Abstract
Brome mosaic virus (BMV), a positive-strand RNA virus in the alphavirus-like superfamily, encodes two RNA replication proteins: 1a, which contains a helicase-like domain and a domain implicated in RNA capping, and 2a, which contains a polymerase-like domain. To further explore their functions, we expressed 1a and 2a individually and together in yeast also expressing replicatable transcripts of BMV genomic RNA3. Complementing prior results that 1a and 2a are required jointly for positive-strand RNA synthesis, both also were required for negative-strand RNA synthesis. Nevertheless, in the absence of 2a, 1a expression increased the accumulation of DNA-derived RNA3 transcripts 8-fold. Increased accumulation was specific for RNA3: none of a diverse set of yeast mRNAs tested showed increased accumulation in the presence of 1a. Increased RNA3 accumulation was not due to increased DNA transcription, but to a 20- to 40-fold increase in the in vivo half-life of RNA3 from 5–10 min in the absence of 1a to more than 3 hr in the presence of 1a. Although (1a+2a)-dependent RNA replication selectively amplified the natural viral 5′ end from among multiple transcription starts of DNA-derived RNA3 transcripts, 1a-induced stabilization affected all RNA3 transcripts, without specificity for the precise viral 5′ end. Increased RNA3 accumulation did not increase expression of a directly translatable, 5′-proximal gene in RNA3, implying that 1a-induced stabilization blocked rather than facilitated RNA3 translation. These and other results suggest that the striking, 1a-induced stabilization of RNA3 may reflect an interaction involved in recruiting viral RNA templates into RNA replication while diverting them from the competing processes of translation and degradation.
After virion uncoating in a cell, the genomic RNAs of positive-strand RNA viruses serve as mRNAs. One or more early viral translation products then induce formation of a membrane-bound complex of viral and, in at least some cases, host proteins that direct RNA-dependent replication of the viral genomic RNA. For many viruses, the same or a similar complex also directs synthesis of one or more subgenomic mRNAs from the negative-strand genomic RNA replication intermediate. Identifying the roles and interactions of viral replication factors in the assembly and function of these replication/mRNA synthesis complexes is thus crucial to understanding viral RNA replication and gene expression.
Brome mosaic virus (BMV), a member of the alphavirus-like superfamily of animal and plant positive-strand RNA viruses, has a genome divided among 5′-capped RNA1, RNA2, and RNA3 (1). RNA3 encodes the 3a protein, which is required for cell-to-cell movement of BMV infection in its natural plant hosts (2), and the coat protein, which is expressed from a subgenomic mRNA, RNA4. RNA1 and RNA2 encode proteins 1a and 2a, respectively, which are required for genomic RNA replication and subgenomic mRNA synthesis. Proteins 1a and 2a share three regions of sequence conservation with proteins encoded by all other members of the alphavirus-like superfamily: 2a (94 kDa) contains a central polymerase-like domain, whereas 1a (109 kDa) contains a C-terminal helicase-like region and an N-terminal domain implicated in viral RNA capping (1). 1a and 2a interact in vitro (3, 4) and colocalize in vivo in an endoplasmic reticulum-associated replication complex that is the site of BMV-specific RNA synthesis (5). Compatible interaction between 1a and 2a is required in vivo for positive-strand RNA synthesis, but whether 1a and 2a both are required for negative-strand RNA synthesis has not been established previously (6).
Upon shifting to nonpermissive temperature, a temperature-sensitive mutation in the 1a helicase-like domain blocks further synthesis of positive- and negative-strand genomic RNA and subgenomic mRNA, suggesting that 1a is required in the synthesis of all forms of viral RNA (7). In addition, exchanging genes and gene segments between BMV and a related bromovirus shows that 1a controls or contributes to template specificity in RNA replication (8). However, the specific roles of 1a and the steps at which it acts have not been determined.
Expressing 1a and 2a in Saccharomyces cerevisiae allows this yeast to support RNA replication and subgenomic mRNA synthesis by RNA3 derivatives (9). To initiate such replication, suitable RNA3 derivatives can be produced as in vitro transcripts and transfected into yeast (9) or transcribed in vivo from appropriately engineered yeast DNA plasmids or chromosomal insertions (10). In addition to 1a and 2a, BMV RNA replication in yeast depends on multiple yeast genes (11).
To further explore the functions of 1a and 2a and their effects on viral RNA, we expressed 1a and 2a separately in yeast also expressing replicatable BMV RNA3 transcripts. We found that, like positive-strand RNA synthesis, negative-strand RNA synthesis required both 1a and 2a. We also report that, in the absence of 2a, 1a markedly and selectively increased the in vivo stability of RNA3, transforming it from a short-lived mRNA to one of the longest-lived mRNAs known in yeast. However, despite its unusually long half-life, 1a-stabilized RNA3 was translated poorly if at all in vivo. Unlike full viral RNA replication, 1a-induced stabilization of RNA3 was not specific for the natural 5′ end of RNA3. These results are consistent with a possible early role for 1a-RNA3 interaction in selecting viral RNA templates for replication and show that positive-strand RNA viruses can strongly and differentially regulate mRNA stability and translation.
MATERIALS AND METHODS
Plasmids.
1a and 2a were expressed from pB1CT19 and pB2CT15, yeast 2μ plasmids with the HIS3 and LEU2 genes, respectively (9). 1a frameshift derivative pB1CT28 was made by cutting pB1CT19 with MluI, treating with Klenow DNA polymerase, and religating. 1a translation then frameshifts after codon 199 and terminates after 15 additional codons. RNA3 was expressed from pB3 (laboratory designation pB3RQ39; ref. 10), a yeast centromeric TRP1 plasmid. To make pB3–5′CAT, the BamHI site 5′ to the 3a gene in pB3TP10 (12) was introduced into pB3 by PCR, creating pB3MJ23. The 0.9-kb TaqI fragment from pBR325 was cloned between the BamHI and ClaI sites of pB3MJ23, thus replacing the 5′ half of the 3a gene with the chloramphenicol acetyltransferase (CAT) gene.
Yeast.
Strain YPH500 (Matα ura3–52 lys2–801 ade2–101 trp1-Δ63 his3-Δ200 leu2-Δ1) was used throughout. Cultures were grown at 30°C in defined synthetic medium containing 2% glucose (glc) or 2% galactose (gal) (13). His, leu, and trp were omitted as needed to select for pB1CT19, pB2CT15, and pB3.
RNA Analysis.
Yeast cells were passaged three times in gal medium, keeping culture density below OD600 = 0.5–0.75 (20–24 hr per passage). Total RNA was extracted and analyzed by Northern blotting as described (9).
Nuclear Run-On Assays.
Yeast cells were permeabilized with n-lauroyl-sarcosine and transcripts were labeled with [α-32P]UTP (14). RNA was extracted (9) and used to probe nylon membranes on which were immobilized full-length, negative-strand RNA3 in vitro transcripts from pB3RQ1, which contains the EcoRI-PstI fragment of pB3TP8 (15) cloned in pSP65 (Promega).
RESULTS
1a Increases Accumulation of DNA-Derived RNA3 Transcripts.
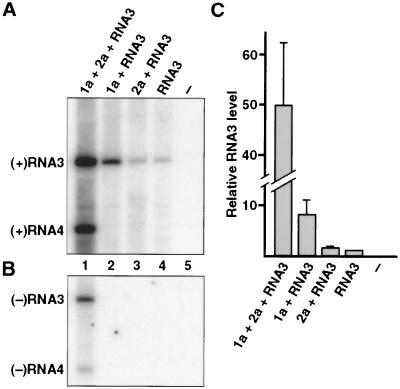
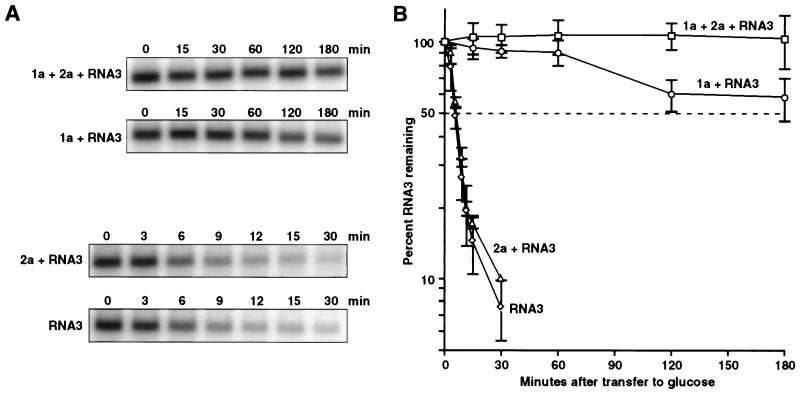
To explore the individual effects of BMV RNA replication proteins 1a and 2a on BMV RNA, we expressed 1a, 2a, and 1a+2a in yeast also expressing wt BMV RNA3 transcripts. 1a and 2a were constitutively expressed from separate yeast DNA plasmids via the ADH1 promoter (9). RNA3 cDNA was inserted between the gal-inducible, glc-repressible yeast GAL1 promoter and a hepatitis δ virus ribozyme in yeast DNA plasmid pB3 to allow transcription of full-length, positive-strand RNA3 transcripts that self-cleave to produce the authentic 3′ end of natural viral RNA3 (Fig. 1). As shown previously (10), production of these RNA3 transcripts in yeast coexpressing 1a and 2a initiated RNA3 replication and subgenomic mRNA synthesis via a negative-strand RNA3 intermediate. Such RNA replication led to an average 49-fold increase in positive-strand RNA3 levels over the accumulation of DNA-derived RNA3 transcripts in cells not expressing 1a or 2a (Fig. 2A, lanes 1 and 4 and C). Subgenomic RNA4 was absent unless 1a and 2a were expressed in combination, whereon it accumulated to levels similar to amplified genomic RNA3 (Fig. 2A, lane 1). Negative-strand RNA3 also appeared only in yeast expressing 1a and 2a (Fig. 2B). Thus, not only positive-strand synthesis (6) but also negative-strand RNA synthesis require both 1a and 2a. As found earlier (7, 10), cells expressing 1a, 2a, and RNA3 also accumulated some subgenomic RNA4-sized negative-strand RNA, but much more slowly than negative-strand RNA3 (Fig. 2B, lane 1).
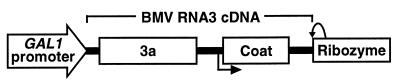
Figure 1.
Schematic of the BMV RNA3 cDNA region in yeast centromeric plasmid pB3. The 3a and coat protein genes are indicated. The GAL1 promoter fused to the 5′ end of RNA3 cDNA allows gal-induced in vivo transcription of RNA3, and the hepatitis δ ribozyme cleaves the transcript at the natural 3′ end of RNA3. The bent arrow below the diagram shows the start of sequences for the subgenomic coat protein mRNA, RNA4, which is transcribed from the negative-strand RNA3 replication intermediate.
Figure 2.
Northern blot analysis of BMV RNA3 accumulation in yeast expressing the indicated BMV components. To ensure full induction and equilibration of RNA3 transcription from pB3, yeast cells were passaged three times in gal-containing medium and total RNA was extracted. Equal amounts of total RNA per sample were denatured with glyoxal, run on a 0.8% agarose gel, transferred to nylon membranes, and hybridized to 32P-labeled in vitro transcript probes. (A) RNA3-derived positive-strand RNAs detected with a probe complementary to the 3′ 200 bases of BMV RNA3. (B) RNA3-derived negative-strand RNAs, as detected with a probe corresponding to 500 bases of coat protein coding sequence. (C) Positive-strand RNA3 levels in the indicated yeast strains were measured with a Molecular Dynamics PhosphorImager and normalized to the level in yeast expressing RNA3 alone. Averages and standard deviations from five independent experiments are shown.
Unexpectedly, despite the absence of any negative-strand RNA3 that could direct RNA replication, yeast expressing 1a showed markedly higher accumulation of RNA3 transcripts than yeast expressing RNA3 alone (Fig. 2A, lanes 2 and 4). Comparison of five independent experiments showed that RNA3 accumulation in 1a-expressing cells averaged eight times higher than in cells lacking 1a (Fig. 2C). Expressing 2a alone had no effect on the accumulation of RNA3 transcripts (Fig. 2A, lane 3). To confirm that increased RNA3 accumulation was due to 1a rather than the HIS3-selectable marker gene or any other feature of the 1a expression plasmid, we inserted a frameshift mutation after codon 199 of the 961-codon 1a gene. This 1a mutant plasmid produced levels of 1a mRNA equivalent to the starting plasmid, but did not increase RNA3 transcript accumulation in the absence of 2a or support RNA3 replication or subgenomic RNA4 synthesis in the presence of 2a (results not shown).
1a Does Not Increase Accumulation of Other Yeast mRNAs.
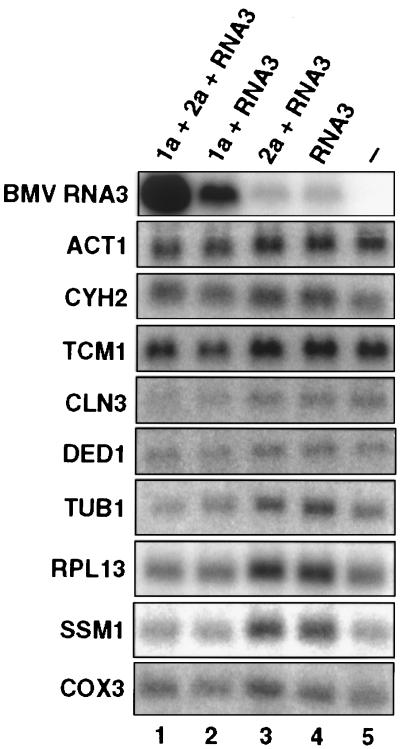
To see whether the effect of 1a on RNA3 transcript accumulation was general, we examined the accumulation of a variety of yeast mRNAs. Wt yeast and yeast expressing relevant combinations of 1a, 2a, and RNA3 were grown in parallel and total RNA extracts were made from each culture. Loading equal amounts of total RNA for each sample, duplicate sets of these samples were electrophoresed and transferred to nylon membranes. Individual membranes were hybridized to probes for the yeast genes ACT1, TUB1, CLN3, DED1, CYH2, TCM1, RPL13, SSM1, and COX3 (Fig. 3). ACT1 encodes actin, whose mRNA is often used as an internal standard for comparing total yeast RNA levels among samples. TUB1 encodes α-tubulin. CLN3 encodes a G1 cyclin regulating the G1-to-S cell cycle transition (16). DED1 encodes a DEAD-box, putative helicase protein implicated in translation initiation (17). CYH2, TCM1, RPL13, and SSM1 encode ribosomal proteins. COX3 is a mitochondrially encoded gene for a cytochrome c oxidase subunit. The top membrane in Fig. 3 was probed for BMV RNA3 and confirms that these RNA samples display the typical, approximately 8-fold stimulation of RNA3 transcript accumulation in the presence of 1a (lane 2 vs. lanes 3–4). The lower panels show that no corresponding 1a-induced increase occurred for any of the nine diverse yeast gene transcripts tested. Rather, on 1a expression, the levels of many of these mRNAs declined approximately 2-fold for unknown reasons (lanes 1–2 vs. lanes 3–5).
Figure 3.
Protein 1a does not increase accumulation of varied yeast mRNAs. Yeast cells expressing the indicated BMV components were grown and total RNA was extracted as in Fig. 2. Equal amounts of total RNA per sample were loaded in parallel on multiple formaldehyde gels (13) and transferred to nylon membranes, and individual membranes were probed for positive-strand BMV RNA3 (Top) or with random-primed, 32P-labeled DNA fragments from yeast genes ACT1, CYH2, TCM1, CLN3, DED1, TUB1, RPL13A, SSM1b, and COX3, as indicated.
1a-Stimulated RNA3 Accumulation Is Not Due to Increased DNA Transcription.
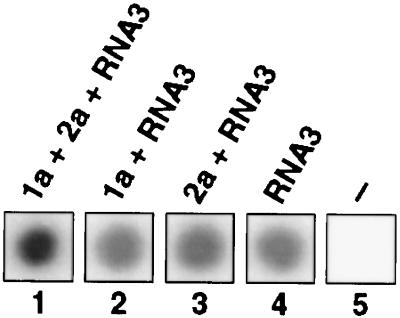
Increased accumulation of RNA3 transcripts in yeast expressing 1a could result from increased transcription of RNA3 from pB3, decreased RNA3 degradation, or both. To compare levels of DNA-dependent RNA3 transcription in yeast with and without 1a, we performed run-on transcription assays (14). Yeast expressing various BMV components were permeabilized with sarkosyl and incubated with ATP, CTP, GTP, and [α-32P]UTP to allow elongation of previously initiated nuclear RNA transcripts. The resulting 32P-labeled RNA samples were hybridized to membranes on which an excess of unlabeled in vitro transcripts of full-length, negative-strand RNA3 had been immobilized.
As shown in Fig. 4, the levels of 32P-labeled positive-strand RNA3 transcripts hybridizing to the immobilized negative-strand RNA3 target were equivalent for yeast expressing RNA3 alone, 1a+RNA3, or 2a+RNA3 (Fig. 4, spots 2–4). Matching results were obtained in independent experiments. The responsiveness of the assay to altered levels of input 32P-labeled RNA3 transcript was confirmed in reconstruction experiments in which parallel membranes were incubated with varying, known amounts of 32P-labeled RNA3 in vitro transcripts, in the presence or absence of run-on transcription products from YPH500 yeast without any BMV components. Thus, transcription of RNA3 from pB3 was not affected by expressing 1a or 2a independently. Two- to 2.5-fold greater synthesis of positive-strand RNA3 was observed in run-on assays of yeast expressing 1a+2a+RNA3 (Fig. 4, spot 1), presumably because of RNA-dependent replication of RNA3. This signal increase may not accurately reflect the level of BMV RNA polymerase in vivo, because the membrane-bound, cytoplasmic BMV RNA replication complex is sensitive to some detergents (18) and may have been inhibited by the sarkosyl permeabilization treatment.
Figure 4.
Nuclear run-on analysis of DNA-dependent RNA3 transcription. Yeast expressing the indicated BMV components were grown in gal-containing medium and permeabilized with sarkosyl, and nuclear RNA transcripts were labeled by incubating the permeabilized cells with [α-32P]UTP for 10 min (14). Total RNA then was extracted and used to probe activated nylon membranes on which equal amounts of full-length, negative-strand, in vitro transcripts of RNA3 had been spotted and immobilized.
1a-stimulated RNA3 Accumulation Is Due to Increased RNA3 Stability.
To compare the stability of RNA3 transcripts in yeast in the presence and absence of 1a and 2a, we analyzed RNA3 decay after selectively inhibiting RNA3 transcription. Yeast expressing RNA3 alone or in combination with 1a, 2a, or both were passaged three times in gal medium to mid-exponential phase to ensure full induction and equilibration of GAL1 promoter-driven RNA3 transcription from pB3. The yeast were then washed with water and transferred to medium containing glc, which represses transcription from the GAL1 promoter within a few minutes (19, 20). After transfer to glc, aliquots of the cells were removed at intervals and frozen at −70°C before RNA extraction and Northern blotting for RNA3.
Fig. 5A shows typical Northern blots from such RNA3 decay analysis. Fig. 5B plots RNA3 decay curves averaged over three independent experiments by using a semilogarithmic plot so that the slope of each decay curve is inversely proportional to the half-life of RNA3. The results show a dramatic effect of 1a on RNA3 stability. In the absence of 1a, RNA3 decayed with a half-life of 5–10 min. This rapid decay was unchanged whether RNA3 was expressed alone or in the presence of 2a. In the presence of 1a alone, RNA3 decay was slowed 20- to 40-fold, yielding a half-life greater than 3 hr. Thus, 30 min after glc addition, less than 10% of the starting RNA3 transcripts survived in yeast lacking 1a, whereas >90% remained in yeast expressing 1a.
Figure 5.
Protein 1a extends RNA3 half-life. (A) Yeast expressing the indicated BMV components were grown in defined, gal-containing medium to induce RNA3 transcription from the pB3 GAL1 promoter. At time zero, these strains were transferred to defined, glc-containing medium to repress RNA3 transcription. For each strain, equal aliquots of cells were removed at the indicated times after transfer to glc medium and frozen at −70°C to stop further decay. Total RNA was then extracted, and equal amounts of total RNA from each sample were analyzed by Northern blotting, as in Fig. 2A, to follow the decay of positive-strand RNA3. Note the different time intervals used for sampling yeast containing or lacking 1a. For comparison, exposures were adjusted to provide similarly intense starting signals for all strains. (B) For three independent experiments as in A, positive-strand RNA3 levels were measured with a Molecular Dynamics PhosphorImager, expressed for each strain as a percentage of the RNA3 level at time zero, and plotted on a semilogarithmic plot vs. time after transfer to glc medium. Averages and standard deviations are shown. RNA3 half-life corresponds to the time at which each curve intersects 50% (dotted line).
In cells expressing 1a+2a+RNA3, RNA3 levels did not decline detectably after transfer to glc, but maintained a steady-state level. This is consistent with Fig. 2, which shows that 98% of the RNA3 in cells expressing 1a+2a+RNA3 is produced by glc-insensitive, RNA-dependent RNA replication rather than glc-repressible, GAL1-promoted DNA transcription.
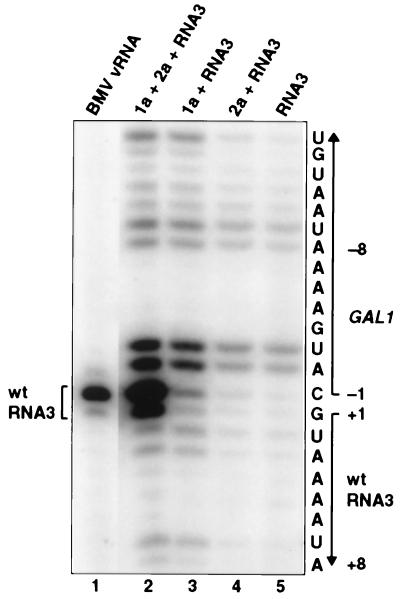
1a Acts on RNA3 Transcripts with Multiple 5′ Start Sites.
The GAL1-RNA3 cassette of pB3 (Fig. 1) produces multiple transcription starts, but 1a- and 2a-dependent RNA replication specifically amplifies the natural 5′ end of RNA3 (10). To see whether 1a-induced stabilization of RNA3 might also show high specificity for the natural RNA3 5′ end, we used primer extension to compare the spectrum of 5′ ends in RNA3 populations from gal-induced, pB3-containing yeast expressing 1a, 2a, both, or neither (Fig. 6). Yeast without 1a or 2a contained pB3-derived RNA3 transcripts with the previously observed, multiple 5′ ends, including doublets at positions −2/−3 and −8/−9 relative to the 5′ end of natural RNA3 and weaker bands at −1 to +3 and −10 to −14 (Fig. 6, lane 5). When 1a and 2a were coexpressed, the pB3-derived RNA3 transcripts were augmented by an intense doublet at +1/−1 (lane 2). This doublet corresponds to the 5′ primer extension pattern of natural BMV RNA3 from virions (lane 1) and results from RNA-dependent replication of RNA3 (10). The band at position +1 represents the 5′ end of the RNA proper whereas the upper, more intense band at position −1 is from cap-dependent incorporation of an additional nucleotide, as shown for RNA3 and other capped RNAs (21).
Figure 6.
Primer extension analysis of the 5′ ends of RNA3 species in yeast expressing the indicated BMV components. Yeast were grown and total RNA was extracted as in Fig. 2, and primer extension was performed with an oligonucleotide complementary to RNA3 bases 30–44 (10). Nucleotide position relative to the RNA3 cDNA sequence is shown at right. Lane 1 shows equivalent analysis of natural BMV virion RNA.
Expressing 2a with RNA3 did not change the primer extension pattern from that of pB3 alone (Fig. 6, lanes 4 and 5). Expressing 1a with RNA3 increased the overall intensity of the band pattern, but the relative intensity of bands in the spectrum of 5′ ends in the RNA3 population showed little or no change from that seen with pB3 alone or pB3+2a (lane 3). Thus, unlike 1a+2a-directed RNA3 replication, 1a-induced stabilization and increased accumulation of RNA3 displayed little or no selectivity for 5′ ends at or near the natural RNA3 5′ end relative to other ends in the pB3-derived RNA3 transcript population.
1a Stimulates Accumulation of RNA3 but Not an RNA3 Translation Product.
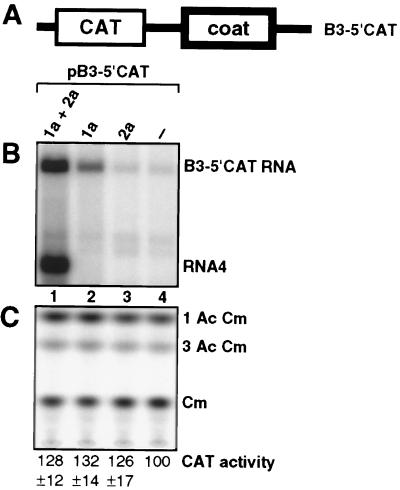
To determine whether the 1a-induced increase in RNA3 stability and accumulation resulted in a comparable increase in the accumulation of a protein translated from RNA3, we replaced the 3a gene in pB3 with the CAT gene (Fig. 7A). Yeast expressing the resulting B3–5′CAT RNA alone or with 1a, 2a, or both were assayed for RNA levels and CAT activity. Northern blotting showed that the effects of 1a and 2a on B3–5′CAT RNA paralleled their effects on wt RNA3 (compare Figs. 7B and 2A): 2a alone had no effect on B3–5′CAT transcript accumulation (lanes 3–4), 1a alone stimulated accumulation of these transcripts 7- to 10-fold (lane 2), and 1a+2a amplified B3–5′CAT RNA 30- to 40-fold higher than starting transcript levels and induced similar levels of subgenomic RNA4 (lane 1).
Figure 7.
1a-stimulated RNA3 accumulation does not increase expression of a gene translated from RNA3. (A) Schematic of B3–5′CAT, an RNA3 derivative in which the 5′ half of the 3a gene has been replaced by the CAT gene. (B) Representative Northern blot analysis of positive-strand B3–5′CAT RNA accumulation in yeast expressing the indicated BMV components, performed as in Fig. 2A. (C) CAT expression in the yeast of Fig. 7B. Total protein was extracted from a portion of each of the same cultures used for RNA analysis in Fig. 7B. Equal aliquots of each sample were analyzed for CAT activity by using 14C-chloramphenicol and thin-layer chromatography, and ensuring that all sample activities were within the linear range of the assay (9, 10). The positions of chloramphenicol (Cm) and its acetylated derivatives (1AcCm, 3AcCm) are shown. CAT activity for each strain was normalized to expression in B3–5′CAT-expressing yeast lacking 1a and 2a, averaged over four, independent experiments, and presented with standard deviation below each lane.
As intended, the 5′-proximal CAT gene was translated from B3–5′CAT transcripts in the absence of 1a, 2a, and BMV subgenomic mRNA synthesis (Fig. 7C, lane 4). However, in repeated experiments, the substantial 1a- and (1a+2a)-induced increases in B3–5′CAT RNA were not accompanied by comparable increases in CAT expression (Fig. 7C, lanes 1 and 2). CAT activity in yeast expressing 1a+B3–5′CAT RNA averaged only 28–32% higher than in yeast expressing B3–5′CAT alone. Moreover, this was not significantly different from CAT activity in 2a+B3–5′CAT yeast, which had the same level of B3–5′CAT RNA as yeast expressing B3–5′CAT alone. Similar CAT activity was found in 1a+2a+B3–5′CAT yeast despite even higher B3–5′CAT RNA accumulation. The failure of CAT expression to increase with B3–5′CAT RNA levels in 1a+2a-expressing yeast may be due solely to 1a-induced effects, as in 1a+B3–5′CAT yeast. Coat protein might also have contributed to suppressing B3–5′CAT translation because recent work shows that production of wt RNA4 in yeast, as in 1a+2a+B3–5′CAT yeast (Fig. 7B), directs BMV coat protein synthesis and encapsidation of RNA3 derivatives (M. Krol and P.A., unpublished results). To allow for a possible lag between accumulation of B3–5′CAT RNA and CAT activity, all four yeast strains of Fig. 7 were assayed for RNA and CAT activity levels at 0, 1, 2, and 3 days after gal induction of B3–5′CAT transcription, but no significant variation in the results was observed. Thus, 7- to 10-fold increased accumulation of B3–5′CAT RNA in the presence of 1a did not lead to significantly increased B3–5′CAT RNA translation.
DISCUSSION
This study tested BMV RNA replication factors 1a and 2a in yeast for individual effects on BMV RNA3 replication templates. Neither 1a nor 2a alone induced negative-strand RNA3 (Fig. 2B). In yeast coexpressing 1a and 2a, negative-strand RNA3 accumulation is higher when RNA-dependent amplification of positive-strand RNA3 occurs, but is easily detected even when RNA-dependent positive-strand RNA3 synthesis is blocked in cis by a 5′ truncation (10, 22). Thus, in the absence of 1a or 2a, negative-strand synthesis did not escape detection as an indirect consequence of inhibiting positive-strand RNA3 amplification (6). Rather, both 1a and 2a must be required during negative-strand synthesis, as in positive-strand genomic and subgenomic RNA synthesis. Because BMV positive-strand RNA synthesis requires some aspect(s) of 1a–2a interaction not required for negative-strand RNA synthesis (6) and negative-strand RNA synthesis terminates early in infection whereas positive-strand synthesis continues (7), it remains possible that positive- and negative-strand synthesis are carried out by functionally distinguishable complexes.
Though unable to direct RNA synthesis on its own, the 1a expression plasmid induced a remarkable increase in the stability and accumulation of DNA-derived RNA3 transcripts in the absence of 2a (Figs. 2 and 5). A 1a frameshift mutation abolished these effects, showing that they were functions of the 1a ORF and suggesting that functional 1a protein is required. The increase in RNA3 stability conferred by 1a is particularly striking when compared with natural yeast mRNA stabilities. The 5- to 10-min RNA3 half-life in the absence of 1a corresponds to that of a moderately unstable yeast mRNA, whereas the >3-hr RNA3 half-life in the presence of 1a represents possibly the longest half-life recorded for an mRNA in yeast (23–25).
1a did not increase the accumulation of any of a varied panel of yeast mRNAs (Fig. 3), suggesting that it does not induce a general block to a major RNA degradation pathway. This selectivity for BMV RNA, the magnitude of RNA3 stabilization, and the established role of 1a in RNA replication suggest that 1a stabilization of RNA3 may result from a function of 1a that normally contributes to BMV infection. Such a 1a function might reflect a direct or indirect interaction between 1a and RNA3 replication templates, between 1a and progeny RNA3 products, or both.
One potential action of 1a on positive-strand RNA3 replication products is RNA capping, because the N-terminal 1a domain shares sequence similarity with alphavirus nsP1 proteins, which are implicated in m7G-methyltransferase and guanylyltransferase activity (26). Decapping, followed rapidly by 5′ to 3′ exonuclease degradation, is an important control point in degrading many yeast mRNAs and at least some mammalian mRNAs (25, 27, 28). Thus, one hypothesis for RNA3 stabilization could be selective recapping of decapped RNA3 by 1a. To produce the observed stabilization, such hypothetical 1a activity in recapping full-length RNA3 would have to outcompete both cell decapping and exonuclease activities. However, BMV RNA capping may normally be linked to BMV RNA synthesis (1, 6), as in vaccinia virus and cellular mRNA transcription complexes (29). More significantly, recapping would not explain the B3–5′CAT results showing that 1a inhibited RNA3 translation as well as degradation. Recapping alone would increase the pool of free, capped RNA3 derivatives and thus translation of those RNAs. For RNA3 derivatives with CAT substituted for the coat protein gene, CAT expression in yeast varies with the level of subgenomic CAT mRNA over at least a 105-fold range (10). In contrast, the 1a-induced, 800% increase in B3–5′CAT RNA accumulation was accompanied by, at most, a 32% increase in CAT expression (Fig. 7), showing that the majority of 1a-stabilized B3–5′CAT RNA was not efficiently translated. Thus, whether capping is involved or not, 1a stabilization of RNA3 must involve other effects inhibitory to translation.
Several observations suggest that 1a-induced RNA3 stabilization might reflect processes involved in recruiting RNA3 templates into replication while removing them from the interfering pathways of translation and degradation. First, as noted in the Introduction, 1a controls at least some aspects of template specificity in RNA replication (8). Second, the poor translation of 1a-stabilized B3–5′CAT RNA is reminiscent of initiation of RNA replication by positive-strand RNA bacteriophage Qβ. Qβ replicase binding to positive-strand viral RNA both leads to initiating RNA synthesis and blocks further translation initiation (30, 31). Mechanisms to clear viral RNA of ribosomes generally may be required to initiate positive-strand RNA virus RNA replication, because viral RNA translation must precede replication but reads RNAs in the opposite direction. Third, for yeast and other eukaryotes, nonsense codon-mediated mRNA degradation and other results demonstrate links between mRNA translation and degradation (27). Thus, a 1a-mediated block to RNA3 translation, increased RNA3 stability, and recruitment of RNA3 replication templates could be linked effects of a single process.
Poliovirus RNA replication factor 3CD and a host factor bind poliovirus RNA at a 5′-proximal cloverleaf implicated in RNA replication, and recent results suggest that these interactions may inhibit viral translation in cis (32). Similarly, viral polymerase binding to a 5′ element in hepadnavirus pregenome RNA is required to initiate reverse transcription and may coordinately suppress translation (33, 34). BMV 1a or a 1a complex also might inhibit RNA3 translation by RNA binding. Such binding might be to the 5′ untranslated region or to distal regions of RNA3, because translational control elements have been found in 3′ untranslated regions of other mRNAs (35, 36). Similarly, stability control elements can be located throughout eukaryotic mRNAs (37).
1a-induced RNA3 stabilization did not require the BMV 2a, 3a, or coat proteins or the 3a coding sequence and, unlike full RNA3 replication, was not inhibited by nonviral sequence extensions at the 5′ end of RNA3. If 1a-induced RNA3 stabilization is related to using RNA3 as a replication template, its 2a-independent nature suggests that it may reflect an early step in template selection. Because of its unusual magnitude, 1a-induced RNA3 stabilization also may prove useful in illuminating general cellular mechanisms controlling mRNA stability and translation, important aspects of gene regulation.
Acknowledgments
We thank R. Quadt for pB3RQ1, D. Ursic, A. Atkins, R. Shirley, and W. Heideman for generously providing DNA used to make yeast in mRNA probes, and J. Ross and M. Sullivan for helpful comments on the manuscript. This research was supported by the National Institutes of Health under Grants GM35072 and GM51301. P.A. is an Investigator of the Howard Hughes Medical Institute.
ABBREVIATIONS
- BMV
brome mosaic virus
- CAT
chloramphenicol acetyltransferase
- gal
galactose
- glc
glucose
- wt
wild type
References
- 1.Ahlquist P. Curr Op Genet Dev. 1992;2:71–76. doi: 10.1016/s0959-437x(05)80325-9. [DOI] [PubMed] [Google Scholar]
- 2.Mise K, Ahlquist P. Virology. 1995;206:276–286. doi: 10.1016/s0042-6822(95)80043-3. [DOI] [PubMed] [Google Scholar]
- 3.Kao C C, Quadt R, Hershberger R P, Ahlquist P. J Virol. 1992;66:6322–6329. doi: 10.1128/jvi.66.11.6322-6329.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kao C C, Ahlquist P. J Virol. 1992;66:7293–7302. doi: 10.1128/jvi.66.12.7293-7302.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Restrepo-Hartwig M, Ahlquist P. J Virol. 1996;70:8908–8916. doi: 10.1128/jvi.70.12.8908-8916.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dinant S, Janda M, Kroner P A, Ahlquist P. J Virol. 1993;67:7181–7189. doi: 10.1128/jvi.67.12.7181-7189.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kroner P A, Young B M, Ahlquist P. J Virol. 1990;64:6110–6120. doi: 10.1128/jvi.64.12.6110-6120.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Traynor P, Ahlquist P. J Virol. 1990;64:69–77. doi: 10.1128/jvi.64.1.69-77.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Janda M, Ahlquist P. Cell. 1993;72:961–970. doi: 10.1016/0092-8674(93)90584-d. [DOI] [PubMed] [Google Scholar]
- 10.Ishikawa M, Janda M, Krol M A, Ahlquist P. J Virol. 1997;71:7781–7790. doi: 10.1128/jvi.71.10.7781-7790.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ishikawa M, Diez J, Restrepo-Hartwig M, Ahlquist P. Proc Natl Acad Sci USA. 1997;94:13810–13815. doi: 10.1073/pnas.94.25.13810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pacha R F, Ahlquist P. J Virol. 1991;65:3693–3703. doi: 10.1128/jvi.65.7.3693-3703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current Protocols in Molecular Biology. New York: Wiley; 1987. [Google Scholar]
- 14.Warner J R. Methods Enzymol. 1991;194:423–428. doi: 10.1016/0076-6879(91)94033-9. [DOI] [PubMed] [Google Scholar]
- 15.Janda M, French R, Ahlquist P. Virology. 1987;158:259–262. doi: 10.1016/0042-6822(87)90265-0. [DOI] [PubMed] [Google Scholar]
- 16.Polymenis M, Schmidt E V. Genes Dev. 1997;11:2522–2531. doi: 10.1101/gad.11.19.2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chuang R-Y, Weaver P L, Liu Z, Chang T-H. Science. 1997;275:1468–1471. doi: 10.1126/science.275.5305.1468. [DOI] [PubMed] [Google Scholar]
- 18.Bujarski J J, Hardy S F, Miller W A, Hall T C. Virology. 1982;119:465–473. doi: 10.1016/0042-6822(82)90105-2. [DOI] [PubMed] [Google Scholar]
- 19.Parker R, Herrick D, Peltz S W, Jacobson A. Methods Enzymol. 1991;194:415–423. doi: 10.1016/0076-6879(91)94032-8. [DOI] [PubMed] [Google Scholar]
- 20.Johnston M. Microbiol Rev. 1987;51:458–476. doi: 10.1128/mr.51.4.458-476.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Allison R F, Janda M, Ahlquist P. J Virol. 1988;62:3581–3588. doi: 10.1128/jvi.62.10.3581-3588.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Quadt R, Ishikawa M, Janda M, Ahlquist P. Proc Natl Acad Sci USA. 1995;92:4892–4896. doi: 10.1073/pnas.92.11.4892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Herrick D, Parker R, Jacobson A. Mol Cell Biol. 1990;10:2269–2284. doi: 10.1128/mcb.10.5.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peltz S W, Jacobson A. In: Control of Messenger RNA Stability. Belasco J, Brawerman G, editors. San Diego: Academic; 1993. pp. 291–328. [Google Scholar]
- 25.Caponigro G, Parker R. Microbiol Rev. 1996;60:233–249. doi: 10.1128/mr.60.1.233-249.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ahola T, Laakkonen P, Vihinen H, Kaariainen L. J Virol. 1997;71:392–397. doi: 10.1128/jvi.71.1.392-397.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jacobson A, Peltz S W. Annu Rev Biochem. 1996;65:693–739. doi: 10.1146/annurev.bi.65.070196.003401. [DOI] [PubMed] [Google Scholar]
- 28.Couttet P, Fromont-Racine M, Steel D, Pictet R, Grange T. Proc Natl Acad Sci USA. 1997;94:5628–5633. doi: 10.1073/pnas.94.11.5628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shuman S. Proc Natl Acad Sci USA. 1997;94:12758–12760. doi: 10.1073/pnas.94.24.12758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weber H, Billeter M A, Kahane S, Weissmann C, Hindley J, Porter A. Nat New Biol. 1972;237:166–169. doi: 10.1038/newbio237166a0. [DOI] [PubMed] [Google Scholar]
- 31.Barrera I, Schuppli D, Sogo J M, Weber H. J Mol Biol. 1993;232:512–521. doi: 10.1006/jmbi.1993.1407. [DOI] [PubMed] [Google Scholar]
- 32.Gamarnik A V, Andino R. RNA. 1997;3:882–892. [PMC free article] [PubMed] [Google Scholar]
- 33.Pollack J R, Ganem D. J Virol. 1994;68:5579–5587. doi: 10.1128/jvi.68.9.5579-5587.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang G H, Seeger C. J Virol. 1993;67:6507–6512. doi: 10.1128/jvi.67.11.6507-6512.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wickens M, Kimble J, Strickland S. In: Translational Control. Hershey J, Mathews M, Sonenberg N, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1996. pp. 411–450. [Google Scholar]
- 36.Izquierdo J M, Cuezva J M. Mol Cell Biol. 1997;17:5255–5268. doi: 10.1128/mcb.17.9.5255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ross J. Trends Genet. 1996;12:171–175. doi: 10.1016/0168-9525(96)10016-0. [DOI] [PubMed] [Google Scholar]