Abstract
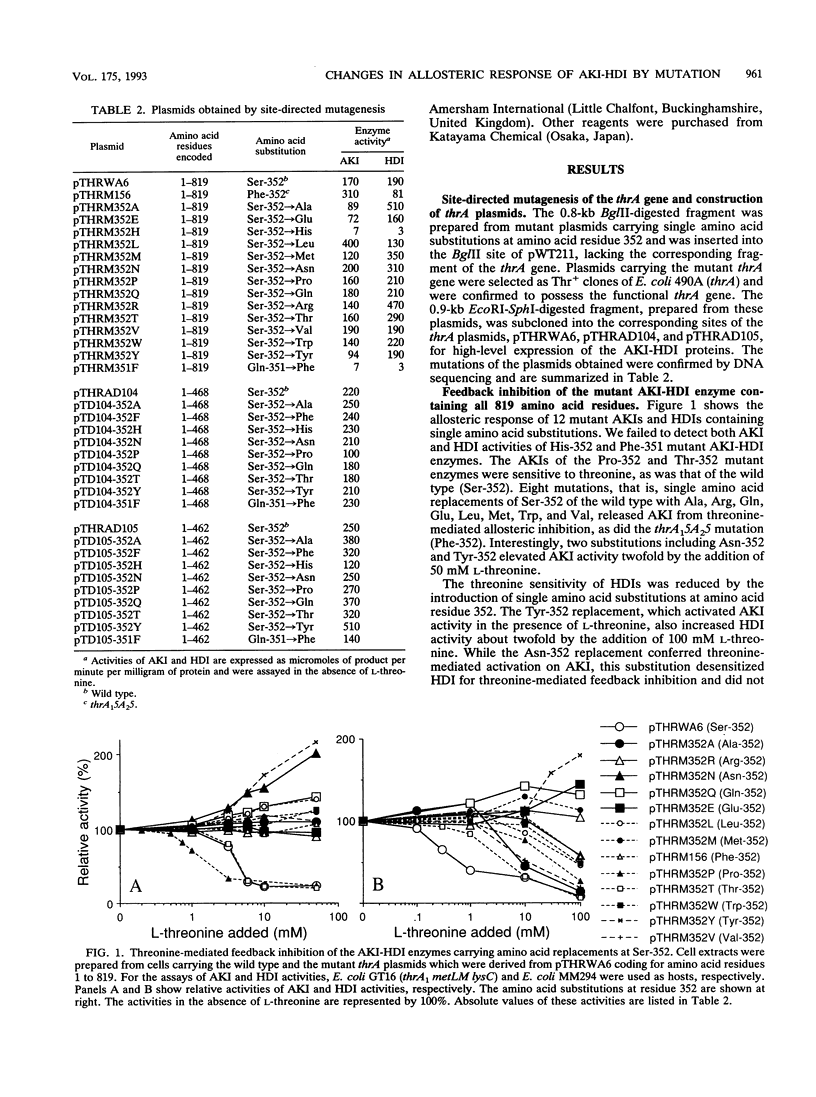
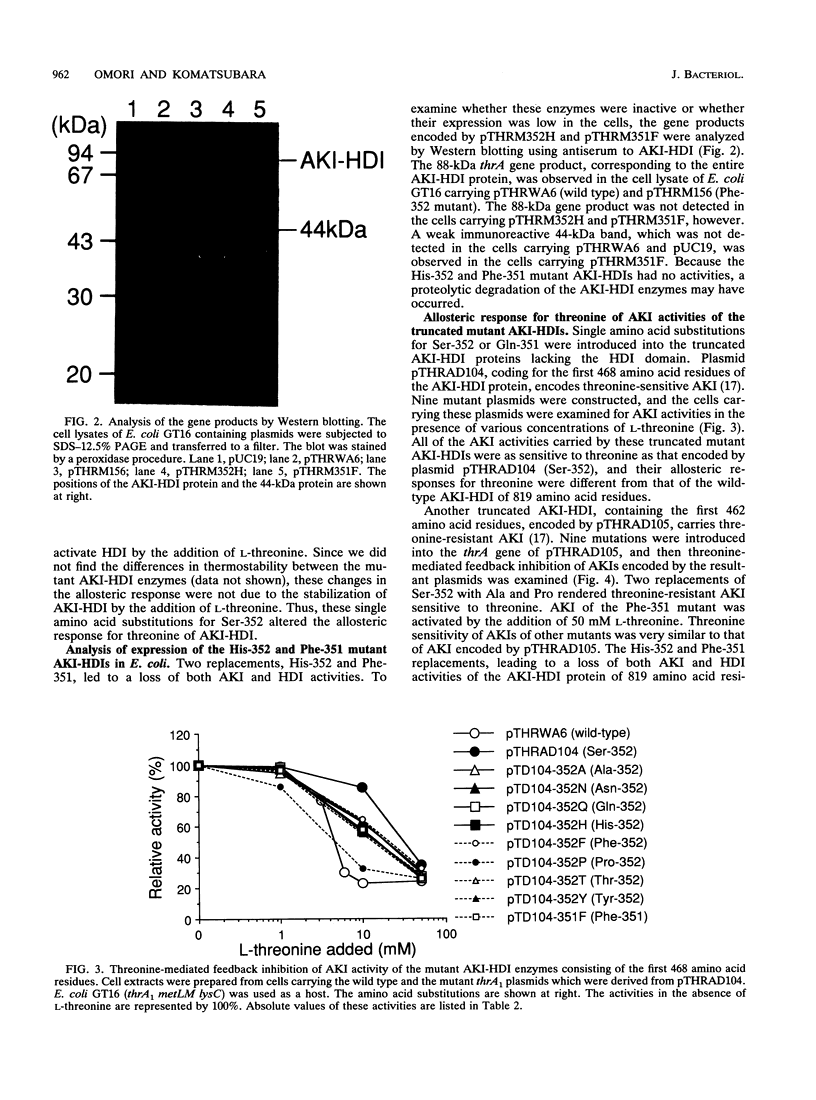
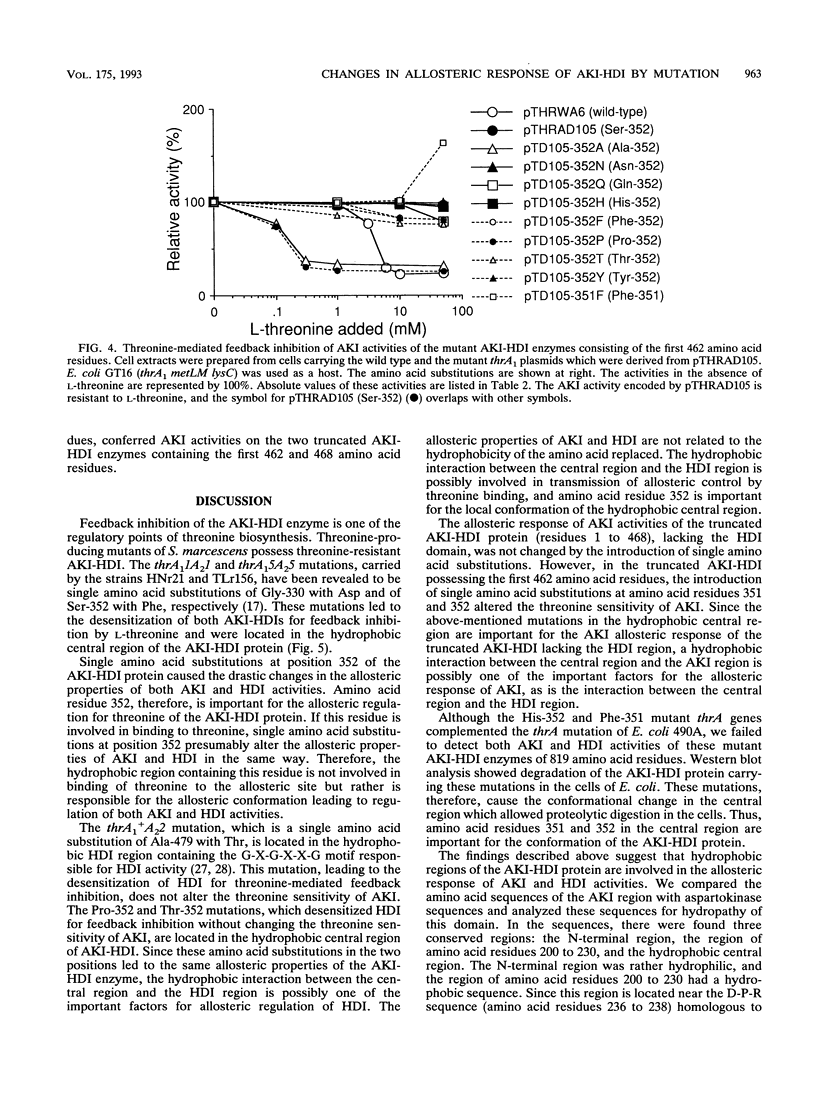
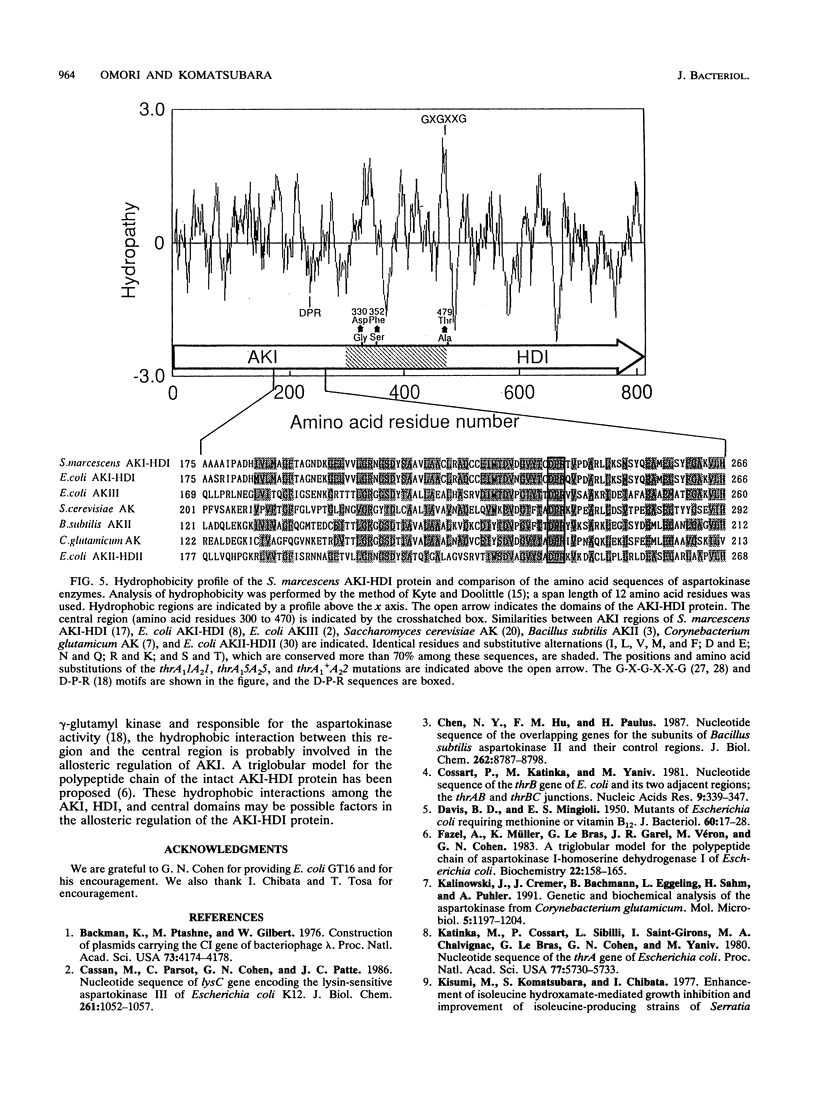
Aspartokinase I and homoserine dehydrogenase I (AKI-HDI) from Serratia marcescens Sr41 are encoded by the thrA gene as a single polypeptide chain. Previously, a single amino acid substitution of Ser-352 with Phe was shown to produce an AKI-HDI enzyme that is not subject to threonine-mediated feedback inhibition. To determine the role of Ser-352 in the allosteric response, the thrA gene was modified by using site-directed mutagenesis so that Ser-352 of the wild-type AKI-HDI was replaced by Ala, Arg, Asn, Gln, Glu, His, Leu, Met, Pro, Thr, Trp, Tyr, or Val. The Thr-352 and Pro-352 replacements rendered AKIs sensitive to threonine. The Tyr-352 and Asn-352 substitutions led to activation, rather than inhibition, of AKI by threonine. The other replacements conferred threonine insensitivity on AKI. The threonine sensitivity of HDI was also changed by the amino acid substitutions at Ser-352. The HDI carried by the Tyr-352 mutant AKI-HDI was activated by threonine. Single amino acid replacements at Ser-352 by Ala, Asn, Gln, His, Phe, Pro, Thr, or Tyr were introduced into truncated AKI-HDIs containing the AKI and the central regions. The AKI activity of the truncated AKI-HDI containing the first 468 amino acid residues was sensitive to threonine, and introduction of the amino acid replacements did not alter the threonine sensitivity of the AKI. Another truncated AKI-HDI containing the first 462 amino acid residues possessed threonine-resistant AKI, whereas the substitutions of Ser-352 with Ala and Pro rendered AKI sensitive to threonine. The replacement of GIn-351 with Phe activated AK1 of the truncated AKI-HDI in the presence of L-threonine. These findings suggest that Ser-352 of the central region of AKI-HDI is possibly a key residue involved with the allosteric regulation of both AKI and HDI activities.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Backman K., Ptashne M., Gilbert W. Construction of plasmids carrying the cI gene of bacteriophage lambda. Proc Natl Acad Sci U S A. 1976 Nov;73(11):4174–4178. doi: 10.1073/pnas.73.11.4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassan M., Parsot C., Cohen G. N., Patte J. C. Nucleotide sequence of lysC gene encoding the lysine-sensitive aspartokinase III of Escherichia coli K12. Evolutionary pathway leading to three isofunctional enzymes. J Biol Chem. 1986 Jan 25;261(3):1052–1057. [PubMed] [Google Scholar]
- Chen N. Y., Hu F. M., Paulus H. Nucleotide sequence of the overlapping genes for the subunits of Bacillus subtilis aspartokinase II and their control regions. J Biol Chem. 1987 Jun 25;262(18):8787–8798. [PubMed] [Google Scholar]
- Cossart P., Katinka M., Yaniv M. Nucleotide sequence of the thrB gene of E. coli, and its two adjacent regions; the thrAB and thrBC junctions. Nucleic Acids Res. 1981 Jan 24;9(2):339–347. doi: 10.1093/nar/9.2.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DAVIS B. D., MINGIOLI E. S. Mutants of Escherichia coli requiring methionine or vitamin B12. J Bacteriol. 1950 Jul;60(1):17–28. doi: 10.1128/jb.60.1.17-28.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazel A., Müller K., Le Bras G., Garel J. R., Véron M., Cohen G. N. A triglobular model for the polypeptide chain of aspartokinase I-homoserine dehydrogenase I of Escherichia coli. Biochemistry. 1983 Jan 4;22(1):158–165. doi: 10.1021/bi00270a023. [DOI] [PubMed] [Google Scholar]
- Kalinowski J., Cremer J., Bachmann B., Eggeling L., Sahm H., Pühler A. Genetic and biochemical analysis of the aspartokinase from Corynebacterium glutamicum. Mol Microbiol. 1991 May;5(5):1197–1204. doi: 10.1111/j.1365-2958.1991.tb01893.x. [DOI] [PubMed] [Google Scholar]
- Katinka M., Cossart P., Sibilli L., Saint-Girons I., Chalvignac M. A., Le Bras G., Cohen G. N., Yaniv M. Nucleotide sequence of the thrA gene of Escherichia coli. Proc Natl Acad Sci U S A. 1980 Oct;77(10):5730–5733. doi: 10.1073/pnas.77.10.5730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisumi M., Komatsubara S., Chibata I. Enhancement of isoleucine hydroxamate-mediated growth inhibition and improvement of isoleucine-producing strains of Serratia marcescens. Appl Environ Microbiol. 1977 Dec;34(6):647–653. doi: 10.1128/aem.34.6.647-653.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsubara S., Kisumi M., Chibata I. Transductional construction of a threonine-producing strain of Serratia marcescens. Appl Environ Microbiol. 1979 Dec;38(6):1045–1051. doi: 10.1128/aem.38.6.1045-1051.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsubara S., Kisumi M., Murata K., Chibata I. Threonine production by regulatory mutants of Serratia marcescens. Appl Environ Microbiol. 1978 May;35(5):834–840. doi: 10.1128/aem.35.5.834-840.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer B., Kramer W., Fritz H. J. Different base/base mismatches are corrected with different efficiencies by the methyl-directed DNA mismatch-repair system of E. coli. Cell. 1984 Oct;38(3):879–887. doi: 10.1016/0092-8674(84)90283-6. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Omori K., Imai Y., Suzuki S., Komatsubara S. Nucleotide sequence of the Serratia marcescens threonine operon and analysis of the threonine operon mutations which alter feedback inhibition of both aspartokinase I and homoserine dehydrogenase I. J Bacteriol. 1993 Feb;175(3):785–794. doi: 10.1128/jb.175.3.785-794.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omori K., Suzuki S., Imai Y., Komatsubara S. Analysis of the Serratia marcescens proBA operon and feedback control of proline biosynthesis. J Gen Microbiol. 1991 Mar;137(3):509–517. doi: 10.1099/00221287-137-3-509. [DOI] [PubMed] [Google Scholar]
- Parsot C., Cossart P., Saint-Girons I., Cohen G. N. Nucleotide sequence of thrC and of the transcription termination region of the threonine operon in Escherichia coli K12. Nucleic Acids Res. 1983 Nov 11;11(21):7331–7345. doi: 10.1093/nar/11.21.7331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafalski J. A., Falco S. C. Structure of the yeast HOM3 gene which encodes aspartokinase. J Biol Chem. 1988 Feb 15;263(5):2146–2151. [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz M., Winoto A., Minard K., Hood L. Clusters of genes encoding mouse transplantation antigens. Cell. 1982 Mar;28(3):489–498. doi: 10.1016/0092-8674(82)90203-3. [DOI] [PubMed] [Google Scholar]
- Sugita T., Komatsubara S., Kisumi M. Cloning and characterization of the mutated threonine operon (thrA(1)5A(2)5BC) of Serratia marcescens. Gene. 1987;57(2-3):151–158. doi: 10.1016/0378-1119(87)90118-1. [DOI] [PubMed] [Google Scholar]
- Thèze J., Margarita D., Cohen G. N., Borne F., Patte J. C. Mapping of the structural genes of the three aspartokinases and of the two homoserine dehydrogenases of Escherichia coli K-12. J Bacteriol. 1974 Jan;117(1):133–143. doi: 10.1128/jb.117.1.133-143.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J., Messing J. Production of single-stranded plasmid DNA. Methods Enzymol. 1987;153:3–11. doi: 10.1016/0076-6879(87)53044-0. [DOI] [PubMed] [Google Scholar]
- Wierenga R. K., Terpstra P., Hol W. G. Prediction of the occurrence of the ADP-binding beta alpha beta-fold in proteins, using an amino acid sequence fingerprint. J Mol Biol. 1986 Jan 5;187(1):101–107. doi: 10.1016/0022-2836(86)90409-2. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Zakin M. M., Duchange N., Ferrara P., Cohen G. N. Nucleotide sequence of the metL gene of Escherichia coli. Its product, the bifunctional aspartokinase ii-homoserine dehydrogenase II, and the bifunctional product of the thrA gene, aspartokinase I-homoserine dehydrogenase I, derive from a common ancestor. J Biol Chem. 1983 Mar 10;258(5):3028–3031. [PubMed] [Google Scholar]



