Abstract
Herpes simplex virus type 1 (HSV-1), the prototypical herpesvirus, has an icosahedral nucleocapsid surrounded by a proteinaceous tegument and a lipoprotein envelope. As in tailed bacteriophages, the icosahedral symmetry of the capsid is broken at one of the twelve vertices, which is occupied by a dodecameric ring of portal protein, UL6, instead of a pentamer of the capsid protein, UL19. The portal ring serves as a conduit for DNA entering and exiting the capsid. From a cryo-EM reconstruction of capsids immuno-gold-labeled with anti-UL6 antibodies, we confirmed that UL6 resides at a vertex. To visualize the portal in the context of the assembled capsid, we used cryo-electron tomography to determine the three-dimensional structures of individual A-capsids (empty, mature capsids). The similarity in size and overall shape of the portal and a UL19 pentamer - both are cylinders of ~ 800 kDa - combined with residual noise in the tomograms, prevented us from identifying the portal vertices directly; however, this was accomplished by a computational classification procedure. Averaging the portal-containing subtomograms produced a structure that tallies with the isolated portal, as previously reconstructed by cryo-EM. The portal is mounted on the outer surface of the capsid floor layer, with its narrow end pointing outwards. This disposition differs from that of known phage portals in that the bulk of its mass lies outside, not inside, the floor. This distinction may be indicative of functional divergence at the level of portal-related functions other than its role as a DNA channel.
Introduction
The HSV-1 capsid initially assembles as a precursor procapsid that subsequently matures, during DNA packaging (Newcomb et al., 1996; Trus et al., 1996). Maturation starts with proteolytic processing of the internal scaffolding protein, UL26.5, which is then expelled: it also involves a major conformational change in the surface shell that stabilizes the fragile procapsid (Heymann et al., 2003). In these highly distinctive behaviors, HSV-1 subscribes to the same paradigm as tailed bacteriophages (Black et al., 1994; King and Chiu, 1997; Hendrix and Duda, 1998; Cerritelli et al., 2002). In packaging, phages translocate their DNA into the capsid via a specialized channel, the portal, that occupies one of the capsid’s twelve icosahedral vertices (Valpuesta and Carrascosa, 1994). After packaging, the tail is attached at this site: the portal is alternatively called the “connector” for that reason. Recently, it has been established that HSV-1 also has a portal, the viral UL6 gene product ( Newcomb et al., 2001; Newcomb et al., 2003; Burch and Weller, 2004; Thurlow et al., 2006), although it has no tail - the viral envelope constituting the infection apparatus of this virus.
The subunits of known bacteriophage portals vary in size from ~ 36 kDa to ~ 83 kDa (Agirrezabala et al., 2005b). Nevertheless, generically, they form oligomeric rings with an axial channel, giving a turbine-like appearance as visualized by electron microscopy (e.g. Dube et al., 1993; Orlova et al., 2003; Agirrezabala et al., 2005a; Tang et al., 2005). In all well documented cases, portals competent to be incorporated into capsids have twelve subunits, although polymorphic variants with different numbers of subunits are encountered in some recombinant expression products (Kocsis et al., 1995; Lurz et al., 2001; Valpuesta et al., 2000; Cingolani et al., 2002). High resolution structures have been determined by X-ray crystallography for the portal/connector of phage φ29 – one of the smallest such molecules, with a 36 kDa subunit (Simpson et al., 2000; Guasch et al., 2002). It is plausible that the other, larger, portal subunits have additional domains grafted onto a common structural core, although there is seldom enough sequence similarity to validate this hypothesis. Some recent studies have succeeded in visualizing the portal in procapsids and/or capsids by electron microscopy and image reconstruction (Tao et al., 1998; Agirrezabala et al., 2005a; Fokine et al., 2005; Jiang et al., 2006). In view of the asymmetric position of the portal vertex, they employed reconstruction schemes based on 5-fold symmetry or no symmetry, instead of the 60-fold icosahedral symmetry commonly applied to capsids (Baker et al., 1999). For the phages studied, the bulk of the portal mass was observed to be located on the interior side of the capsid shell.
Previously, we used cryo-electron microscopy and image reconstruction to characterize the HSV-1 UL6 protein purified from a recombinant baculovirus system (Trus et al., 2004). This expression product turned out to be polymorphic with 11-fold to 14-fold rings: nevertheless, in cryo-electron micrographs, we were able to distinguish the 12-fold rings - which appear to be the physiologically relevant form (see above) - and calculate a three-dimensional structure at 16 Å resolution. Noting many similarities and some differences between capsid assembly as practiced by herpesviruses and the tailed bacteriophages (Steven et al., 2005; Bamford et al., 2005; Baines and Duffy, 2006), we were interested in how the HSV-1 portal is configured in the context of an assembled capsid. However, because the HSV-1 capsid has no tail attached nor a prolate morphology to mark the site of the portal vertex, we could not perform a non- symmetric reconstruction directly; instead, alternative strategies were required.
To address this issue, we performed cryo-electron microscopy and image analysis of capsids immuno-gold labeled with anti-UL6 antibodies, and cryo-electron tomography (Grünewald et al., 2003; Lucic et al., 2005) of unlabeled capsids. To minimize potential ambiguities in interpreting densities close to the inner surface of the capsid shell, the latter experiments were performed with A-capsids, which are essentially empty. (Of the three kinds of capsids that may be recovered from the nuclei of cells infected with wild-type HSV-1, C-capsids contain packaged DNA but not the scaffolding protein, UL26.5; B-capsids lack DNA but retain the scaffolding protein; and A-capsids contain neither DNA nor the scaffolding protein. All three capsids have the mature conformation of the surface shell). To confirm the findings of the tomographic analysis, we performed complementary experiments by freeze-fracture EM.
Results and Discussion
Cryo-EM reconstruction of B-capsids labeled with anti-UL6 antibodies and colloidal gold
Our initial strategy was to reconstruct capsids using lower than icosahedral symmetry, i.e. 5-fold symmetry or no symmetry. To do so, we first had to identify the portal vertex on each capsid. For this purpose, we used immuno-gold labeling with anti-UL6 antibodies. These experiments were performed with B-capsids because we could obtain them in greater abundance than A-capsids or C-capsids. As a consequence of the many grid washings required in the immuno-labeling protocol, we could only obtain a few capsids per micrograph. Nevertheless, a total of 634 capsids were collected from digitized micrographs and analyzed. The majority (73%) of labeled capsids had only one gold bead, and the remainder, two or three beads. In view of the ambiguity of multiple labels, the analysis was restricted to the 460 singly labeled capsids.
We determined the orientations of these capsids by projection-matching relative to a pre-existing density map (Methods). To prevent the strongly scattering gold beads from interfering with this analysis, they were painted out to the local background density. After the orientation of a given capsid was determined, the corresponding projection of the reference map – in which the positions of the twelve vertices are specified - was used to identify the labeled vertex. In some orientations, two or more vertices project close together. Such ambiguous cases had to be discarded. The remaining capsids (N=156) were reconstructed with 5-fold symmetry and with no symmetry. However, the resulting density maps turned out to be too noisy to yield conclusive information about the portal vertex (data not shown). Based on this experience and the numbers of particles (> 104) that were used to obtain adequate no-symmetry reconstructions of (smaller) capsids (Jiang et al., 2006; Lander et al., 2006), we concluded that a prohibitively large number of micrographs would be needed in this case. However, these data did provide positive confirmation that UL6 resides at a vertex. A reconstruction in which icosahedral symmetry was imposed is presented in Figure 1b. It establishes that the gold beads – and hence, the UL6 molecules - are vertex-associated.
Figure 1.
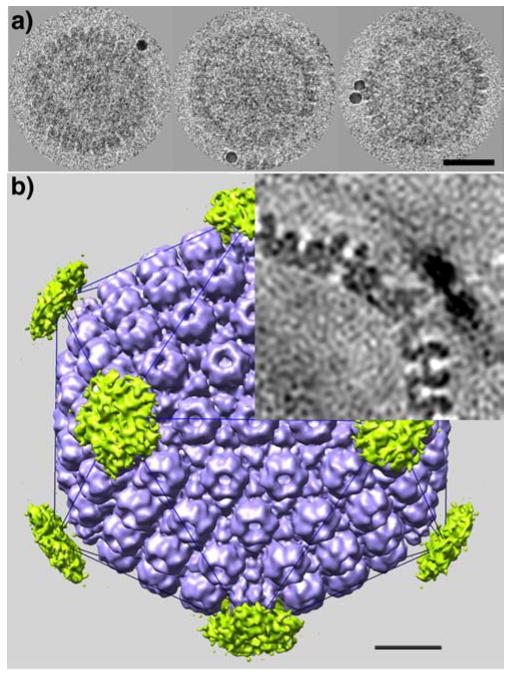
(a) Cryo-electron microscopy of HSV-1 B-capsids, labeled with anti-UL6 polyclonal antiserum and 10 nm gold particles. Bar = 50 nm. (b) Cryo-EM reconstruction based on 156 capsids (blue). A difference map calculated between the reconstruction in which the gold beads were included and one in which they were erased from the projection images (the same origins and orientation angles were used in both cases and were determined from the latter data), shows density of the gold beads concentrated over the vertices (green pentagons). This difference map confirms that UL6 is vertex-resident. The top right quadrant shows a central section that emphasizes the much greater density of the gold beads, even after 12-fold dilution and local smearing in the reconstruction. Bar = 20 nm.
Cryo-electron tomography of A-capsids
In a second attempt to visualize the portal complex in situ, we analyzed A-capsids by cryo-electron tomography. A-capsids were used, as we anticipated that their tomograms should be more readily interpretable in the absence of the partially ordered scaffold material present in B-capsids. Seven tilt series were recorded and 150 capsids were extracted from the resulting tomograms. Their resolution was ~ 5.5 nm in-plane, according to the NLOO2D criterion (Cardone et al., 2005). A central slice through one of these tomograms is shown in Figure 2.
Figure 2.
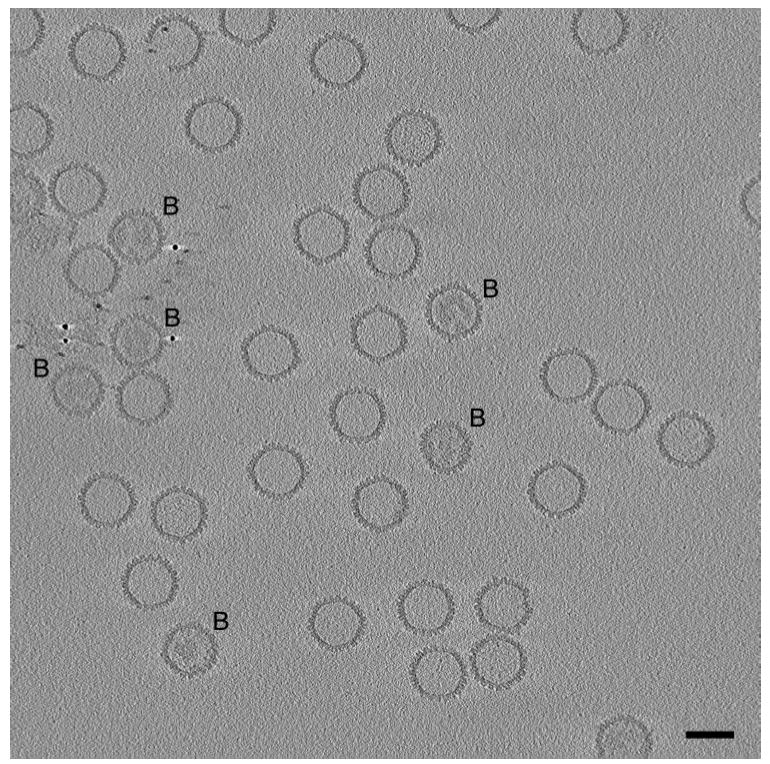
Central slice through a cryo-electron tomogram of a field of HSV-1 capsids. The slice is 3 nm thick. No denoising was applied to the tomogram. Bar=100 nm. Most of the capsids present are A-capsids, i.e. empty capsids. A few are B-capsids (marked B), with more-or-less ordered internal material, representing the scaffolding protein, UL26.5. Several of the gold particles used as fiducial markers are seen in the left/middle of the field. One of the A-capsids (top left) is missing part of its shell. The fact that this slice samples all of the capsids centrally or almost so indicates that they are aligned quite precisely in the z-dimension. Presumably, this alignment results from their being confined within an ice film whose thickness, ~ 150 nm, as measured from the tomogram, is not much greater than the capsid diameter of 125 nm. Most of the capsids are oriented so that they are viewed along or close to a threefold axis of icosahedral symmetry; accordingly, they appear hexagonal in outline, and their icosahedral facets are sampled in slices through the tomogram (not shown) parallel to this one and offset by ~ 50 nm.
Our strategy was to consistently align capsids from the tomograms and then average them, and our initial hope was that the noise level would be low enough to allow the portal vertices to be identified directly by visual inspection. A modeling experiment comparing the UL6 dodecamer with a UL19 pentamer from a B-capsid reconstruction – both, essentially noise-free – showed that the two structures should be readily distinguishable at 5.5 nm resolution (data not shown; see also Frangakis et al., 2002). However, residual noise, combined with the resolution anisotropy generated by the “missing wedge” effect (Frank, 1992), prevented us from discriminating the portal vertices with confidence by visual criteria.
Identification of the portal vertices by template-matching
Next, we conducted a computational classification based on correlation analysis. The capsids were rotated into a standard orientation and twelve vertex-containing volumes were excised from each. In a classification experiment, all twelve vertices of a given capsid were compared with a reference structure (see below), and the one that gave the best match was taken to be the portal vertex. Thus identified, the portal vertices of all 150 capsids, already aligned, were then averaged. Such an experiment can have only three outcomes: the UL19 pentamer; the portal structure (assuming it to be significantly different from the UL19 pentamer); or some linear combination thereof. Since 92% (11/12) vertices are UL19 pentamers, a random classification will give a result indistinguishable from the UL19 pentamer. Only if the reference is capable of picking out the portal vertex with quite high efficiency – say, 75–80%-do we expect to obtain a reliable result, clearly discriminated from the UL19 pentamer.
We performed classifications using as reference, the cryo-EM reconstruction of UL6 in both orientations (up/down) and at various radial positions relative to the plane of the capsid shell (see Materials and Methods). Only for one orientation – with the narrow end of the UL6 dodecamer pointing outwards – did the classification return a result consistent with the starting model. At the optimal radial setting for the portal protein, its wider end docks snugly into the cavity in the floor layer created by the removal of a UL19 pentamer. Classification with the portal in the other orientation did not return the reference structure, but instead, a UL19-like structure. This result provided an important confirmation that the outcome of the previous classification did not simply result from imprinting noise on a template, i.e. recovering a replica of the starting model. As a further control, classification was performed with a geometrical model that recapitulated, in simplified form, the main features of the portal. It also yielded the portal structure from the tomographic data set (and not the synthetic model). When this reference was shifted up or down by more than 2.5 nm from the optimal position, the result switched to a UL19 pentamer-like structure.
Structure of the portal-discriminated A-capsid
The classification described above allowed us to average the 150 capsid tomograms in a consistent orientation, yielding an asymmetric representation of the A-capsid at 6 nm resolution (Fig. 3). The portal complex is mounted on the outer surface of the capsid floor (Fig. 4), and it essentially replaces a pentamer at one vertex. It closely resembles the cryo-EM structure when the latter is limited to the same resolution except that it has an additional, internal, axial density. We cannot determine whether this density is residual noise, or an additional protein component, or results from a conformational change that accompanies assembly of the portal into the capsid. The adjacent UL19 hexamers do not seem to be affected by the insertion of the portal protein. Like the UL19 pentamers at the other vertices, the portal is surrounded by five triplexes (Fig. 4). The protruding portion of the UL6 portal is somewhat smaller than the UL19 pentamer protrusion (cf. Figs. 4a and 4b), but is generally similar in shape and dimensions. The main differences between the two protrusions are in their respective internal structures (axial cavities).
Figure 3.
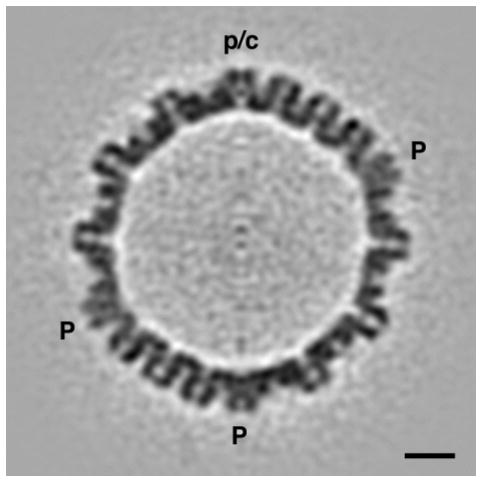
Central section through an averaged tomographic reconstruction of A-capsids. 150 individual tomograms were mutually aligned, with their portal/connector vertex (p/c) at the top, and averaged. Noise was further reduced by 5-fold averaging about the symmetry axis running vertically in the center of this plane. The 12-fold rotational symmetry of the portal is amplified to 60-fold symmetry by this operation: at the current resolution (5 nm), this is tantamount to cylindrical averaging about the local symmetry axis. Nevertheless, the average distribution of density along this axis and perpendicular to it, at this vertex site, are correctly conveyed. In these respects, this vertex site differs markedly from the vertices (P) occupied by pentamers of UL19. Bar = 20 nm.
Figure 4.

Structures of (a) portal and (b) non-portal vertices of HSV-1 A-capsids from cryo-electron tomography. The top panels compare sagittal sections (A) through the respective vertices. The adjacent hexamers of UL19 (H) and triplexes (T) are the same in both cases, but the structures around the symmetry axis are markedly different. Below are shown tangential sections at levels B and C (as marked on the top panels). Note that the axial channel at the inner (floor) layer of the capsid (level B and below) is wider in the portal (a) than in the UL19 pentamer (b), and vice versa at the external surface (level C). Moreover, the external protrusion is narrower in the portal. Bars = 10 nm. Bottom panels - surface renderings of the regions around the two kinds of vertex are compared. The portal is surrounded by five triplexes, as is the UL19 pentamer. Two triplexes are designated with arrows (left panel).
Reconstruction versus tomography
It is noteworthy that although almost the same numbers of capsids, 156 vs. 150, were used in the reconstruction as in the tomographic analysis, the latter gave a better result. (The tomograms used 3–4-fold higher electron doses, but this should give, at most, a 2-fold reduction in noise and it is not clear that the additional dose is entirely beneficial, in view of incipient radiation damage - Conway et al., 1993). This outcome is somewhat surprising in view of the “dose fractionation therorem” (Hegerl and Hoppe, 1976; McEwen et al., 1995) which holds that the two approaches should yield similar results. It has the encouraging implication that current reconstruction procedures do not fully exploit the information present in the cryo-micrographs and significant improvements are feasible.
The HSV-1 portal is externally mounted
Our analysis places the bulk of the UL6 portal mass on the external surface of the floor layer, where it forms a protrusion of approximately the same size as those of UL19 capsomers (Fig. 4). Our confidence in this assignment is based on the following considerations: (i) if the UL6 protrusion were, in fact, on the inner surface, we should see it there consistently in individual tomograms, as we readily see the similar-sized UL19 protrusions on the outer surface (Fig. 2); (ii) our correlation analysis did not detect the portal at an interior radial position. In this respect, the HSV-1 portal differs from the phage portals that have been characterized to date, the bulk of whose masses are located interior to the capsid shell (see Introduction). In view of this trend and the many points of resemblance between the HSV-1 capsid and those of tailed phages (see Introduction; and Wikoff et al, 2000; Baker et al., 2005), we carried out two further experiments to confirm our localization of the UL6 portal.
First, we performed a classification of the tomographic vertex data, using as reference a solid cylinder of about the same height and diameter as UL6 or phage portals of similar size, mounted with one end flush with the floor layer and pointing inwards (Fig. 5a). If the capsids were to have a vertex occupied by an inwards-pointing portal, we expected this template to pick them out. However, because the principal feature of the resulting averaged vertex is a large, UL19 pentamer-like, external protrusion and there is only a faint annulus of density on the inner surface of the floor (arrow in Fig. 5b, cf. Fig. 5c), this experiment indicates that the portal is not internally mounted, confirming the previous result. To explain the annulus, we infer that some A-capsids were not entirely empty and this classification template tended to pick out vertices with small amounts of material – most likely, residual scaffolding protein - deposited on the capsid inner surface in their general vicinity.
Figure 5.
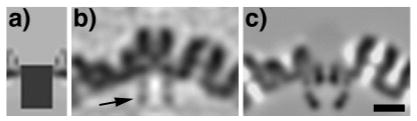
A UL19 pentamer-like external protrusion is present on the structure yielded by a tomographic averaging experiment in which vertices were selected, one per capsid, on the basis of their having density underlying the floor layer at the vertex site. The reference template (a) was a uniform solid cylinder of 14 nm in height and 10 nm in diameter, mounted with its top flush with the outer surface of the floor layer. In each capsid, it should detect the vertex with the most density in this region, regardless of what density there may be outside the floor. (b) The resulting averaged vertex structure (central section). (c) Corresponding section of a model with the UL6 portal seated in the floor layer and pointing inwards. In panel (b), there is a faint annulus under the bloor (arrow). This density is much weaker than that in the shell, indicating substoichiometric occupancy, and its apparent symmetry (left/right) results from the cylindrical averaging that was performed to boost the signal-to-noise ratio. Bar = 20 nm.
In a second control experiment, we used a different approach that also allows one to visualize individual capsomers – freeze-fracture electron microscopy (Figure 6). The hypothesis that the HSV-1 portal points inwards predicts that one vertex per capsid should have on its outer surface a depression, lower than the surrounding triplexes, instead of a protrusion. Metal replicas were prepared in two ways: from fracturing pelleted capsids, and from capsids adsorbed to mica flakes. The former preparation should depict capsids in random orientations whereas there is, in principle, a possibility that the portal might adhere preferentially to the mica and therefore not be represented in the replicas. Of 77 vertices that were clearly visualized in the former preparations, all were occupied by protrusions comparable in size to adjacent hexon protrusions. Similarly, 13 vertices visualized in the latter preparations all had protrusions. Given random sampling of portal and non-portal vertices in these experiments, the a priori probability that this data set would fail to include a single portal vertex is 4·10−4, i.e. (11/12)77+13. This result further supports the conclusion that the portal vertex does indeed have a protrusion, as inferred from the tomographic analysis.
Figure 6.
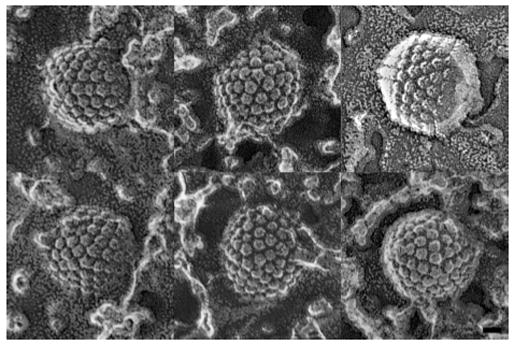
Gallery of HSV-1 capsids presenting clearly defined vertices in electron micrographs of preparations made by the quick freeze/deep etch/rotary shadowing technique. Vertex sites are identifiable from being surrounded by five, rather than six, neighbors. There were protrusions at all the vertices observed in these micrographs. Bar = 25 nm.
Structural and functional diversification of portals
As the floor layer of the HSV-1 capsid is structurally similar to tailed phage capsid shells, it is plausible that the portions of the respective portals that traverse the capsid shells are also structurally similar; however, other domains may differ. Noting that HSV-1, unlike the phages, does not affix a tail at this site, we conjecture that the externally exposed domains of the UL6 portal may be involved in other functions, such as docking of the terminase for packaging (Yu and Weller, 1998; Wills et al., 2006), or the binding of other proteins after packaging to ensure that the genome remains stably encapsidated. Conversely, internal domains of some phage portals afford platforms for the assembly of core proteins (Cerritelli et al., 2003; Agirrezabala et al., 2005a; Jiang et al., 2006; Lander et al., 2006) which, in some cases, are ejected from the phage head to participate in the infection process (Kemp et al., 2005). There is, to date, no evidence for such core proteins in herpesvirus capsids and, consequently, no need for portal domains to accommodate them.
Materials & Methods
Immuno-labeling of capsids
B-capsids were isolated from Vero cells infected with HSV-1 KOS, as previously described (Sheaffer et al., 2000). Immuno-labeling was performed according to (Newcomb et al., 2001): basically, rabbit polyclonal antibodies raised against a chimeric protein consisting of MBP fused to the C-terminal 298 amino acids of UL6 (a gift of J. C. Baines, Cornell University), were used as a primary UL6-specific antibody, followed by a gold-labeled secondary antibody.
Cryo-electron microscopy and image reconstruction
Capsids were adsorbed to grids bearing continuous carbon films and immuno-labeled as described above, and then vitrified and observed on a CM200-FEG electron microscope (FEI, Mahwah, NJ), according to (Cheng et al., 2002). Micrographs were recorded on film using low-dose procedures, at a magnification of 38,000x. The defocus settings were such that the first zeros of the contrast transfer function were at ~ (2.5 nm)−1. Negatives were digitized on a SCAI scanner (Z/I Imaging, Huntsville, Ala) and binned three-fold to give pixels of 0.55 nm at the specimen.
Particles were extracted and preprocessed with the X3D program (Conway et al., 1993). Two versions of each file were maintained: one original, and one with the gold beads painted out to the local background density. The gold beads were detected automatically in bshow (Heymann and Belnap, 2006). Geometrical parameters (Euler angles and origin coordinates) were determined by applying EMPFT (Baker and Cheng, 1996) to the gold bead-erased data. Icosahedral reconstructions were calculated using EM3DR for both the original and the gold bead-erased data, using the same geometrical parameters in each case. Bsoft (Heymann and Belnap, 2006) was used for calculating asymmetric reconstructions.
Cryo-electron tomography
A-capsids isolated from Vero cells infected with the KOS strain of HSV-1 according to (Newcomb et al., 1993) were mixed with a suspension of 10-nm BSA-gold particles (Aurion, Wageningen, the Netherlands). These gold particles were intended to serve as fiducial markers for aligning the projections in tomographic tilt series. 4 μl drops were applied to grids covered with holey carbon film, thinned by blotting with filter paper, then plunged into liquid ethane and transferred into a Gatan 626 cryoholder (Pleasanton, CA). Vitrified samples were examined using a Tecnai-12 electron microscope (FEI, Hillsboro, OR) equipped with a Gatan energy filter (GIF 2002). Tilt series were acquired at a defocus of ~ 4 μm and a final magnification of 38,500 (pixel size, 0.78 nm) on a charge-coupled device camera (2,048 × 2,048 pixels, Gatan). The angular range covered was typically from −57 to +57° in increments of 1°. Data were recorded under low-dose conditions, using SerialEM (Mastronarde, 2005) to perform automatic tilting, predictive tracking, focusing and image acquisition. The cumulative electron doses for complete tilt series were from 35 to 80 electrons/Å2.
Projections were aligned and tomograms calculated, using IMOD (Kremer et al., 1996). Tomogram resolution was assessed by NLOO2D method as implemented in ELECTRA (Cardone et al., 2005). For each tomogram, three particles were selected and their resolutions were assessed at the threshold 0.3. These three values were then averaged.
Image classification
After the gold particles were used to align the projections, they were painted out to prevent them from degrading the tomogram and the subsequent classification procedures. Their locations in the tomograms were determined by cross-correlation with a synthetic model of a gold particle, filtered in the Fourier domain to take into account the missing wedge, and their positions on projections were specified by appropriately projecting these coordinates. The gold particles were then painted out in the projections and a filtered tomogram was calculated.
Capsids excised from the tomograms were brought into a standard orientation by correlation alignment relative to a cryo-EM density map of the A-capsid at 2 nm resolution (Cheng et al., 2002). Twelve subvolumes, centered on the vertices and including the five surrounding hexons, were then extracted from each capsid. The orientations of these subvolumes were refined by aligning them relative to the corresponding subvolume from the cryo-EM map. The cross-correlation function used took into account the missing wedge in Fourier space. Finally, a subvolume of 25 × 25 × 35 voxels centered on the vertex capsomer, was extracted in each case. These data were analyzed to identify the portal vertices by a template-matching method. As reference, we used the cryo-EM structure of the portal (Trus et al., 2004) in both orientations (up/down, relative to the capsid surface) and in various radial positions relative to the capsid surface. Cross-products between a vertex volume and the model were calculated from 2D representations obtained by cylindrically averaging them around their central (trans-vertex) axis. The latter operation (rotational averaging) was performed in the Fourier domain, to exclude the spatial frequencies in the missing wedge. For each capsid, the vertex that gave the highest correlation was assigned as the portal vertex. This procedure was performed for both orientations of the model, and with the models at different radial positions relative to the center of the capsid (N=15, radius from 55.4 to 66.3 nm). The radial position of the portal was specified by the density map with the highest variance (Cantele et al., 2003). We performed a control experiment with a computer-generated geometric model composed of two separated solids that summarized the overall shape of the portal: a hollow cylinder with length 5 nm and radius 5 nm, mounted above a truncated cone, 8 nm in length and 5 to 12 nm in radius. It led to essentially the same structure. After the portal vertices were identified, the capsids were rotated to bring the portal vertex into a standard position, and averaged. To further improve the signal-to-noise ratio, five-fold symmetry around the portal channel axis was applied. The resolution of this map was determined by Fourier Shell Correlation (0.3 threshold). Alignment and classification procedures were performed with routines from Bsoft (Heymann and Belnap, 2006), modified as needed and wrapped into Python scripts.
Freeze-fracture electron microscopy
Replicas were prepared either from cross-fractured capsid pellets (Heuser, 1981) or from capsids adsorbed to mica flakes (Heuser, 1989). Briefly, two drops of a suspension of finely ground mica flakes are added to 0.5 ml of capsids at ~ 2 mg/ml. After 30 sec incubation to allow adsorption, the mica flakes were pelleted, washed twice with 70 mM KCl, 30 mM Hepes pH 7.2, 5 mM MgCl2, and 3mM EGTA, then layered on to an 800-μm-thick slice of aldehyde-fixed lung for support during ‘quick-freezing’, which was accomplished by “smashing” the samples onto a liquid helium-cooled copper block. The frozen slurry of mica flakes was fractured in a Balzers freeze etching unit and immediately etched for ~ 3 min at −100°C; then rotary-replicated with ~2 nm of platinum applied from an angle of 11° above the horizontal and “backed” with a ~10 nm film of carbon. Replicas are separated from the mica flakes by overnight flotation on a solution of concentrated hydrofluoric acid and washed several times in distilled water, picked up on 75-mesh formvar-coated microscope grids and viewed in a JEOL transmission electron microscope. Stereo images (not shown) were obtained using ±10° of tilt.
Acknowledgments
We thank Dr. B. Heymann for help with programming and the provision of software, and Ms. L. You for scanning micrographs. This study was supported in part by the Intramural Research Program of NIAMS, by NIH grant No. AI41644-10 to JCB, and by USPHS grant number GM29647-23 to JEH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agirrezabala X, Martin-Benito J, Caston JR, Miranda R, Valpuesta JM, Carrascosa JL. Maturation of phage T7 involves structural modification of both shell and inner core components. Embo J. 2005a;24(21):3820–9. doi: 10.1038/sj.emboj.7600840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agirrezabala X, Martin-Benito J, Valle M, Gonzalez JM, Valencia A, Valpuesta JM, Carrascosa JL. Structure of the connector of bacteriophage T7 at 8A resolution: structural homologies of a basic component of a DNA translocating machinery. J Mol Biol. 2005b;347(5):895–902. doi: 10.1016/j.jmb.2005.02.005. [DOI] [PubMed] [Google Scholar]
- Baines JD, Duffy C. Nucleocapsid assemby and envelopment of Herpes Simplex Virus. In: Sandri-Goldin R, editor. Alpha Herpesviruses: Molecular and Cell Biology. Horizon Press; Norwich, UK: 2006. [Google Scholar]
- Baker TS, Cheng RH. A model-based approach for determining orientations of biological macromolecules imaged by cryoelectron microscopy. J Struct Biol. 1996;116:120–130. doi: 10.1006/jsbi.1996.0020. [DOI] [PubMed] [Google Scholar]
- Baker TS, Olson NH, Fuller SD. Adding the third dimension to virus life cycles: three-dimensional reconstruction of icosahedral viruses from cryo-electron micrographs. Microbiological and Molecular Biology Reviews. 1999;63(4):862–922. doi: 10.1128/mmbr.63.4.862-922.1999. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker ML, Jiang W, Rixon FJ, Chiu W. Common ancestry of herpesviruses and tailed DNA bacteriophages. J Virol. 2005;79(23):14967–70. doi: 10.1128/JVI.79.23.14967-14970.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamford DH, Grimes JM, Stuart DI. What does structure tell us about virus evolution? Curr Opin Struct Biol. 2005;15(6):655–63. doi: 10.1016/j.sbi.2005.10.012. [DOI] [PubMed] [Google Scholar]
- Black LW, Showe MK, Steven AC. Morphogenesis of the T4 head. In: Karam J, editor. Molecular Biology of Bacteriophage T4. Am. Soc. Microbiol; Washington,D.C: 1994. pp. 218–258. [Google Scholar]
- Burch AD, Weller SK. Nuclear sequestration of cellular chaperone and proteasomal machinery during herpes simplex virus type 1 infection. J Virol. 2004;78(13):7175–85. doi: 10.1128/JVI.78.13.7175-7185.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantele F, Lanzavecchia S, Bellon PL. The variance of icosahedral virus models is a key indicator in the structure determination: a model-free reconstruction of viruses, suitable for refractory particles. J Struct Biol. 2003;141(1):84–92. doi: 10.1016/s1047-8477(02)00577-4. [DOI] [PubMed] [Google Scholar]
- Cardone G, Grunewald K, Steven AC. A resolution criterion for electron tomography based on cross-validation. J Struct Biol. 2005;151(2):117–29. doi: 10.1016/j.jsb.2005.04.006. [DOI] [PubMed] [Google Scholar]
- Cerritelli ME, Conway JF, Cheng N, Trus BL, Steven AC. Molecular mechanisms in bacteriophage T7 assembly, maturation and DNA containment. In: Chiu W, Johnson JE, editors. Advances in Protein Chemistry: Virus Structure. Academic Press; San Diego: 2002. [DOI] [PubMed] [Google Scholar]
- Cerritelli ME, Trus BL, Smith CS, Cheng N, Conway JF, Steven AC. A second symmetry mismatch at the portal vertex of bacteriophage T7: 8-fold symmetry in the procapsid core. J Mol Biol. 2003;327(1):1–6. doi: 10.1016/s0022-2836(03)00117-7. [DOI] [PubMed] [Google Scholar]
- Cheng N, Trus BL, Belnap DM, Newcomb WW, Brown JC, Steven AC. Handedness of the herpes simplex virus capsid and procapsid. J Virol. 2002;76(15):7855–9. doi: 10.1128/JVI.76.15.7855-7859.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani G, Moore SD, Prevelige PE, Jr, Johnson JE. Preliminary crystallographic analysis of the bacteriophage P22 portal protein. J Struct Biol. 2002;139(1):46–54. doi: 10.1016/s1047-8477(02)00512-9. [DOI] [PubMed] [Google Scholar]
- Conway JF, Trus BL, Booy FP, Newcomb WW, Brown JC, Steven AC. The effects of radiation damage on the structure of frozen hydrated HSV-1 capsids. J Struct Biol. 1993;111:222–233. doi: 10.1006/jsbi.1993.1052. [DOI] [PubMed] [Google Scholar]
- Dube P, Tavares P, Lurz R, van Heel M. The portal protein of bacteriophage SPP1: a DNA pump with 13-fold symmetry. EMBO J. 1993;12:1303–1309. doi: 10.1002/j.1460-2075.1993.tb05775.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fokine A, Kostyuchenko VA, Efimov AV, Kurochkina LP, Sykilinda NN, Robben J, Volckaert G, Hoenger A, Chipman PR, Battisti AJ, Rossmann MG, Mesyanzhinov VV. A three-dimensional cryo-electron microscopy structure of the bacteriophage phiKZ head. J Mol Biol. 2005;352(1):117–24. doi: 10.1016/j.jmb.2005.07.018. [DOI] [PubMed] [Google Scholar]
- Frangakis AS, Bohm J, Forster F, Nickell S, Nicastro D, Typke D, Hegerl R, Baumeister W. Identification of macromolecular complexes in cryoelectron tomograms of phantom cells. Proc Natl Acad Sci U S A. 2002;99(22):14153–8. doi: 10.1073/pnas.172520299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank Je. Electron Tomography. Plenum Press; New York: 1992. [Google Scholar]
- Grunewald K, Desai P, Winkler DC, Heymann JB, Belnap DM, Baumeister W, Steven AC. Three-dimensional structure of herpes simplex virus from cryo-electron tomography. Science. 2003;302(5649):1396–8. doi: 10.1126/science.1090284. [DOI] [PubMed] [Google Scholar]
- Guasch A, Pous J, Ibarra B, Gomis-Ruth FX, Valpuesta JM, Sousa N, Carrascosa JL, Coll M. Detailed architecture of a DNA translocating machine: the high-resolution structure of the bacteriophage phi29 connector particle. J Mol Biol. 2002;315(4):663–76. doi: 10.1006/jmbi.2001.5278. [DOI] [PubMed] [Google Scholar]
- Hegerl R, Hoppe W. Influence of electron noise on three-dimensional image reconstruction. Zeitschrift Naturforsch A. 1976;314(12):1717–21. [Google Scholar]
- Hendrix RW, Duda RL. Bacteriophage HK97 head assembly: a protein ballet. Adv Virus Res. 1998;50:235–88. doi: 10.1016/s0065-3527(08)60810-6. [DOI] [PubMed] [Google Scholar]
- Heuser J. Preparing biological samples for stereomicroscopy by the quick-freeze, deep-etch, rotary-replication technique. Methods Cell Biol. 1981;22:97–122. doi: 10.1016/s0091-679x(08)61872-5. [DOI] [PubMed] [Google Scholar]
- Heuser J. Protocol for 3-D visualization of molecules on mica via the quick-freeze, deep-etch technique. J Electron Microsc Tech. 1989;13(3):244–63. doi: 10.1002/jemt.1060130310. [DOI] [PubMed] [Google Scholar]
- Heymann JB, Belnap DM. Bsoft: Image processing and molecular modeling for electron microscopy. J Structl Biol. 2006 doi: 10.1016/j.jsb.2006.06.006. in press. [DOI] [PubMed] [Google Scholar]
- Heymann JB, Cheng N, Newcomb WW, Trus BL, Brown JC, Steven AC. Dynamics of herpes simplex virus capsid maturation visualized by time-lapse cryo-electron microscopy. Nat Struct Biol. 2003;10(5):334–41. doi: 10.1038/nsb922. [DOI] [PubMed] [Google Scholar]
- Jiang W, Chang J, Jakana J, Weigele P, King J, Chiu W. Structure of epsilon15 bacteriophage reveals genome organization and DNA packaging/injection apparatus. Nature. 2006;439(7076):612–6. doi: 10.1038/nature04487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Li Z, Zhang Z, Baker ML, Prevelige PE, Jr, Chiu W. Coat protein fold and maturation transition of bacteriophage P22 seen at subnanometer resolutions. Nat Struct Biol. 2003;10(2):131–5. doi: 10.1038/nsb891. [DOI] [PubMed] [Google Scholar]
- Kemp P, Garcia LR, Molineux IJ. Changes in bacteriophage T7 virion structure at the initiation of infection. Virology. 2005;340(2):307–17. doi: 10.1016/j.virol.2005.06.039. [DOI] [PubMed] [Google Scholar]
- King J, Chiu W. The procapsid-to-capsid transition in double-stranded DNA bacteriophages. In: Chiu W, Burnett RM, Garcea R, editors. Structural Biology of Viruses. Oxford University Press; New York: 1997. pp. 288–311. [Google Scholar]
- Kocsis E, Cerritelli ME, Trus BL, Cheng N, Steven AC. Improved methods for determination of rotational symmetries in macromolecules. Ultramicroscopy. 1995;60:219–228. doi: 10.1016/0304-3991(95)00070-2. [DOI] [PubMed] [Google Scholar]
- Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J Struct Biol. 1996;116(1):71–6. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- Lander GC, Tang L, Casjens SR, Gilcrease EB, Prevelige P, Poliakov A, Potter CS, Carragher B, Johnson JE. The structure of an infectious P22 virion shows the signal for headful DNA packaging. Science. 2006;312(5781):1791–5. doi: 10.1126/science.1127981. [DOI] [PubMed] [Google Scholar]
- Lucic V, Forster F, Baumeister W. Structural studies by electron tomography: from cells to molecules. Annu Rev Biochem. 2005;74:833–65. doi: 10.1146/annurev.biochem.73.011303.074112. [DOI] [PubMed] [Google Scholar]
- Lurz R, Orlova EV, Gunther D, Dube P, Droge A, Weise F, van Heel M, Tavares P. Structural organisation of the head-to-tail interface of a bacterial virus. J Mol Biol. 2001;310(5):1027–37. doi: 10.1006/jmbi.2001.4800. [DOI] [PubMed] [Google Scholar]
- Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol. 2005;152(1):36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- McEwen BF, Downing KH, Glaeser RM. The relevance of dose-fractionation in tomography of radiation-sensitive specimens. Ultramicroscopy. 1995;60(3):357–73. doi: 10.1016/0304-3991(95)00082-8. [DOI] [PubMed] [Google Scholar]
- Newcomb WW, Homa FL, Thomsen DR, Booy FP, Trus BL, Steven AC, Spencer JV, Brown JC. Assembly of the herpes simplex virus capsid: characterization of intermediates observed during cell-free capsid formation. Journal of Molecular Biology. 1996;263(3):432–46. doi: 10.1006/jmbi.1996.0587. [DOI] [PubMed] [Google Scholar]
- Newcomb WW, Juhas RM, Thomsen DR, Homa FL, Burch AD, Weller SK, Brown JC. The UL6 gene product forms the portal for entry of DNA into the herpes simplex virus capsid. J Virol. 2001;75(22):10923–32. doi: 10.1128/JVI.75.22.10923-10932.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newcomb WW, Thomsen DR, Homa FL, Brown JC. Assembly of the herpes simplex virus capsid: identification of soluble scaffold-portal complexes and their role in formation of portal-containing capsids. J Virol. 2003;77(18):9862–71. doi: 10.1128/JVI.77.18.9862-9871.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newcomb WW, Trus BL, Booy FP, Steven AC, Wall JS, Brown JC. Structure of the herpes simplex virus capsid. Molecular composition of the pentons and the triplexes. J Mol Biol. 1993;232(2):499–511. doi: 10.1006/jmbi.1993.1406. [DOI] [PubMed] [Google Scholar]
- Orlova EV, Gowen B, Droge A, Stiege A, Weise F, Lurz R, van Heel M, Tavares P. Structure of a viral DNA gatekeeper at 10 A resolution by cryo-electron microscopy. EMBO J. 2003;22(6):1255–62. doi: 10.1093/emboj/cdg123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheaffer AK, Newcomb WW, Brown JC, Gao M, Weller SK, Tenney DJ. Evidence for controlled incorporation of herpes simplex virus type 1 UL26 protease into capsids. J Virol. 2000;74(15):6838–48. doi: 10.1128/jvi.74.15.6838-6848.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson AA, Tao YZ, Leiman PG, Badasso MO, He YN, Jardine PJ, Olson NH, Morais MC, Grimes S, Anderson DL, Baker TS, Rossmann MG. Structure of the bacteriophage phi 29 DNA packaging motor. Nature. 2000;408(6813):745–750. doi: 10.1038/35047129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steven AC, Heymann JB, Cheng N, Trus BL, Conway JF. Virus maturation: dynamics and mechanism of a stabilizing structural transition that leads to infectivity. Curr Opin Struct Biol. 2005;15(2):227–36. doi: 10.1016/j.sbi.2005.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang L, Marion WR, Cingolani G, Prevelige PE, Johnson JE. Three-dimensional structure of the bacteriophage P22 tail machine. Embo J. 2005;24(12):2087–95. doi: 10.1038/sj.emboj.7600695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y, Olson NH, Xu W, Anderson DL, Rossmann MG, Baker TS. Assembly of a tailed bacterial virus and its genome release studied in three dimensions. Cell. 1998;95(3):431–7. doi: 10.1016/s0092-8674(00)81773-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurlow JK, Murphy M, Stow ND, Preston VG. Herpes simplex virus type 1 DNA-packaging protein UL17 is required for efficient binding of UL25 to capsids. J Virol. 2006;80(5):2118–26. doi: 10.1128/JVI.80.5.2118-2126.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trus BL, Booy FP, Newcomb WW, Brown JC, Homa FL, Thomsen DR, Steven AC. The herpes simplex virus procapsid: Structure, conformational changes upon maturation, and roles of the triplex proteins VP19c and VP23 in assembly. J Mol Biol. 1996;263:447–462. doi: 10.1016/s0022-2836(96)80018-0. [DOI] [PubMed] [Google Scholar]
- Trus BL, Cheng N, Newcomb WW, Homa FL, Brown JC, Steven AC. Structure and polymorphism of the UL6 portal protein of herpes simplex virus type 1. J Virol. 2004;78(22):12668–71. doi: 10.1128/JVI.78.22.12668-12671.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valpuesta JM, Carrascosa JL. Structure of viral connectors and their function in bacteriophage assembly and DNA packaging. Q Rev Biophys. 1994;27:107–155. doi: 10.1017/s0033583500004510. [DOI] [PubMed] [Google Scholar]
- Valpuesta JM, Sousa N, Barthelemy I, Fernandez JJ, Fujisawa H, Ibarra B, Carrascosa JL. Structural analysis of the bacteriophage T3 head-to-tail connector. J Struct Biol. 2000;131(2):146–155. doi: 10.1006/jsbi.2000.4281. [DOI] [PubMed] [Google Scholar]
- Wikoff WR, Liljas L, Duda RL, Tsuruta H, Hendrix RW, Johnson JE. Topologically linked protein rings in the bacteriophage HK97 capsid. Science. 2000;289(5487):2129–33. doi: 10.1126/science.289.5487.2129. [DOI] [PubMed] [Google Scholar]
- Wills E, Scholtes L, Baines JD. Herpes Simplex Virus 1 DNA Packaging Proteins Encoded by UL6, UL15, UL17, UL28, and UL33 Are Located on the External Surface of the Viral Capsid. J Virol. 2006;80(21):10894–9. doi: 10.1128/JVI.01364-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Weller SK. Herpes simplex virus type 1 cleavage and packaging proteins UL15 and UL28 are associated with B but not C capsids during packaging. J Virol. 1998;72(9):7428–39. doi: 10.1128/jvi.72.9.7428-7439.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]


