A band with the same Rf factor is also labeled in core PSII particles (23) from which the LHC has been removed by ion chromatography in the presence of excess detergent (data not shown). The same band is labeled in cyanobacterial PSII (25), which lacks these LHC proteins (data not shown). These data, obtained on PSII after further purification or as isolated from a prokaryotic source, show that this 14C-labeled band corresponds to the D2 and/or D1 polypeptides. These data also indicate that the labeling does not arise from an enzymatic contaminant, because evolutionarily divergent enzymes and highly purified PSII samples give 14C incorporation. The amount of labeling is decreased at pH 6.0, when compared with pH 7.5 (data not shown). This is the expected result if the free base of methylamine is the reactive species. In Fig. 1, data indicate that 14C labeling from methylamine is not dependent on the presence of a functional Mn cluster, because 14C labeling of CP47 and D2/D1 (Fig. 1 A and B, lane 3) is observed in PSII from which Mn, MSP, and other extrinsic subunits have been removed (PSII-3).
Quantitation of the Amount of 14C Incorporation from [14C]Methylamine.
The amount of radioactive label incorporated into subunits of PSII-3 (Fig. 1B, lane 3) is qualitatively similar to the amount retained on BSAO (Fig. 1B, lane AO). The amount of 14C incorporated into PSII-2 is similar, if incubations are extended (see time course in Fig. 1C). A quantitative estimate of 14C incorporation into PSII-3 from [14C]methylamine was obtained by comparison to a standard curve of 14C-methylated BSA (specific activity: 12.5 μCi/mg); these standard curves were run on the same gel. In PSII-3, the percent 14C incorporation into CP47 was 3.6% (mole/mole) and into D2/D1 was 6.6%, with a 30-min incubation time. The percent 14C incorporation into BSAO, run on the same gel, was 5%. Thus, the amount of 14C bound to PSII-3 subunits is indistinguishable from the amount trapped in BSAO, suggesting that a similar covalent adduct is formed. The lack of reaction of methylamine with PSII-1 subunits may be due to decreased access to the active site (15, 16).
PAGE Control Experiments.
PSII contains noncovalently bound PQ (plastoquinone), which may react with primary amines. PSII-1 does not show 14C labeling (Fig. 1B, lane 1), suggesting that comigration of PQ with subunits is not the basis for 14C incorporation. The data in Fig. 1D support this conclusion, because mixing a stoichiometric excess of purified PQ (Hoffman–LaRoche) with PSII before addition of [14C]methylamine did not result in a change in the pattern of 14C labeling (Fig. 1D, compare lanes 1 and 2). When purified PQ is mixed with [14C]methylamine, all the detectable 14C label migrates at the dye front (Fig. 1D, lane 3).
MSP (24) and LHC (23) were released from PSII and purified, and the potential of these proteins to react with methylamine was assessed. Also, PSI (25), another chlorophyll-containing reaction center, was tested. Fig. 1 (A and B) demonstrates that treatment of isolated MSP (lane 4), LHC (lane 5), and PSI (lane 6) with [14C]methylamine does not result in significant incorporation of 14C into these proteins. The low, background-level labeling, observed with purified LHC (lane 5), may be due to amine interaction with residual pigments retained by LHC. A similar result is observed when non-fatty-acid-free BSA is employed in the molecular weight marker set. As expected, fatty-acid-free BSA gave no reaction (data not shown). Because PSII-1, MSP, and LHC do not label to significant levels, we can rule out nonspecific modifications of amino acid side chains, which may occur during gel electrophoresis, as the basis for the incorporation of radiolabel in PSII-2 and PSII-3. Note that unmodified amino acid side chains and the peptide carbonyl group are not expected to react with primary amines (43).
Experiments were performed in strict darkness to investigate the effect of illumination on formation on the 14C adduct. The results of experiments, which employed PSII-3 and [14C]methylamine, showed that the CP47 and D2/D1 labeling was observed in the dark, but at decreased levels.
EPR Control Experiments Show that Methylamine Does Not Reduce or Bind to Tyrosyl Radicals D and Z.
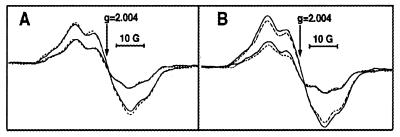
An intact Mn cluster is not necessary to observe labeling of protein subunits from [14C]methylamine. Manganese-depleted preparations exhibit EPR signals from the tyrosyl radicals, D⋅ and Z⋅. In contrast to radical Mox, D⋅ and Z⋅ are known to form unmodified radicals (44). To rule out the possibility that the reactions observed are due to the reaction of methylamine at D⋅ and Z⋅, we performed EPR control experiments on PSII-2 and PSII-3. The addition of methylamine has no effect on the line shapes or yields of D⋅ and Z⋅ in either preparation (Fig. 2). A similar result was obtained at 1 mg Chl/ml and with no potassium ferricyanide. These results are inconsistent with the one-electron oxidation of methylamine at either D⋅ and Z⋅, because electron donors reduce the radical yield under continuous illumination (for example, see ref. 23). In PSII-3 at 1 mg Chl/ml, addition of 1 mM benzylamine-hydrochloride also had no significant effect on either the EPR yields or lineshapes (data not shown).
Figure 2.
Room-temperature EPR spectra of tyrosyl radicals D⋅ and Z⋅ in Cl−-depleted PSII-2 (A) and PSII-3 (B). Spectra were recorded in the absence (dotted lines) or presence (solid lines) of 4 mM methylamine-hydrochloride. In both A and B, spectra with larger amplitudes were recorded under illumination and correspond to D⋅ and Z⋅; spectra with smaller amplitudes were recorded in the dark after illumination and correspond to D⋅.
PAGE, [14C]Formaldehyde, and PSII.
Labeling of PSII subunits from radioactive methylamine may be due to binding of methylamine and stabilization of a covalent adduct through the addition of reductant. Alternatively, if PSII acts as an amine oxidase, as we will show below, labeling of subunits with 14C may be a result of the production of a radioactive aldehyde, which can modify lysine residues (45). Once released from PSII, MSP shows no reaction with [14C]methylamine (Fig. 1 A and B, lane 4), and yet labeling of this subunit is observed when PSII-2 is treated with [14C]methylamine (Fig. 1 A and B, compare lanes 2 and 3). This result is consistent with labeling of MSP with a radioactive product, such as formaldehyde, that is produced from an amine oxidase activity intrinsic to PSII. If MSP is incubated in the presence of [14C]methylamine and BSAO, MSP labeling is observed (data not shown). This experiment demonstrates that it is possible to label MSP with 14C-labeled formaldehyde, which is produced as a result of an enzymatic amine oxidation.
To further explore the mechanism of labeling of subunits in PSII-3 and PSII-2, these preparations were incubated with [14C]formaldehyde (specific activity: 30 mCi/mmol) (Figs. 1 E and F). In PSII-3, incubation with this radiolabeled compound results in 14C incorporation into bands in the D2/D1 region (Fig. 1E, lanes 4 and 6). There is little detectable labeling of CP47, however (Fig. 1E, lanes 4 and 6), even with this high concentration of formaldehyde and in the absence of MSP. This result is significant, because removal of MSP is known to allow access of bulky reagents to CP47 (reviewed in ref. 46). Controls employing [14C]formaldehyde and [14C]methylamine in the absence of protein (Fig. 1E, lanes 1–3) show that the 14C-labeled compound in the stacking gel results from the reaction of [14C]formaldehyde with other buffer components. In PSII-2, Fig. 1F shows that MSP and bands in the D2/D1 region are labeled from [14C]formaldehyde (lanes 1–3). The amount of 14C incorporated into CP47, when treated with 0.5 mM, 1 mM, or 2 mM [14C]formaldehyde (Fig. 1F, lanes 1–3), is less than the amount of 14C incorporated into CP47 from [14C]methylamine, at a free base concentration of 2.8 μM (lane 4), even after correction for specific activity.
These experiments are consistent with the conclusion that radioactive methylamine binds to a site in CP47 and is oxidized to produce a radioactive product, such as [14C]formaldehyde, which modifies MSP. However, experiments employing exogenous [14C]formaldehyde cannot rule out the possibility that amine oxidation occurs at another site, which is in close proximity to CP47 and can label this subunit in situ. At this time, our data on D2/D1 labeling are consistent with the explanation that D2/D1 can be modified with [14C]methylamine or [14C]formaldehyde.
For PSII-2 and PSII-3, the addition of NaCNBH3 instead of DTT resulted in incorporation of an increased amount of 14C label nonspecifically into PSII. Similar results were obtained in the absence of any reductant. We conclude that NaCNBH3 is ineffective as a reductant in PSII, either for thermodynamic or kinetic reasons. We attribute the increased labeling observed in the absence of reductant to increased production of formaldehyde (see section below) and to increased access to reactive sites when the protein is denatured in the presence of ongoing amine oxidation.
Anion Effects on the Formation of the 14C-Labeled Covalent Adduct.
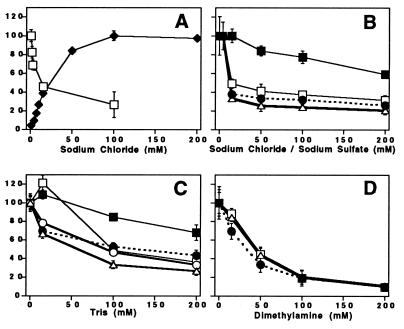
Cl− is known to be a required cofactor for photosynthetic O2 evolution (47). The influence of Cl− on O2 evolution has been attributed to binding of Cl− at or near the Mn cluster, and Cl− plays an important role in facilitating the turnover of the enzyme (reviewed in ref. 7). Fig. 3A (solid diamonds) confirms that, as expected (13, 14), O2 evolution in PSII-2 depends on the addition of NaCl. Also as expected (13, 14), Na2SO4 was not effective as an activator of O2 evolution. At pH 7.5 (Fig. 3A, solid diamonds) there is a requirement of 19 mM Cl− to achieve one-half the maximum rate of O2 evolution.
Figure 3.
Effect of various compounds on O2 evolution and 14C labeling of subunits in PSII at pH 7.5. The incubation time with the 14C-labeled amine was 30 min. In some cases, the error bars are smaller than the symbols used to represent the data. In A, solid diamonds, the O2 evolution activity of PSII-2 at pH 7.5 was measured as a function of added NaCl. Sample buffer contained: 0.4 M sucrose/50 mM Hepes-NaOH, pH 7.5/10 mM Ca(OH)2/1 mM potassium ferricyanide/0.25 mM recrystallized 2,6-dichlorobenzoquinone. The maximum (100%) corresponds to 350 μmol O2 (mg Chl-hr)−1. In A, open squares, the amount of 14C bound to CP47 in PSII-2 was measured as a function of added NaCl. Samples were incubated with 0.19 mM [14C]benzylamine-hydrochloride (free base concentration, 2.8 μM). In B, the amount of bound 14C was measured as a function of added NaCl. PSII-2 was incubated with 4 mM [14C]methylamine-hydrochloride (free base concentration, 2.8 μM). Because [14C]methylamine was only available as the hydrochloride, the first data point was obtained at 4 mM Cl−. Data shows labeling of CP47 (open squares), MSP (open triangles), and the D2/D1 polypeptides (solid circles). In B, the amount of bound 14C in CP47 was also measured as a function of added sodium sulfate after incubation of PSII-2 with 4 mM [14C]methylamine-hydrochloride (solid squares). MSP and the D2/D1 polypeptides behaved similarly on the addition of sodium sulfate (data not shown). In C and D, the amount of bound 14C from 4 mM [14C]methylamine-hydrochloride was measured as a function of added (unlabeled) Tris and dimethylamine, respectively. Data shows labeling of CP47 (open squares), MSP (open triangles), and the D2/D1 polypeptides (solid circles) in PSII-2. Data in C also shows the Tris competition with labeling of CP47 (solid squares) and the D2/D1 (open circles) polypeptides in PSII-3.
Treatment with 7-[14C]benzylamine hydrochloride (specific activity: 59 mCi/mmol) labels subunits of PSII-2 in a Cl−-dependent manner (Fig. 3A, open squares). Given the differences in the employed techniques and given the signal-to-noise ratio, the Cl− dependence parallels the activation of O2 evolution (Fig. 3A, solid diamonds).
Fig. 3B shows that the binding and oxidation of [14C]methylamine to PSII-2 subunits is also Cl−-dependent. The amount of 14C labeling from [14C]benzylamine and [14C]methylamine is similar. Na2SO4 (Fig. 3B, solid squares) competes less effectively with 14C incorporation from [14C]methylamine, suggesting that Cl− is the competitive agent. For example, note the second data point, where Cl− suppresses 14C binding from methylamine, but there is no significant suppression observed with SO4−2 (Fig. 3B). At anion concentrations between 100 and 200 mM, a significant SO4−2-induced effect on the formation of the 14C adduct is observed. However, these are SO4−2 concentrations that are higher than the Cl− concentrations required for stimulation of oxygen evolution and that exert a slight (14–23%) inhibitory effect on enzymatic activity. We attribute these changes to a nonspecific ionic strength effect. Because the anion dependence of amine binding parallels the requirements for O2 evolution, these results support the conclusion that binding and oxidation of amines occurs at the catalytic site of PSII-2. We ascribe the small amount of residual labeling at high Cl− concentrations (for example, Fig. 3B) to amine binding at the second, Cl−-insensitive amine binding site of PSII (8–10).
The effect of addition of Cl− on 14C adduct formation in PSII-3 was assessed, as well. The results obtained with Cl− are indistinguishable from the effects observed when SO4−2 is added to PSII-2 (Fig. 3B; data not shown). This is the expected result, because previous work has demonstrated that removal of the Mn cluster eliminates the Cl− binding site in PSII (48).
Competition Experiments with Other Amines.
Fig. 3C demonstrates that Tris, another primary amine, is competitive with binding and oxidation of [14C]methylamine in PSII-2. Fig. 3D shows that a secondary amine, dimethylamine, also competes with 14C labeling from methylamine. On a free base basis, dimethylamine (pKa 10.8) is a more effective competitor with [14C]methylamine, as compared with Tris (pKa 8.1).
Manganese Removal Does Not Change the Results of Tris Competition with D2/D1 Labeling.
Tris competition experiments show that the concentration dependence of D2/D1 labeling is similar in PSII-3 and PSII-2 (Fig. 3C, open and solid circles). The D and Z tyrosine radicals are located in the D2 and D1 polypeptides, and removal of manganese changes the lifetime of Z⋅ (reviewed in ref. 2). Therefore, the competition data in Fig. 3C reinforce the conclusion that methylamine is not interacting with redox-active tyrosines D and Z or with the manganese cluster, but with one or more separate, amino acid-derived site(s). However, CP47 (Fig. 3C, open and solid squares) shows a different concentration dependence in the Tris competition experiment, when PSII-2 and PSII-3 are compared. This result suggests that removal of manganese or MSP changes the relative binding affinities of CP47 for Tris and methylamine. Other evidence for interactions between CP47 and MSP has been obtained and described previously (reviewed in ref. 46).
Isolation of the Aldehyde Product and Determination of the Specific Activity of Amine Oxidation in PSII.
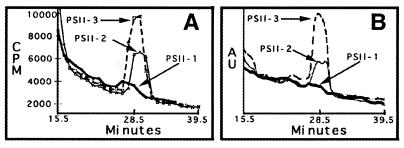
To obtain direct evidence for the oxidation of amines in PSII, the aldehyde product of benzylamine oxidation was isolated and quantitated. Fig. 4A (solid line) demonstrates that incubation of PSII-2 with 7-[14C]benzylamine hydrochloride results in the extraction of a 14C-labeled peak, which does not migrate with [14C]benzylamine, but has the same retention time as benzaldehyde. The 14C-labeled peak also gave rise to an increase in UV absorbance at 254 nm (Fig. 4B, solid line). Pooling and reinjection of this labeled material gave a 14C-labeled peak, which again coeluted with the same retention time as a benzaldehyde standard (data not shown). The addition of 50 mM NaCl suppressed the production of benzaldehyde in PSII-2.
Figure 4.
Chromatogram showing the purification of [14C]benzaldehyde from PSII. In A and B, the distribution of 14C and the absorbance at 254 nm are plotted versus elution time, respectively. The elution time for the benzaldehyde standard was 28.5 ± 1.0 min; 14C (A) and UV (B) peaks coeluted with the benzaldehyde standard run on that same day. A and B show PSII-1 (boldface line), PSII-2 (solid line), and PSII-3 (dashed line).
A [14C]benzaldehyde product also was isolated from PSII-3; the compound was produced in higher yield (Fig. 4, dashed line). As expected (48), the addition of 50 mM Cl− had no detectable effect on the production of [14C]benzaldehyde in PSII-3.
The amount of [14C]benzaldehyde produced in PSII-2 and PSII-3 is four to seven times the background amount of 14C obtained when PSII-1 is incubated with labeled benzylamine (Fig. 4, boldface line). Controls conducted with [14C]benzylamine in the absence of PSII also gave a small 14C peak with the same retention time; we attribute this background level to a small amount of autooxidation of benzylamine during the manipulation of the sample (38). The background level of autooxidation was not changed by the addition of 20 μM Mn+2 to [14C]benzylamine.
Quantitation of the amount of [14C]benzaldehyde produced by PSII-2 and PSII-3 was performed through scintillation counting; correction for the efficiency of counting was performed with a standard curve of [14C]benzylamine. In PSII-2, in a 4.5-hr experiment, the specific activity for benzylamine oxidation is 0.40 ± 0.07 μmol (μmol reaction center-hr)−1 or 0.0016 μmol benzaldehyde (mg Chl-hr)−1. By contrast, the maximum rate of oxygen evolution in this preparation is 350 μmol O2 (mg Chl-hr)−1 when electron acceptors are employed. In PSII-3, the specific activity for benzylamine oxidation is 0.64 ± 0.04 μmol (μmol reaction center-hr)−1; this preparation is not active in oxygen evolution. In PSII-2 and PSII-3, the yields of benzaldehyde are 1.8 ± 0.3 mol per mol reaction center and 2.9 ± 0.2 mol per mol reaction center, respectively. Correcting the yields for the small amount of benzaldehyde produced by autooxidation in PSII-1 gives yields of 1.4 mol/mol and 2.5 mol/mol for PSII-2 and PSII-3. These values for the yield and specific activity are lower limits, because we have not accounted for nonquantitative extraction of benzaldehyde.
Phenylhydrazine Binding.
The results described above are consistent with amine binding and oxidation at a cofactor containing an activated carbonyl group. However, [14C]Phenylhydrazine hydrochloride (2.65 mCi/mmol, California Bionuclear Corporation, Los Angeles), which is known to bind to the topaquinone in BSAO (32), does not result in a significant amount of 14C incorporation into PSII at concentrations up to 57 μM (2.8 μM free base). These low concentrations of phenylhydrazine were sufficient to label amine oxidase (data not shown).
At 4 mM [14C]phenylhydrazine, labeling of CP47 and D2/D1 is observed. However, significant 14C labeling of other PSII subunits occurs, as well as labeling of protein components of the molecular weight marker set. Phenylhydrazine can interact with PSII via one-electron reactions, as well as react via nucleophilic reactions. Thus, we explain these results by proposing that one-electron oxidation reactions in PSII compete with binding of phenylhydrazine to any putative carbonyl-containing group. Such competition would necessitate the use of high concentrations of phenylhydrazine and would give rise to nonspecificity in the pattern of 14C labeling.
DISCUSSION
We have reported evidence that covalent adducts, derived from primary amines, can be trapped on PSII protein subunits. By purification and quantitation of the 14C-labeled product, we have demonstrated that amine oxidation occurs to give the corresponding aldehyde. Anion competition experiments support the interpretation that the binding and oxidation reactions occur at the catalytic site of water oxidation. However, control experiments show that these reactions do not depend on redox activity involving high-valence manganese or involving the D· and Z· tyrosyl radicals.
We have determined the specific activity of amine oxidation in PSII and have found that this process has a rate that is many orders of magnitude slower than the optimal rate of water oxidation. Interestingly, the rate of amine oxidation, the yield of aldehyde product, and the amount of 14C covalent labeling, as assessed by PAGE, all increase in the absence of the manganese cluster and MSP. These are conditions where the water oxidation activity of the enzyme has been inactivated. The observation of a covalent 14C adduct derived from amines, in both plant and cyanobacterial PSII, may indicate that the interaction with amines has been evolutionarily conserved. Amine oxidation may be a vestigial activity, which the enzyme has been optimized to avoid, and may be a necessary consequence in the design of an efficient, enzymatic water oxidase.
Multiple turnovers of the amine oxidase in PSII must occur to explain our measured aldehyde stoichiometries. We speculate that the reoxidation mechanism is mediated by light-induced electron transfer reactions, which operate at very positive potentials in PSII, and would be thermodynamically capable of reoxidizing intermediates. Because the rate of amine oxidation in PSII is slow, the reoxidation of the electron acceptors by O2 may be rate limiting.
To explain our results, we propose that the active site of PSII contains one or more amino acid residues, which have been posttranslationally modified to contain an activated carbonyl group. This species may be the same cofactor that gives rise to the Mox EPR signal (3–6) and may be located in CP47 and/or the D2/D1 subunits. Whatever its structure, this cofactor stabilizes a covalent intermediate under reducing conditions in a manner quantitatively similar to the carbonyl containing topa of BSAO (40). In addition, this putative PSII cofactor can catalyze the same reaction as a topa-type quinocofactor (38, 39), i.e., the formation of an aldehyde from an amine. Such a species may play one of two roles in O2 evolution; these two roles are not mutually exclusive. The first possibility is that such a species might provide ligation to manganese atoms in the catalytic site. The second possibility is that such a species might function directly in the chemistry of water oxidation (49).
Our results suggest that an activated carbonyl group is present at the active site for photosynthetic water oxidation and that there may be evolutionary similarities between the enzymatic mechanism of amine oxidation and the mechanism of water oxidation.
Acknowledgments
This work was supported by National Institutes of Health Grant GM43272. We thank Profs. Charles Yocum, Terry Bricker, Judith Klinman, and JoAnne Stubbe and her research group for helpful discussions. We are also grateful to Drs. Klinman and Sophie Wang for the gift of BSAO and to Prof. Anath Das for the use of equipment.
ABBREVIATIONS
- BSAO
bovine serum amine oxidase
- Chl
chlorophyll
- CP47
chlorophyll-binding protein of approximate molecular mass of 47,000
- D2/D1
intrinsic subunits that bind most of the prosthetic groups in photosystem II
- LHC
light-harvesting complex
- MSP
manganese stabilizing protein
- PQ
plastoquinone
- PSI and PSII
photosystems I and II
- PSII-1
PSII membranes
- PSII-2
salt-washed PSII membranes that are depleted of the 24- and 18-kDa proteins and other extrinsic subunits
- PSII-3
PSII membranes that are depleted of the manganese cluster, the manganese stabilizing protein, and other extrinsic subunits
References
- 1.Yocum C F. Biochim Biophys Acta. 1991;1059:1–15. [Google Scholar]
- 2.Barry B A, Boerner R J, de Paula J C. In: The Molecular Biology of the Cyanobacteria. Bryant D, editor. I. Dordrecht, The Netherlands: Kluwer; 1994. pp. 215–257. [Google Scholar]
- 3.Noren G H, Barry B A. Biochemistry. 1992;31:3335–3342. doi: 10.1021/bi00128a005. [DOI] [PubMed] [Google Scholar]
- 4.Boerner R J, Bixby K A, Nguyen A P, Noren G H, Debus R J, Barry B A. J Biol Chem. 1993;268:1817–1823. [PubMed] [Google Scholar]
- 5.Boerner R J, Barry B A. J Biol Chem. 1994;269:134–137. [PubMed] [Google Scholar]
- 6.Ma C, Barry B A. Biophys J. 1996;71:1961–1972. doi: 10.1016/S0006-3495(96)79394-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yocum C F. In: Manganese Redox Enzymes. Pecoraro V L, editor. New York: VCH; 1992. pp. 71–83. [Google Scholar]
- 8.Sandusky P O, Yocum C F. FEBS Lett. 1983;162:339–343. [Google Scholar]
- 9.Sandusky P O, Yocum C F. Biochim Biophys Acta. 1984;766:603–611. [Google Scholar]
- 10.Sandusky P O, Yocum C F. Biochim Biophys Acta. 1986;849:85–93. [Google Scholar]
- 11.Adelroth P, Lindberg K, Andreasson L-E. Biochemistry. 1995;34:9021–9027. doi: 10.1021/bi00028a010. [DOI] [PubMed] [Google Scholar]
- 12.Lindberg K, Wydrzynski T, Vanngard T, Andreasson L-E. FEBS Lett. 1990;264:153–155. [Google Scholar]
- 13.Ghanotakis D F, Topper J N, Babcock G T, Yocum C F. FEBS Lett. 1984;170:169–173. [Google Scholar]
- 14.Miyao M, Murata N. FEBS Lett. 1984;168:118–120. [Google Scholar]
- 15.Ghanotakis D F, Topper J N, Yocum C F. Biochim Biophys Acta. 1984;767:524–531. [Google Scholar]
- 16.Tamura N, Cheniae G. Biochim Biophys Acta. 1985;809:245–259. [Google Scholar]
- 17.Achee F M, Chervenka C H, Smith R A, Yasunobu K T. Biochemistry. 1968;7:4329–4335. doi: 10.1021/bi00852a027. [DOI] [PubMed] [Google Scholar]
- 18.Janes S J, Klinman J P. Biochemistry. 1991;30:4599–4605. doi: 10.1021/bi00232a034. [DOI] [PubMed] [Google Scholar]
- 19.Janes S M, Klinman J P. Methods Enzymol. 1995;258:20–34. doi: 10.1016/0076-6879(95)58034-4. [DOI] [PubMed] [Google Scholar]
- 20.Berthold D A, Babcock G T, Yocum C F. FEBS Lett. 1981;134:231–234. [Google Scholar]
- 21.Miyao M, Murata N. Biochim Biophys Acta. 1983;725:87–93. [Google Scholar]
- 22.Yamomoto Y, Doi M, Tamura N, Nishimura N. FEBS Lett. 1981;133:265–268. [Google Scholar]
- 23.MacDonald G M, Barry B A. Biochemistry. 1992;31:9848–9856. doi: 10.1021/bi00155a043. [DOI] [PubMed] [Google Scholar]
- 24.Kuwabara T, Murata T, Miyao M, Murata N. Biochim Biophys Acta. 1986;850:146–155. [Google Scholar]
- 25.Noren G H, Boerner R J, Barry B A. Biochemistry. 1991;30:3943–3950. doi: 10.1021/bi00230a020. [DOI] [PubMed] [Google Scholar]
- 26.Barry B A. Methods Enzymol. 1995;258:303–319. doi: 10.1016/0076-6879(95)58053-0. [DOI] [PubMed] [Google Scholar]
- 27.Xu Q, Bricker T M. J Biol Chem. 1993;267:25816–25821. [PubMed] [Google Scholar]
- 28.Patzlaff J S, Barry B A. Biochemistry. 1996;35:7802–7811. doi: 10.1021/bi960056z. [DOI] [PubMed] [Google Scholar]
- 29.Piccioni R, Bellemare G, Chua N. In: Methods in Chloroplast Molecular Biology. Edelman H, Hallick R B, Chua N-H, editors. Amsterdam: Elsevier; 1982. pp. 985–1014. [Google Scholar]
- 30.Bollag D M, Edelstein S J. Protein Methods. New York: Wiley–Liss; 1991. pp. 114–116. [Google Scholar]
- 31.Chamberlain J P. Anal Biochem. 1979;98:132–135. doi: 10.1016/0003-2697(79)90716-4. [DOI] [PubMed] [Google Scholar]
- 32.Janes S M, Mu D, Wemmer D, Smith A J, Kaus S, Maltby D, Burlingame A L, Klinman J P. Science. 1990;248:981–987. doi: 10.1126/science.2111581. [DOI] [PubMed] [Google Scholar]
- 33.Klinman J P, Mu D. Annu Rev Biochem. 1994;63:299–344. doi: 10.1146/annurev.bi.63.070194.001503. [DOI] [PubMed] [Google Scholar]
- 34.Wang S W, Mure M, Medzihradszky K F, Burlingame A L, Brown D E, Dooley D M, Smith A J, Kagan H M, Klinman J P. Science. 1996;273:1078–1084. doi: 10.1126/science.273.5278.1078. [DOI] [PubMed] [Google Scholar]
- 35.Parsons M R, Convery M A, Wilmot C M, Yadav K D, Blakeley V, Corner A S, Phillips S E, McPherson M J, Knowles P F. Structure. 1995;3:1171–1184. doi: 10.1016/s0969-2126(01)00253-2. [DOI] [PubMed] [Google Scholar]
- 36.Anthony C. Biochem J. 1996;320:697–711. doi: 10.1042/bj3200697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kumar V, Dooley D M, Freeman H C, Guss J M, Harvey I, McGuirl M A, Wilce M C J, Zubak V M. Structure. 1996;4:943–955. doi: 10.1016/s0969-2126(96)00101-3. [DOI] [PubMed] [Google Scholar]
- 38.Sayre L M, Lee Y. Methods Enzymol. 1985;258:53–69. doi: 10.1016/0076-6879(95)58037-9. [DOI] [PubMed] [Google Scholar]
- 39.Mure M, Klinman J P. Methods Enzymol. 1995;258:39–52. doi: 10.1016/0076-6879(95)58036-0. [DOI] [PubMed] [Google Scholar]
- 40.Hartmann C, Klinman J P. J Biol Chem. 1987;262:962–965. [PubMed] [Google Scholar]
- 41.Stacy G W, Day R I, Morath R J. J Am Chem Soc. 1955;77:3869–3872. [Google Scholar]
- 42.Dunahay T G, Staehelin L A, Seibert M, Ogilvie P D, Berg S P. Biochim Biophys Acta. 1984;764:179–193. [Google Scholar]
- 43.Streitweiser A J, Heathcock C H. Introduction to Organic Chemistry. New York: Macmillan; 1976. p. 378. [Google Scholar]
- 44.Barry B A, El-Deeb M K, Sandusky P O, Babcock G T. J Biol Chem. 1990;265:20139–20143. [PubMed] [Google Scholar]
- 45.Means G E. Methods Enzymol. 1977;XLVII:469–479. doi: 10.1016/0076-6879(77)47047-2. [DOI] [PubMed] [Google Scholar]
- 46.Bricker T M. Photosynth Res. 1990;24:1–13. doi: 10.1007/BF00032639. [DOI] [PubMed] [Google Scholar]
- 47.Izawa, S., Heath, R. L. & Hind, G. (1969) Biochim. Biophys. Acta 388–398. [DOI] [PubMed]
- 48.Lindberg K, Vanngard T, Andreasson L-E. Photosynth Res. 1993;38:401–408. doi: 10.1007/BF00046767. [DOI] [PubMed] [Google Scholar]
- 49.Roberts J L, Jr, Sugimoto H, Barrette W C, Jr, Sawyer D T. J Am Chem Soc. 1985;107:4556–4557. [Google Scholar]