Abstract
Mutations in the adenomatous polyposis coli gene (which encodes a protein called APC) are associated with the formation of intestinal polyps and colon cancers. To facilitate the functional study of APC we have isolated its Drosophila homolog (D-APC) by screening an expression library with an antibody against human APC. The isolated cDNA encodes a predicted 2416-amino acid protein containing significant homology to multiple domains of mammalian APCs. D-APC has seven complete armadillo repeats with 60% identity to its human homolog, one β-catenin binding site, and up to 7 copies of a 20-amino acid repeat with the average of 50% identity to human APC at amino acid level. D-APC, like its human counterpart, also contains a basic domain. Expression of the domain of D-APC homologous to the region required for β-catenin down-regulation resulted in down-regulation of intracellular β-catenin in a mammalian cell line. This same region bound to the Armadillo (Arm) protein, in vitro, the Drosophila homolog of β-catenin. D-APC RNA and protein expression is very low, if detectable at all, during stages when Arm protein accumulates in a striped pattern in the epidermis of the Drosophila embryos. Removing zygotic D-APC expression did not alter Arm protein distribution, and the final cuticle pattern was not affected significantly. As observed in the rodent, high levels of D-APC expression have been detected in the central nervous system, suggesting a role for D-APC in central nervous system formation.
In both humans and mice, mutations in the adenomatous polyposis coli gene (which encodes a protein called APC) predispose heterozygotes to inherited colonic polyps and cancer (1, 2). Mutations in the remaining wild-type allele are observed in the earliest recognizable benign tumors, suggesting that loss of wild-type APC activity plays an early role in tumorigenesis (3, 4). How APC functions in tumor suppression is not known. Wild-type APC blocks cells cycle progression from the G0–G1 and G1 to S (5) and causes some level of apoptosis when induced in a cell line lacking endogenous APC (6). Biochemical characterizations of the protein have identified a number of possible pathways and interacting proteins, such as β-catenin (7, 8), microtubules (9, 10), EB1 (11), and the human homolog of Drosophila discs-large tumor suppressor (DLG) (12). In a colon carcinoma cell line lacking wild-type APC, transient expression of wild-type, but not mutant, APC has been shown to down-regulate cytoplasmic β-catenin (13). In addition, higher levels of β-catenin has been observed in APC(−/−) tumors than normal neighboring APC(+/−) cells (14). The sequences in APC required for this down-regulation are located in the middle of the protein and consist of several copies of a 20-amino acid repeat (13). The vast majority of clinically important mutations in APC predict truncated proteins lacking this region, as well as the domains carboxy terminal to it that interact with microtubules and DLG (15).
The interaction between β-catenin and APC suggested that APC may play a role in the wingless (Wg)/Wnt signaling pathway. This pathway has been well characterized in Drosophila embryos where it controls anterior posterior pattern within each segment. In this system, secreted Wg protein counters a negative signal involving zeste-white 3 (Zw3), the homolog of glycogen synthetase kinase 3β (GSK), and results in the accumulation of cytoplasmic Armadillo (Arm), the homolog of β-catenin. Wnt-1 expression has a similar effect on β-catenin level in vertebrates (16, 17), an effect which can be counteracted by the presence of APC (18).
Most of the results linking APC and the β-catenin/Wnt-1 pathway involve artificial manipulations of APC levels and may not reflect the normal role of the gene. To augment the study of human APC, we have identified a Drosophila homolog of APC (D-APC). It is capable of down-regulating β-catenin in a colon carcinoma cell line and of interacting with the Arm protein in vitro. However, its expression pattern and a preliminary genetic analysis suggest that it may not be an essential component of Arm regulation in the Wg pathway.
MATERIALS AND METHODS
Antibody Production.
Antibodies were generated by the method described elsewhere (19). Glu–tag fusion protein of human APC (amino acid 1034–2130; APC2) (8) or S–tag fusion protein of pSH7 (see below; amino acids 648-1376) was used as the antigen. pSH7 was subcloned into pET29 (Novogen) using EcoRI site (pSH7b). Freund’s complete and incomplete adjuvant (GIBCO/BRL) were used for the primary immunization and booster injections, respectively. The antiserum was purified by using an affinity column where the antigen was coupled to CNBr-activated Sepharose 4B CL beads (Pharmacia), according to the method described in ref. 19.
Cloning of D-APC cDNA.
A Drosophila melanogaster 5′ Stretch Plus λ gt11 library (CLONTECH) of 2.5 × 105 independent plaques were screened with the α-APC2 polyclonal antibody. Screening and subcloning of positive clones into Bluescript sk (Stratagene) were carried out by the methods described in the manufacturer’s protocol. One partial cDNA (pSH7) of 2.3 kb that has homology to human APC was identified by DNA sequencing. To obtain the remaining region of the cDNA, the library mentioned above was rescreened with a 472-bp PCR probe (generated with oligonucleotides 5′-CCACTCTGGAGGCACGAA-3′ and 5′-CTGGTCCTGATATGTCCAC-3′) for the 5′ end. The 3′ end of the cDNA was isolated using the 3′ rapid amplification of the cDNA end system (GIBCO/BRL). Two primers were designed within 300- to 400-bp upstream region from the 3′ end of the original clone (5′-GAGGACAGTCCCTGTAAC-3′ and 5′-CTTCTCAGTGGTCTCTGG-3′) as a nested primer and used for the PCR. Multiple partial D-APC clones were isolated by rescreening the cDNA libraries (Stratagene and CLONTECH). The nucleotide sequence of the D-APC was determined by sequencing at least three overlapping clones. Inconsistencies of the nucleotide sequences between the multiple clones were at nucleotides 6426 (C/T), 6486 (C/G), 6515–6517 (ATC/CTC/CCA), 6590–6591 (AT/CA), and 6594 (T/G).
DNA Sequencing.
DNA sequencing was carried out by dideoxy chain termination method with Sequenase (United States Biochemical) as described in the manufacturer’s protocol and automated sequencing (SeqWright, Houston). DNA sequence analysis was carried out with the University of Wisconsin Genetics Computer Group sequence analysis programs. GenBank and EMBL data bases were searched by use of the tfasta and blastp programs. Homology searches between the two species were carried out by the gap and bestfit programs, and the multiple sequence alignment was done by the pileup program.
β-Catenin Down-Regulation and in Vitro Binding Assay.
pD-APC was subcloned into the plasmid cDNA/AMP vector (Invitrogen). The assay for down-regulation of β-catenin was done as described previously (13). 35S radiolabeled Arm was produced by the TnT in vitro translation system (Invitrogen) using arm cDNA as described (20) and kindly provided by M. Peifer (University of North Carolina). S-tag pD-APC protein was produced from pSH7b. The binding assay was carried out by the methods described elsewhere (21).
Drosophila Stocks, Genetics, and Chromosomal in Situ Hybridization.
Df(3R)3450 and DpB152 were obtained from the Bloomington stock center (Bloomington, IN). The absence of D-APC gene in Df(3R)3450 was confirmed by in situ hybridization on polytene chromosomes of Df(3R)3450/Oregon R and whole-mount in situ hybridization in embryos.
The chromosomal in situ hybridization was done by the method described previously (22). pSH7 was used to generate the DNA probe.
Df(3R)3450 and DpB152 were generated by crossing DpB152 virgin females to Df(3R)3450 males.
Individual male carrying Df(3R)3450 was back-crossed to Df(3R)3450 females. Individual female from this cross was back-crossed to the male carrying Df(3R)3450. A fraction of the offspring from this cross produced D-APC null embryos.
In Situ Hybridization on Whole-Mount Embryos.
In situ hybridization on whole-mount embryos was performed by the method described in ref. 23, with an antisense RNA probe of pSH7.
Immunohistochemistry and Confocal Microscopy.
The method for immunohistochemistry was described elsewhere (24). For immunostaining, embryos were fixed by the heat methanol fixation method (25). Signals were detected by fluorescence labeling, either rhodamine- or fluorescein-conjugated antibody (Boehringer Mannheim). Double labeling was performed by combination of rhodamine and fluorescein labeling, and embryos were viewed with a laser scanning confocal microscope (Bio-Rad).
RESULTS AND DISCUSSION
Isolation of D-APC Gene.
To identify a Drosophila homolog of APC, a Drosophila cDNA expression library was screened using a polyclonal antibody made against a fragment of the human APC protein containing the β-catenin binding sites. Western blots of embryonic extracts indicated several cross-reacting proteins in Drosophila, at least one of which could coimmunoprecipitate the Drosophila β-catenin homolog, Armadillo (data not shown). Consistent with the cross reactive nature of the antisera, only 1 of the 16 positive clones from the expression library that were sequenced showed convincing homology to APC.
Northern blots probed with this 2.3-kb cDNA detected an ≈8-kb transcript, suggesting that the initial isolate was a partial cDNA (data not shown). The remaining portion was isolated by rescreening libraries, and rapid amplification of cDNA ends. Several overlapping clones were obtained that covered an 8.3-kb cDNA region and collectively defined a complete ORF. The start codon of the ORF was identified by the presence of stop codons in all reading frames upstream of the first methionine. The ORF is terminated by a stop codon TAG at nucleotides 7456–7458 that is followed by multiple stop codons. The cDNA clones terminate in an oligo(dA) stretch shortly after the common polyadenylylation signal. The ORF thus defined comprises 7251 nucleotides and encodes a protein of 2416 amino acids with a predicted molecular mass of 260 kDa, which is smaller than the APC proteins of mouse, rat, and human.
To determine the genetic location of D-APC, the cloned gene was hybridized to polytene chromosomes of larval salivary glands. A single site of hybridization was detected near the tip of the right arm of the third chromosome (position 98F). D-APC is deleted in Df(3R)3450, which extends from 98E3 to 99A4-6 (26). It is not present in a duplication of the tip of the third chromosome [Dp(1;3)B152] covering all bands distal to 98F14 (27). This positions D-APC between bands 98E3 and 98F14. Df(3R)3450 deletes a number of known genes including Darkener of apricot, sponge, and string.
D-APC Shows Conserved Domains of Mammalian APCs An analysis of the predicted D-APC amino acid sequence revealed a striking similarity between D-APC and mammalian APCs (m-APCs).
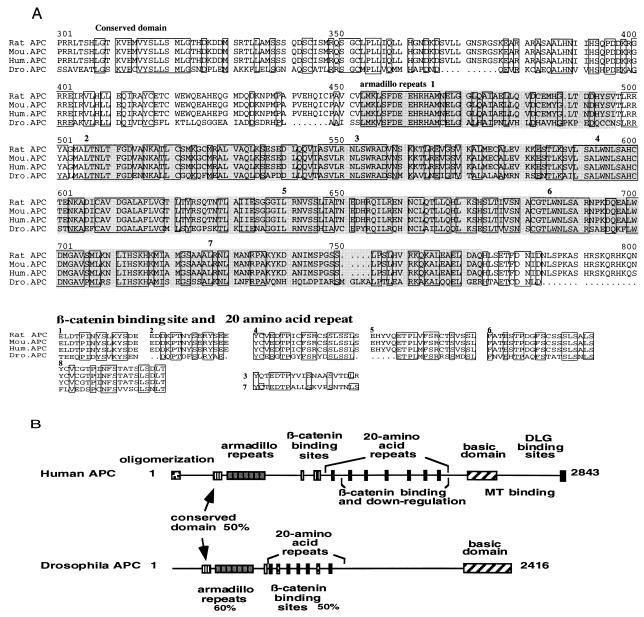
Similar to m-APCs, D-APC contains several potential glycosylation sites and phosphorylation sites and lacks a signal sequence or transmembrane domain. The deduced D-APC protein has 27% identity and 46% similarity over all to human APC. Furthermore, there is striking homology in domains previously identified in m-APCs. First, like m-APCs, the amino-terminal third of D-APC contains seven armadillo repeats with ≈60% identity to those observed in the human protein (Fig. 1). The repeats lie outside the region recognized by the human antibody used to screen the expression library; their presence provides further support that the gene described here is an APC homolog.
Figure 1.
Comparison of APC proteins from different species. (A). Multiple sequence alignment of predicted amino acid of APCs. Only highly conserved regions are shown here. Conserved amino acids in all of the species are boxed. Seven armadillo repeats are marked in gray and designated in order. (B) Comparison of the domains between D-APC and human APC. D-APC conserves most of the domains in human APC. The two stripped boxes are the homologous regions to human APC, which lack alignment by multiple sequence analysis.
In addition, D-APC has at least 6 of the 10 β-catenin binding sites previously identified in human APC, with ≈50% identity at the amino acid level (Fig. 1). D-APC contains both types of β-catenin binding sites observed in the human protein, one 15-amino acid repeat with the PXXYS motif thought to bind β-catenin, and 5 of the 20-amino acid repeat with PXXFS motif thought to have β-catenin down-regulating activity. Such repeats are not found in other β-catenin binding proteins, such as E-cadherin or α-catenin, and are thought to be hallmarks of m-APCs. There are two additional regions in D-APC that contain a PXXXS motif (Fig. 1). One of these regions shows 40–50% identity to various 20-amino acid repeats, and the other region shows 40–50% identity to various β-catenin binding sites and 20-amino acid repeats in the human protein, though they do not align with any particular mammalian repeat (Fig. 1). One of the regions with the PXXFS motif in D-APC could be a 15-amino acid repeat, based on its alignment to the second 15-amino acid repeat of human APC (Fig. 1). Because their exact function is not known, one of the PXXFS motif and two PXXXS motif sites could raise the number of β-catenin binding sites (15-amino acid repeat) from one to three, or the number of 20-amino acid repeats from five to seven. A potential GSK phosphorylation site (28) has been observed in five out of seven 20-amino acid repeats. These results also suggest that overall homology between D-APC and m-APCs could be underestimated due to lack of alignment in some conserved domains.
The carboxy terminus of the Drosophila and human APC do not show significant homology; 21% amino acid identity in the carboxyl-terminal 72 amino acids containing the DLG binding site. At carboxyl-terminal end, D-APC lacks the S/T(X)V motif, a sequence required for the binding to disks-large homology repeat domain (12). However, the carboxy terminal 270 amino acids of D-APC show a charged basic character as observed in m-APCs between amino acids 2200 and 2400 (29). This basic region has been implicated in the binding of m-APCs to microtubules (3, 9). The basic domain of D-APC has weak homology to the microtubule binding domain of microtubule-associated protein 4. This finding suggests that the carboxy terminus of the D-APC may function as that of human APC in binding to microtubules. In addition to the domains mentioned above, a well conserved region of amino acids not previously identified as a separate entity is located slightly amino terminal to the armadillo repeats between amino acids 308 and 417. This conserved domain has 50% amino acid identity between D- and m-APCs, indicating an interesting region yet to be studied functionally. Very limited homology of D-APC to m-APCs was observed in oligomerization domain, as well as DLG binding site. This finding does not exclude the possibility that D-APC may dimerize or interact using a different site. It remains possible that alternative splicing might produce as-yet-undetected D-APC variants with high homology to m-APCs in this region. Alternative splicing forms of the cDNA in 5′ end of human APC have been reported (30). Altogether, this similarity of domain structure of D-APC to m-APCs, including armadillo repeats, β-catenin binding sites, and basic domain suggests that D-APC may have significant functional similarity to m-APCs.
D-APC Down-Regulates Cytoplasmic β-Catenin in a Colon Carcinoma Cell Line.
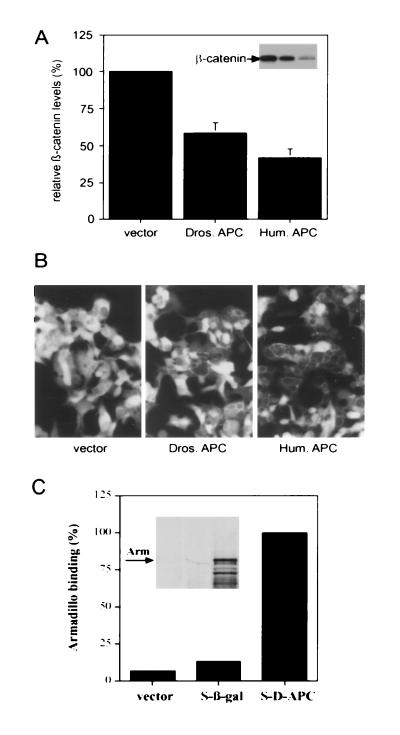
Munemitsu et al. (13) have shown that the 20-amino acid repeat region in the central part of human APC down-regulates cytoplasmic β-catenin levels in a colon carcinoma cell line (SW480), which lacks endogenous wild-type APC. To test whether D-APC might have a similar function, an expression construct with a partial D-APC cDNA containing the β-catenin binding sites was generated and transiently transfected it into the colon carcinoma cell line SW480. Forty-eight hours after transfection, the protein level and cellular localization of β-catenin were measured by Western blot analysis and immunostaining, respectively. D-APC significantly reduced the concentration of β-catenin protein, with an efficiency ≈60% of that of human APC (Fig. 2A). Immunostaining showed that intracellular cytoplasmic β-catenin decreased significantly, after introduction of the D-APC fragment, but β-catenin localizing at plasma membrane was not significantly altered (Fig. 2B). These results demonstrate that D-APC can down-regulate intracellular β-catenin levels similar to that of human APC, suggesting that D-APC is also a functional homolog of the human APC.
Figure 2.
Functional analysis of D-APC. Regulation of β-catenin levels by D-APC (A and B). SW480 cells were transfected with either empty vector or vector containing cDNAs coding for a fragment of D-APC encompassing codons 648-1376 (Dros. APC) or human APC codons 1034–2130 (Hum. APC). (A) Quantitative immunoblot. Cell lysates were normalized for protein amount and 75 μg of each sample were analyzed for β-catenin level by SDS/PAGE and immunoblotting. The β-catenin level is presented as a percentage relative to that of the cells transfected with the empty vector. The data shown are the mean of three independent experiments. (B) Immunostaining of β-catenin. The transfected SW480 cells were stained with an antibody specific to β-catenin. Each frame represents a 230 × 350 μm field. (C) Interaction of D-APC with Arm in vitro. Bacterially expressed fusion tagged protein of the D-APC fragment codons 648-1376 (S-D-APC), β-galactosidase (S-β-gal), or fusion tag vector alone was captured to S protein-agarose, which recognizes the tag region. [35S]Methionine labeled in vitro translated Arm protein was then mixed with each of the tagged proteins captured on the beads. Bound Arm protein is represented as a percentage relative to the Arm protein bound to the S-D-APC. The data shown are the mean of two independent experiments.
The existence of homologous β-catenin binding sites in D-APC raised a question whether D-APC interacts with the Drosophila homolog of β-catenin, the Arm protein. To test this possibility an in vitro binding assay was carried out using a bacterially expressed D-APC fusion protein containing β-catenin binding sites and Arm protein translated in vitro. Arm bound to the D-APC fragment containing the β-catenin binding sites, but not to a control composed of a β galactosidase fusion protein (Fig. 2C), suggesting that binding between Arm and the D-APC fragment is specific. Altogether these results indicate that the β-catenin binding sites in D-APC can substitute for human APC in β-catenin down-regulation and that the same region interacts directly with Arm.
Expression of D-APC During Stages of wingless (wg) Signaling in the Embryo.
The ability of D-APC to down-regulate β-catenin levels in a mammalian cell line raised the possibility that D-APC might regulate its Drosophila homolog Arm in vivo. The best-characterized modulation in Arm levels occurs during embryonic development, when Arm protein accumulates in a striped pattern in response to expression of the segment polarity gene wg (31). To address whether D-APC might be a component of that system, we examined D-APC expression during stages when Arm localizes in a striped pattern.
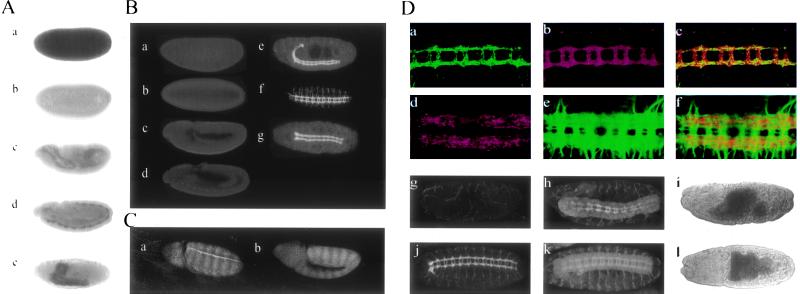
When embryos collected from heterozygous Df(3R)3450 parents are hybridized with probes for D-APC RNA, all preblastoderm stages show large amounts of RNA (Fig. 3Aa), likely derived from the wild-type allele present during oogenesis in the heterozygous mother and stored in the egg. Transcripts began to decline at the syncytial blastoderm stage, and little RNA could be detected in embryos by the end of cellularization (Fig. 3Ab). This condition persists through gastrulation and the extended germ-band stages when wg is first transcribed and Arm proteins are expressed in the striped pattern (Fig. 3Ac). Transcripts of D-APC are observed only after the completion of germ-band shortening (Fig. 3Ad), but not seen in D-APC null embryos (data not shown). This late zygotic transcription is concentrated in the CNS (Fig. 3Ae), although some RNA was detected in epidermis after stage 13.
Figure 3.
Characterization of D-APC in Drosophila embryos (A) mRNA expression of D-APC during Drosophila embryogenesis. (a–e) D-APC mRNA expression detected by in situ hybridization in Drosophila embryos. (a) One hour after fertilization D-APC mRNA is supplied from mother. (b) Cellularization stage. No significant amount of D-APC mRNA. (c) Gastrulation stage. D-APC mRNA is the same as background level. (d) Germ-band shortening stage. D-APC is expressed in developing central nervous system (CNS). (e) Stage 17. D-APC is strongly expressed in CNS as well as in the epidermis at some level. (B) Protein localization of D-APC during embryogenesis in wild-type embryos. (a–e) Lateral view of the embryos at various stages. (a) One hour after fertilization. (b) Cellularization stage. (c) Gastrulation stage. There is not significant amount and difference in D-APC level in the a–c embryos. (d) Germ-band shortening stage. D-APC protein starts localizing in the developing CNS. (e) Postgerm-band shortening stage. D-APC protein localizes in the CNS as well as in the epidermis at low levels. (f and g) Ventral views of the embryo at stage 14 and 17, respectively. D-APC localizes strongly in the CNS. D-APC level in the commissure tends to be higher in stage 14 than in stage 17. (C) The loss of zygotic D-APC expression did not significantly alter Arm striping pattern. (a and b) Arm staining of wild-type and Df(3R)3450 homozygous [APC(−/−)] embryo, respectively, at stage 10. (D) Characteristic of D-APC in CNS. (a–c) Close association of D-APC and Arm in CNS. Ventral view of the embryo double-stained for D-APC (green) (a) and Arm (red) (b). (c) Superimposition of images a and b. There are certain levels of colocalization but not in the entire axon fiber tracts. (d–f) Close association of D-APC and tubulin. Ventral view of the embryo double-stained for D-APC (red) (d) and tubulin (green) (e). (f) Superimposition of the images of d and e. (g–l) Comparison of CNS structure between Df(3R)3450 and B152 [APC(−/−)] (g–i) and wild-type (j–l) embryos. Both the wild-type and the APC(−/−) embryos (16–22 hr collection) were double-stained for D-APC protein (g and j) with anti-D-APC antibody and for CNS structure (h and k) by anti-horseradish peroxidase staining. Morphology of the embryos (i and l) were visualized by direct transmission of the laser. In APC(−/−) embryos, the fiber tracts (h) are thinner than those in wild-type embryos (k). All the images were obtained by confocal microscopy.
If D-APC protein plays a role in Wg mediated Arm regulation, the absence of detectable D-APC RNA during extended germ-band stages would require that any D-APC protein be derived from the maternal RNA, which is degraded in late cleavage embryos. To examine this possibility, polyclonal antibodies to D-APC were prepared in rabbits. The specificity of these antibodies for D-APC was confirmed by their failure to stain late stages of homozygous deficiency embryos (Fig. 3Dg). In contrast to the early maternal RNA distributions, little protein staining was observed in wild-type embryos until germ-band shortening (Fig. 3B a–d). At this point the protein distribution parallels the distribution of zygotic D-APC transcripts and is highly concentrated in the nervous system (Fig. 3B e and f). Because little D-APC protein was detected in stages prior to shortening, the maternally supplied RNAs observed during cleavage does not appear to contribute a significant amount of protein to the embryo. Failure to detect D-APC protein at stages when Wg and Arm protein localize in a striped pattern suggests that D-APC may not play a role in Arm regulation at those stages.
Arm regulation plays a direct role in the establishment of a segmental pattern of the embryonic cuticle. Failure to down-regulate Arm protein results in an uniform “naked cuticular” fate, whereas uniform low levels result in a homogenous lawn of denticles (32). To further investigate the function of D-APC, we examined the cuticle pattern and Arm protein expression in D-APC homozygous null embryos. Df(3R)3450 embryos lacking zygotic D-APC show a normal Arm stripe pattern (Fig. 3Cb). When such embryos contain the duplication DpB152, they differentiate a normal cuticle pattern, even though they lack all zygotic APC (data not shown). These observations are consistent with the view that zygotic D-APC does not play a role in Arm regulation in ventral cuticle pattern formation.
Although the possibility cannot be excluded that a low but significant maternal contribution of D-APC functions in wg signaling it seems likely that D-APC is not an essential component of that pathway during this extended germ-band stage. Before these experiments, it appeared that GSK (=Zw3) mediated its effect on Arm via an interaction with the Drosophila homolog of m-APC (28). That possibility seems less likely now, given that the cuticle phenotype of maternally null zw3 embryos can be partially rescued by zygotic zw3 expression at stages when no D-APC appears to be present (33). Recently, in Xenopus, the homolog of Zw3, GSK has been shown to directly interact with and phosphorylate β-catenin (34). This phosphorylation status correlates with the stability of β-catenin (34). These observations support the idea that Wg and APC may regulate Arm levels at distinct stages of development possibly using different mechanisms of regulation. This would explain their largely nonoverlapping expression patterns. In this view, low levels of D-APC during midembryonic stages may be essential for an unencumbered response of epidermal cells to the wg signal.
D-APC Expression Pattern in the CNS.
During late stages, D-APC is expressed in the CNS. The RNA expression is first seen slightly after the onset of germ-band shortening and its level increases as germ-band shortening proceeds (Fig. 3Ad). High levels of RNA persist at least until cuticle formation (Fig. 3Ae). No significant expression was observed in neuroblasts; instead, the expression was restricted to the postmitotic population in the CNS. This result is similar to that observed in the rat in which APC was expressed in postmitotic neurons in the CNS (35).
The D-APC protein derived from this zygotic expression localizes in axon fiber tracts and motor neurons (Fig. 3 B e–g, and D a and d). When staining is first observed, the levels are similar in both commissural and longitudinal tracts, but after completion of germ-band shortening, it is significantly more intense in the longitudinal tracts (Fig. 3 B f and g, and D a and d). Within the tracts the pattern is not regular but appears distributed in patches or streaks within specific fibers (Fig. 3D a and d). This fibrous pattern contrasts with the more uniform distributions of other CNS markers such as Arm, tubulin, and horseradish peroxidase (Fig. 3D b, e, and k). In double-immunofluorescence staining for D-APC and Arm, the two proteins show a general colocalization (Fig. 3D a–c). Highest levels of D-APC, however, are not associated with high levels of Arm, since superimposition of digitized D-APC (green) and Arm (red) images give patches of green and red coloration (Fig. 3Dc). A similar close association was observed in the fiber tracts between D-APC and tubulin staining, but again colocalization was not complete (Fig. 3D d–f).
A possible interaction between D-APC and Arm is supported by the late expression of both proteins in the CNS. If D-APC down-regulates Arm protein, one might have expected to see relatively high levels of Arm protein where the D-APC protein level is low and vice versa. The generally uniform Arm expression argues that such regulation would have to be very transient if it exists at all. Very high levels of the Arm protein were not observed in the axon fiber tracts of D-APC null embryos. The high level of D-APC RNA expression in the Drosophila CNS parallels similar observations in the rat (35). In both organisms, expression of APC appears to be restricted to postmitotic neurons. In Drosophila the pattern of APC expression in fiber tracts changes with time. APC protein can be observed clearly in the commissural fibers of stage 13 embryos, but it does not increase as the CNS develops. This contrasts with the increases in commissural staining levels observed with antibodies to Arm, tubulin, and horseradish peroxidase, and with the increases in D-APC observed in the longitudinal fiber tracts during the same period. This changing pattern of D-APC protein expression during the CNS development suggests a possible role in the patterning of the fiber tracts. To test this possibility, CNS development of Df(3R)3450 and DpB152 embryos was examined. As mentioned above, such embryos lack D-APC and an undefined number of adjacent genes but form normal cuticle. Df(3R)3450 and DpB152 embryos showed no dramatic morphological disorganization of the CNS (Fig. 3 D h and k). The only defect observed was in the longitudinal fiber tracts, which appear thinner and less developed than those of wild-type embryos at a similar stage (Fig. 3D h, i, k, and l). The specificity of the defects is consistent with the generally higher expression of D-APC in longitudinal fibers at these stages. Therefore, loss of D-APC may result in abnormalities or retardation of their development, although a role for other unidentified gene(s) normally present in the deleted region cannot be excluded.
Given the late timing of its expression, a role for APC in the CNS would be temporally distinct from any well characterized process involving wingless signaling. In Drosophila CNS development, wg functions largely in specification of neuroblasts (36), at stages much earlier than the postmitotic stages when D-APC is expressed. One other Wnt family gene (D-wnt3) is expressed in the CNS at late stages. However, it does not appear to regulate Arm at least in an in vitro tissue culture system (37) and thus, would be a poor candidate for countering a D-APC-based effect on the Arm protein. Thus, Wg and D-APC might have distinct functions in the CNS development as well.
Acknowledgments
We are grateful to E. Villareal and L. Gelbert for help during initial cDNA screening, to A. A. Salyer and A. K. Teresky for generation of human APC antibody, and to R. P. Ray for demonstrating the chromosome squash experiment. We would like to thank J. Goodhouse for training and assistance with confocal microscopy. We would also like to thank Y. Amed for critical reading of the manuscript, and Y. Hiromi, M. Lu, N. Horikoshi, and all of our colleagues in the Levine and Wieschaus laboratories for suggestions, comments, and stimulating discussions.
Footnotes
Abbreviations: APC, adenomatous polyposis coli; m-APC, mammalian APC; D-APC, Drosophila APC; Arm, Armadillo; Wg, wingless; Zw3, zeste-white 3; GSK, glycogen synthetase kinase; CNS, central nervous system.
Data deposition: The sequence reported in this paper has been deposited in the GenBank data base (accession no. U77947U77947).
References
- 1.Ichii S, Horii A, Nakatsuru S, Furuyama J, Utsunomiya J, Nakamura Y. Hum Mol Genet. 1992;1:387–390. doi: 10.1093/hmg/1.6.387. [DOI] [PubMed] [Google Scholar]
- 2.Levy D B, Smith K J, Beazer-Barclay Y, Hamilton S R, Vogelstein B, Kinzler K W. Cancer Res. 1994;54:5953–5958. [PubMed] [Google Scholar]
- 3.Su L K, Kinzler K W, Vogelstein B, Preisinger A C, Moser A R, Luongo C, Gould K A, Dove W F. Science. 1992;256:668–670. doi: 10.1126/science.1350108. [DOI] [PubMed] [Google Scholar]
- 4.Moser A R, Luongo C, Gould K A, McNeley M K, Shoemaker A R, Dove W F. Eur J Cancer. 1995;31:1061–1064. doi: 10.1016/0959-8049(95)00181-h. [DOI] [PubMed] [Google Scholar]
- 5.Baeg G H, Matsumine A, Kuroda T, Bhattacharjee R N, Miyashiro I, Toyoshima K, Akiyama T. EMBO J. 1995;14:5618–5625. doi: 10.1002/j.1460-2075.1995.tb00249.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morin P J, Vogelstein B, Kinzler K W. Proc Natl Acad Sci USA. 1996;93:7950–7954. doi: 10.1073/pnas.93.15.7950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rubinfeld B, Souza B, Albert I, Muller O, Chamberlain S H, Masiarz F R, Munemitsu S, Polakis P. Science. 1993;262:1731–1734. doi: 10.1126/science.8259518. [DOI] [PubMed] [Google Scholar]
- 8.Su L K, Vogelstein B, Kinzler K W. Science. 1993;262:1734–1737. doi: 10.1126/science.8259519. [DOI] [PubMed] [Google Scholar]
- 9.Munemitsu S, Souza B, Muller O, Albert I, Rubinfeld B, Polakis P. Cancer Res. 1994;54:3676–3681. [PubMed] [Google Scholar]
- 10.Smith K J, Levy D B, Maupin P, Pollard T D, Vogelstein B, Kinzler K W. Cancer Res. 1994;54:3672–3675. [PubMed] [Google Scholar]
- 11.Su L K, Burrell M, Hill D E, Gyuris J, Brent R, Wiltshire R, Trent J, Vogelstein B, Kinzler K W. Cancer Res. 1995;55:2972–2977. [PubMed] [Google Scholar]
- 12.Matsumine A, Ogai A, Senda T, Okumura N, Satoh K, Baeg G H, Kawahara T, Kobayashi S, Okada M, Toyoshima K, Akiyama T. Science. 1996;272:1020–1023. doi: 10.1126/science.272.5264.1020. [DOI] [PubMed] [Google Scholar]
- 13.Munemitsu S, Albert I, Souza B, Rubinfeld B, Polakis P. Proc Natl Acad Sci USA. 1995;92:3046–3050. doi: 10.1073/pnas.92.7.3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Inomata M, Ochiai A, Akimoto S, Kitano S, Hirohashi S. Cancer Res. 1996;56:2213–2217. [PubMed] [Google Scholar]
- 15.Miyoshi Y, Nagase H, Ando H, Horii A, Ichii S, Nakatsuru S, Aoki T, Miki Y, Mori T, Nakamura Y. Hum Mol Genet. 1992;1:229–233. doi: 10.1093/hmg/1.4.229. [DOI] [PubMed] [Google Scholar]
- 16.Bradley R S, Cowin P, Brown A M. J Cell Biol. 1993;123:1857–1865. doi: 10.1083/jcb.123.6.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hinck L, Nelson W J, Papkoff J. J Cell Biol. 1994;124:729–741. doi: 10.1083/jcb.124.5.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Papkoff J, Rubinfeld B, Schryver B, Polakis P. Mol Cell Biol. 1996;16:2128–2134. doi: 10.1128/mcb.16.5.2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harlow E & D L. Antibodies: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1988. [Google Scholar]
- 20.Riggleman B, Wieschaus E, Schedl P. Genes Dev. 1989;3:96–113. doi: 10.1101/gad.3.1.96. [DOI] [PubMed] [Google Scholar]
- 21.Horikoshi N, Usheva A, Chen J, Levine A J, Weinmann R, Shenk T. Mol Cell Biol. 1995;15:227–234. doi: 10.1128/mcb.15.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ashburner M. Drosophila: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 23.Tautz D, Pfeifle C. Chromosoma. 1989;98:81–85. doi: 10.1007/BF00291041. [DOI] [PubMed] [Google Scholar]
- 24.Muller H A, Wieschaus E. J Cell Biol. 1996;134:149–163. doi: 10.1083/jcb.134.1.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peifer M, Orsulic S, Sweeton D, Wieschaus E. Development (Cambridge, UK) 1993;118:1191–1207. doi: 10.1242/dev.118.4.1191. [DOI] [PubMed] [Google Scholar]
- 26.Bilder D, Scott M P. Genetics. 1995;141:1087–1100. doi: 10.1093/genetics/141.3.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kongsuwan K, Dellavalle R P, Merriam J R. Genetics. 1986;112:539–550. doi: 10.1093/genetics/112.3.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rubinfeld B, Albert I, Porfiri E, Fiol C, Munemitsu S, Polakis P. Science. 1996;272:1023–1026. doi: 10.1126/science.272.5264.1023. [DOI] [PubMed] [Google Scholar]
- 29.Groden J, Thliveris A, Samowitz W, Carlson M, Gelbert L, et al. Cell. 1991;66:589–600. doi: 10.1016/0092-8674(81)90021-0. [DOI] [PubMed] [Google Scholar]
- 30.Thliveris A, Samowitz W, Matsunami N, Groden J, White R. Cancer Res. 1994;54:2991–2995. [PubMed] [Google Scholar]
- 31.Riggleman B, Schedl P, Wieschaus E. Cell. 1990;63:549–560. doi: 10.1016/0092-8674(90)90451-j. [DOI] [PubMed] [Google Scholar]
- 32.Klingensmith J, Nusse R. Dev Biol. 1994;166:396–414. doi: 10.1006/dbio.1994.1325. [DOI] [PubMed] [Google Scholar]
- 33.Siegfried E, Chou T B, Perrimon N. Cell. 1992;71:1167–1179. doi: 10.1016/s0092-8674(05)80065-0. [DOI] [PubMed] [Google Scholar]
- 34.Yost C, Torres M, Miller J R, Huang E, Kimelman D, Moon R T. Genes Dev. 1996;10:1443–1454. doi: 10.1101/gad.10.12.1443. [DOI] [PubMed] [Google Scholar]
- 35.Bhat R V, Baraban J M, Johnson R C, Eipper B A, Mains R E. J Neurosci. 1994;14:3059–3071. doi: 10.1523/JNEUROSCI.14-05-03059.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chu-LaGraff Q, Doe C Q. Science. 1993;261:1594–1597. doi: 10.1126/science.8372355. [DOI] [PubMed] [Google Scholar]
- 37.Fradkin L G, Noordermeer J N, Nusse R. Dev Biol. 1995;168:202–213. doi: 10.1006/dbio.1995.1072. [DOI] [PubMed] [Google Scholar]