Abstract
The syndrome of resistance to thyroid hormone is characterized by elevated serum free thyroid hormones, failure to suppress pituitary thyrotropin secretion, and variable peripheral refractoriness to hormone action. Here we describe a novel leucine to valine mutation in codon 454 (L454V) of the thyroid hormone β receptor (TRβ) in this disorder, resulting in a mutant receptor with unusual functional properties. Although the mutant protein binds ligand comparably to wild-type receptor and forms homo- and heterodimers on direct repeat, everted repeat, or palindromic thyroid response elements, its ability to activate transcription via these elements is markedly impaired. The hydrophobic leucine residue lies within an amphipathic α-helix at the carboxyl terminus of TRβ and the position of the homologous residue in the crystal structure of TRα indicates that its side chain is solvent-exposed and might interact with other proteins. We find that two putative transcriptional mediators (RIP140 and SRC-1) exhibit hormone-dependent association with wild-type TR. In comparison, the interaction of this natural mutant (L454V) and artificial mutants (L454A, E457A) with RIP140 and SRC-1 is markedly reduced. Furthermore, coexpression of SRC-1 is able to restore the transcriptional activity of the L454V mutant receptor, indicating that the interaction of this residue with accessory proteins is critical for transcriptional activation. Finally, the occurrence of the L454V mutation in resistance to thyroid hormone, together with impaired negative regulation of the thyroid-stimulating hormone α promoter by this mutant, suggests that the amphipathic α-helix also mediates hormone-dependent transcriptional inhibition, perhaps via interaction with these or other accessory factors.
Keywords: resistance to thyroid hormone, hormone-dependent transactivation, coactivator
Resistance to thyroid hormone (RTH) is characterized by elevated serum-free thyroid hormones, failure to suppress pituitary thyroid-stimulating hormone (TSH) secretion, and variable refractoriness to hormone action in peripheral tissues. Following the observation that familial RTH was tightly linked to the thyroid hormone receptor β (TRβ) gene locus (1), a growing number of β-receptor mutations have been identified in kindreds with this disorder (2, 3). In keeping with other members of the nuclear receptor superfamily, the TR is organized into distinct functional domains (4). The carboxyl-terminal (D/E) region of TR mediates hormone binding and nuclear localization functions and also contains sequences that mediate homodimeric TR interactions and the formation of heterodimers with the retinoid X receptor (RXR) (5). All the RTH mutations described to date localize to the D/E domains and the majority cluster within two hot spots (6, 7). Their properties have provided valuable insights into structure–function relationships in the receptor. Most mutant receptors exhibit reduced hormone binding (6–8), whereas their ability to localize to the nucleus is not impaired (8). With a few exceptions (9–11), mutant receptors retain the ability to form homodimers and they all heterodimerize with the RXR (9, 12). These properties correlate with the location of mutation clusters outside regions that are involved in heterodimerization (5) and which are less important for hormone binding (13).
The D/E domains of TR also confer the ability to regulate target gene transcription in a hormone-dependent manner. A nine amino acid sequence at the receptor carboxyl terminus has been shown to be essential for this ligand-inducible transactivation function (AF-2) and is deleted in v-erbA, the oncogenic counterpart of TR (14). This region is highly conserved among nuclear receptors and mutational analyses in a number of them have demonstrated its importance in transactivation (15–20). Here we describe a natural mutation substituting valine for leucine at codon 454 within this sequence motif in TRβ. Although it retains DNA binding, hormone binding, and dimerization functions, this mutant receptor transactivates target genes poorly and is a powerful dominant negative inhibitor of wild-type (WT) receptor action. Comparison with the crystal structure of the ligand-binding domain of TRα (21) indicates that this leucine residue is solvent-exposed with the potential to interact with other proteins. To test this hypothesis, we examined the interaction of WT and mutant TRs with two putative transcriptional mediators, RIP140 and SRC-1, which have recently been isolated (22–24). RIP140 is a 140-kDa protein that exhibits hormone-dependent binding to the estrogen receptor (ER) and was identified by Far Western blot analysis. SRC-1 is a 125-kDa protein that was identified by its binding to the progesterone receptor in the presence of ligand in a yeast two-hybrid assay system.
MATERIALS AND METHODS
Clinical and Genetic Studies.
The proband (AJ) presented at the age of 20 with palpitations and was noted to have a goiter and raised serum thyroid hormones. Following initial treatment with carbimazole she underwent thyroid surgery and was free of symptoms for 10 years. The goiter and tachycardia recurred, necessitating radioiodine treatment, following which she developed hypothyroidism requiring thyroxine replacement. Despite 200 μg thyroxine daily, the TSH response to TRH was exaggerated, rising from a basal value of 35 mu/liter to a peak of 275 mu/liter 30 min after TRH. The TSH response was reduced (95 mu/liter), but preserved following T3 administration (120 μg/4 days), suggesting a diagnosis of resistance to thyroid hormone. The daughter of the proband (CJ) had low weight, tachycardia, tremor, and a small goiter at birth. A diagnosis of neonatal thyrotoxicosis was made initially and carbimazole treatment instituted. Symptoms improved during infancy and thyroxine therapy to control her goiter was discontinued at puberty. Current symptoms include palpitations, muscle weakness, and sensitivity to bright light. The sister (ES) of the index case showed abnormal thyroid function on biochemical screening and was asymptomatic for many years, but recently developed a persistent tachycardia. Serum-free T4 and serum-free T3 levels were measured with fluoroimmunometric assays using Delfia technology (Wallac, Milton Keynes, U.K). TSH measurements were performed using a sensitive “second generation” assay (Wallac). Current thyroid function tests in the affected individuals are as follows: proband (AJ) free T4 75 pmol/liter (normal 9–20), free T3 16 pmol/liter (normal 3–7.5), and TSH 0.4 mu/liter (normal 0.4–4); daughter (CJ) free T4 49 pmol/liter, free T3 14 pmol/liter, and TSH 1.3 mu/liter; sister (ES) free T4 54 pmol/liter, free T3 18 pmol/liter, and TSH 1.7 mu/liter.
Leukocyte DNA from each affected individual was extracted using standard techniques. Coding exons of the human (h)TRβ gene were amplified using PCR and then sequenced directly as described (7). The presence of a receptor mutation was verified by at least two independent PCRs and sequencing reactions in each individual. The nomenclature used to describe the receptor mutation conforms to a consensus statement from the First Workshop on Thyroid Hormone Resistance (25).
Hormone- and DNA-Binding Assays.
Mutations were introduced into the receptor by site-directed mutagenesis of the hTRβ1 cDNA in M13mp18 as described (9) and verified by sequencing. WT and mutant receptor proteins were synthesized by coupled in vitro transcription and translation with rabbit reticulocyte lysate (TNT, Promega) and the T3-binding affinity of each was determined using a filter-binding assay (7).
Receptor binding to DNA was determined by electrophoretic mobility-shift assay using in vitro translated receptors and oligonucleotide duplexes corresponding to an optimized palindromic thyroid response element (TRE), a direct thyroid response element TRE from the malic enzyme gene promoter, and an everted repeat TRE from the chicken lysozyme gene (9). The ability of WT and mutant TR-RXR heterodimers to bind T3 was tested using a reverse gel mobility-shift assay. The procedure was identical to the conventional assay except that the in vitro translated receptor was labeled with [35S]methionine and incubated with a non-labeled oligonucleotide duplex in the presence or absence of 1 nM 125I-labeled T3. Receptor-associated 125I-T3 was selectively detected by autoradiography in the presence of a 35S filter.
Transfection Assays.
WT and mutant receptor function was assayed by cotransfection with reporter genes into JEG-3 human choriocarcinoma cells as described (9). WT and mutant hTRβ cDNAs were expressed using a vector containing the Rous sarcoma virus (RSV) enhancer and promoter. RSVCAT, driving the expression of chloramphenicol acetyltransferase, was used as control expression vector. PALTKLUC, MALTKLUC, and F2TKLUC contain two copies of a palindromic TRE or a single copy of a direct repeat or everted repeat TRE, respectively, upstream of the thymidine kinase promoter and luciferase cDNA (9). TSHαLUC contains the 5′-flanking region of the human TSHα-subunit gene from −846 to +44 bp, coupled to luciferase (9). The internal control plasmid Bos–β-galactosidase (β-gal), containing the human elongation factor 1α gene promoter driving β-gal, was used to correct for variations in transfection efficiency. SRC-1 was amplified from a human B-cell cDNA library with primers based on the previously reported SRC-1 sequence (24) and cloned into a eukaryotic expression vector (pSG5).
Protein–Protein Interaction Assays.
Fusion proteins containing the ligand-binding domain of WT of mutant receptors linked to glutathione S-transferase were expressed in Escherichia coli and purified as described (18). The vector containing RIP140 has been described elsewhere (23). The SRC-1 cDNA described above was cloned into pBluescript SK+. [35S]Methionine-labeled proteins were synthesized by coupled in vitro transcription and translation of each cDNA (TNT system, Promega). Aliquots of immobilized fusion protein were incubated with either in vitro translated 35S-labeled RIP140 or SRC-1 proteins in PD buffer (50 mM Tris·HCl/0.1 M KCl/0.14 M NaCl/0.5% Nonidet P-40/10% glycerol, pH 8.0) in the presence or absence of 5 μM ligand for one hr at room temperature, then washed four times with NETN buffer (20 mM Tris·HCl/0.1 M NaCl/1 mM EDTA/0.5% Nonidet P-40, pH 8.0) and analyzed by SDS/PAGE (18). Gels were Coomassie stained to check for equal loading of fusion protein, then exposed to autoradiography.
RESULTS
Genetic Analyses.
Direct sequencing of exon 10 of the TRβ gene in the proband (AJ) indicated that she was heterozygous for a single nucleotide substitution (1645 TTG to GTG), which corresponded to a leucine to valine mutation at codon 454 (L454V) in the predicted amino acid sequence. Exons 4–9, encoding all but the first eight amino acids of the β receptor, were also sequenced in this individual and no other abnormalities were detected. Subsequent analyses showed that her daughter (CJ) and sister (ES) were also heterozygous for this mutation, demonstrating concordance between the presence of a receptor defect and biochemical abnormalities that are typical of RTH.
Hormone and DNA Binding.
To determine its functional consequences, the L454V mutation was introduced into the WT TRβ1 cDNA and mutant protein generated by coupled transcription and translation in vitro. Hormone-binding assays showed comparable binding of 125I-T3 to WT and mutant receptor proteins and Scatchard analysis indicated that their ligand-binding affinities were similar [Mean and SEM of at least three determinations: WT, Ka = 1.8 × 1010 M−1 (±0.3); L454V, Ka = 1.2 × 1010 M−1 (±0.1)].
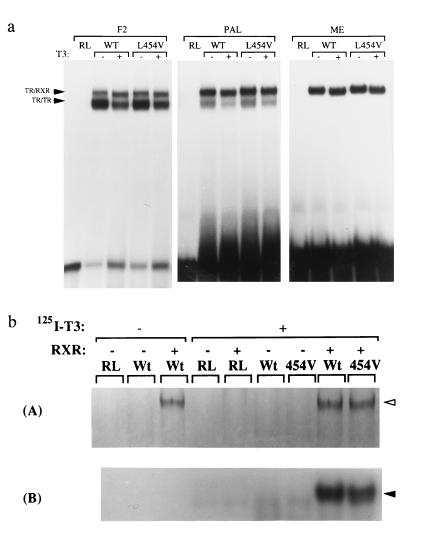
Previous studies (26, 27) indicate that TR can bind to DNA either as a homodimer or as a heterodimer with RXR on different TRE configurations. Accordingly, the DNA-binding properties of WT and mutant receptors were studied using everted repeat (F2), palindromic (PAL), and direct repeat (ME) TREs in the presence of hRXRα (Fig. 1a). Under these conditions the L454V mutant formed homodimer and heterodimer complexes on F2 and PAL and heterodimers on the direct repeat (MAL) comparably with WT. The WT and mutant homodimer complexes were equally attenuated following ligand binding (28), whereas the TR–RXR complexes were quantitatively unaffected but showed a small change in mobility. Recent studies have shown that in some contexts, heterodimerization can induce allosteric changes, which preclude the binding of ligand to either receptor partner (29, 30). To confirm that the L454V receptor mutant was still able to bind ligand while complexed with RXR, reverse electrophoretic mobility-shift assays were performed using MAL TRE. [35S]Methionine-labeled TR and RXR were incubated with unlabeled TRE in the presence or absence of 125I-T3 and analyzed by PAGE (Fig. 1b). Fig. 1b (Upper) displays the formation of [35S]methionine-labeled TR–RXR complexes (lanes 3, 8, and 9). Exposure of the same gel to detect 125I-T3 (Fig. 1b Lower) showed that both WT and mutant heterodimer complexes were labeled with radioligand (lanes 8 and 9).
Figure 1.
(a) DNA binding and dimerization of WT and mutant (L454V) receptors. Electrophoretic mobility-shift assays show the formation of TRβ homodimers and TR/RXRα heterodimers (arrowheads) in the presence (+) or absence (−) of 10 nM T3 on everted repeat (F2), palindromic (PAL), and direct repeat (ME) configurations of TRE. (b) Both WT and mutant (454V) receptors retain the ability to bind T3 when bound to DNA as a heterodimer with RXRα. 35S-labeled receptors were incubated with unlabeled malic enzyme TRE oligonucleotide duplexes in the presence or absence of 125I-labeled T3. (A) Formation of heterodimers with RXRα by 35S-labeled TRβ. (B) A 125I-selective exposure of the same gel showing retention of ligand binding by both WT and L454V TR-RXR heterodimers. RL denotes reticulocyte lysate control.
Functional Activity and Dominant Negative Inhibition.
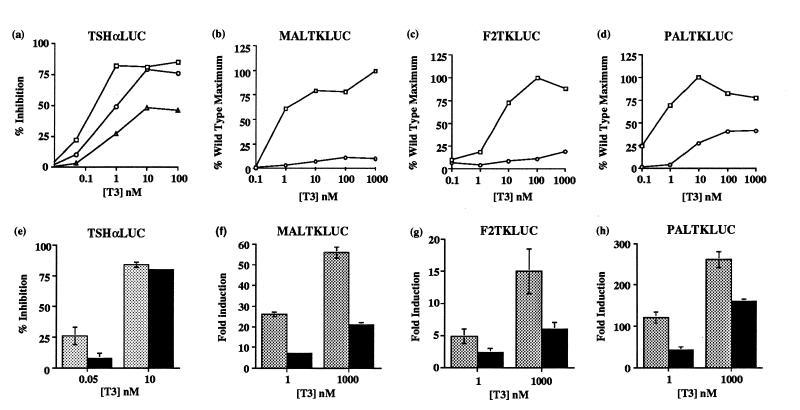
The function of WT and L454V mutant receptors was studied in transient transfection assays using a reporter gene (TSHαLUC) containing the negatively regulated human TSHα promoter (Fig. 2a). Inhibition of TSHαLUC by mutant receptor was impaired, exhibiting a right-shifted profile compared with WT, with maximal inhibition attained at higher T3 concentrations. This is particularly evident when the significant background inhibition of TSHαLUC activity in RSVCAT-transfected cells, which may be mediated by endogenous TR, is taken into account.
Figure 2.
(a–d) Modulation of reporter gene expression by WT (□) or L454V mutant receptors (○). Six-well plates of JEG-3 cells were transfected with 2 μg (TSHαLUC, MALTKLUC, PALTKLUC) or 4 μg (F2TKLUC) of reporter gene, 100 ng of receptor expression vector, and 200 ng of Bos–β-gal. Following incubation with T3, luciferase activity was normalized for β-gal activity. Hormone-dependent inhibition of TSHαLUC is expressed relative to values in non-T3 treated cells. Inhibition of TSHαLUC in control cells transfected with 100 ng RSVCAT is also shown (Δ). Activation of MAL/PAL/F2 reporters by L454V is expressed as a percentage of the maximal induction by WT receptor. The results shown are the mean of at least three independent transfections in triplicate and the SEM was less than 5% (TSHαLUC) or 10% (MAL/F2/PAL). (e–g) Dominant negative activity of L454V mutant receptor. JEG-3 cells were transfected with reporter and reference plasmids as in a–d and 100 ng of WT receptor expression vector together with an equal amount of RSVCAT (shaded) or L454V mutant (solid) expression vectors. Inhibition or induction of reporter activity was calculated following incubation with low or high T3 concentrations as indicated. The results are the mean of at least three transfections in triplicate.
Similar studies were performed using the positively regulated direct repeat (MALTKLUC), everted repeat (F2TKLUC), and palindromic (PALTKLUC) TRE-containing reporter genes. In comparison with significant hormone-dependent activation seen with WT on MAL and F2 (Fig. 2 b and c), the L454V mutant was unable to induce significant reporter gene activity, even with saturating concentrations of T3. With a palindromic TRE (Fig. 2d) the mutant receptor retained some hormone-dependent activation potential, but this was markedly reduced (≈30%) compared with its WT counterpart.
The dominant negative potential of the L454V receptor mutant was tested. Either WT receptor alone or WT plus an equal amount of L454V mutant was cotransfected with reporter genes (TSHα, MAL, F2, PAL) at low (0.05 nM TSHα; 1nM MAL/F2/PAL) or high (10 nM TSHα; 1000 nM MAL/F2/PAL) T3 concentrations, and luciferase activity was measured. In the presence of mutant receptor, induction of all three reporter genes by WT receptor was markedly inhibited (MAL, 31%; F2, 50%; PAL, 32% versus 100% with WT alone) at low T3 concentrations (Fig. 2 f–h). Even at higher T3 concentrations, the L454V mutant continued to exert a significant inhibitory effect (MAL, 40%; F2, 34%; PAL, 68%). The mutant receptor also exerted a dominant negative effect with the negatively regulated TSHα promoter at low T3 concentrations, which was reversed at higher levels (Fig. 2e).
Location of Homologous Leucine in Structure of the rTRα Ligand-Binding Domain and Protein–Protein Interaction Assays.
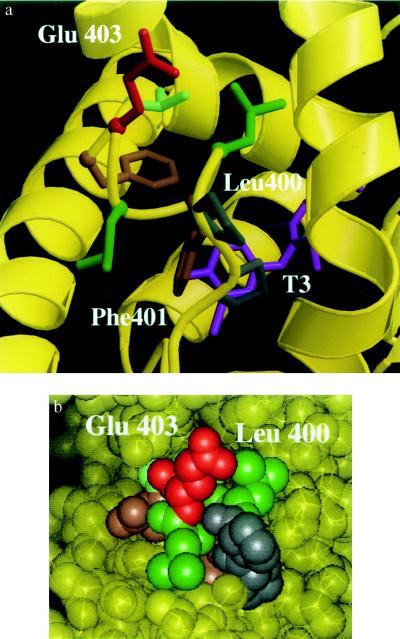
The crystal structure of the ligand-binding domain of TRα has recently been elucidated (21), enabling us to examine the position of the residue homologous to Leu-454 of TRβ, Leu-400. As shown in Fig. 3a, this hydrophobic residue is not directly involved in interaction with ligand, in keeping with its preserved hormone-binding affinity. Furthermore, the residue is solvent accessible (Fig. 3b), suggesting that it could be involved in protein–protein interactions.
Figure 3.
(a) Ribbon drawing of the amphipathic α-helix (399-PLFLEVF-405) from the crystal structure of rat TRα ligand-binding domain, showing the interactions of residues that are important for transactivation. A charged residue, Glu-403 (red), projects toward solvent, whereas a hydrophobic residue, Phe-401 (brown), contacts the ligand T3 (magenta). The residue homologous to Leu-454 in hTRβ, Leu-400 (green), is also solvent accessible. The gray residues (Pro-398, Pro-399) correspond to Pro-452 and Pro-453 in hTRβ. (b) Space-filling representation of the rat TRα ligand-binding domain emphasizing the solvent accessibility of Leu-400 and Glu-403. Phe-401 is not visible, since it is buried within the ligand-binding cavity. The view and color scheme are identical to that of Fig. 3a.
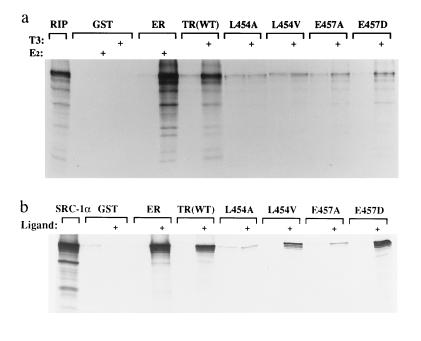
To test this proposal, we examined the interaction of WT and mutant TRs with RIP140, a protein known to interact with the ER. In addition to the natural L454V mutant, we tested artificial mutations of this (L454A) and an adjacent charged residue (E457A, E457D). We have shown previously that these mutations also have little effect on ligand binding (18). However, the L454A mutant exhibits negligible ligand-dependent transactivation and the transcriptional activity of the E457A mutant is also markedly attenuated. The E457D mutant does retain modest transcription activation function (18). In a protein–protein interaction assay the ligand-binding domain of WT TRβ exhibited strong T3-dependent interaction with 35S-labeled RIP140, comparable to hormone-dependent interaction of this protein with ER (Fig. 4a). In comparison, the ligand-dependent binding of the transactivation mutants (L454A, L454V, E457A) to RIP140 was abolished or markedly attenuated, whereas the E457D mutant retained weak association with this protein. The relative interaction of different mutants with RIP140 was too low to be reliably quantitated.
Figure 4.
Interaction of ER, WT TRβ, and transactivation mutants with in vitro synthesized RIP140 and SRC-1. 35S-labeled in vitro translated RIP140 (a) and SRC-1 (b) were incubated with equal amounts of glutathione S-transferase (GST), GST-ER, or GST-TR fusion proteins in the presence (+) or absence (−) of 1 μM estradiol or T3, as appropriate.
In a further study, the association of these mutant receptors with the putative transcriptional coactivator SRC-1 was also examined (Fig. 4b). As with RIP140, the L454A and E457A mutations severely reduced ligand-dependent interaction of the receptor with SRC-1 (2 ± 1% and 6 ± 2%, respectively, relative to WT), in accordance with the minimal transcriptional activity of these mutants. The L454V mutant, which is less transcriptionally impaired than L454A and E457A, retained moderate interaction with SRC-1 (39 ± 4%). In contrast to its binding to RIP140, interaction of the E457D mutant with SRC-1 was comparable to that of WT receptor (96 ± 13%).
Effect of Coexpressed SRC-1 on Activity of WT and L454V Mutant Receptors.
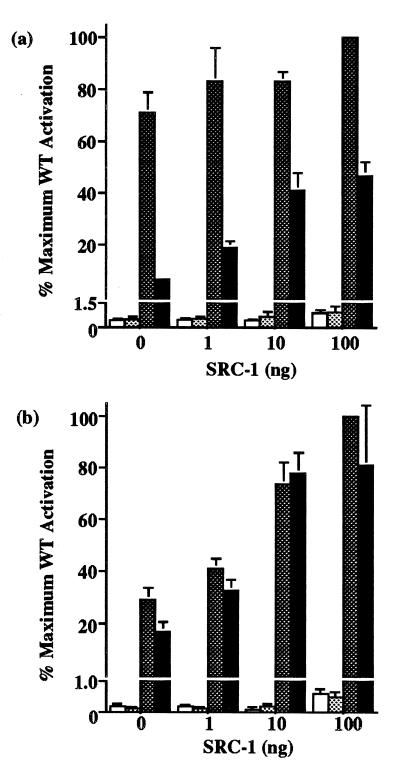
As the L454V mutant receptor exhibits a significantly reduced affinity for SRC-1 in vitro, we hypothesized that enhancing the level of SRC-1 expression might alleviate its loss of transactivation function. WT or L454V mutant receptors were tested with MAL or PAL reporter genes together with increasing amounts of cotransfected SRC-1 expression vector, in the absence and presence of a maximal T3 concentration. With MALTKLUC, hormone-dependent activation by WT receptor was not significantly enhanced by SRC-1 (Fig. 5a). In contrast, the transcriptional activity of the L454V mutant was markedly augmented, increasing from 7% to 47% of WT levels with incremental amounts of SRC-1. The addition of SRC-1 had little effect on basal reporter activity with either WT or mutant receptor. With the PALTKLUC reporter, cotransfection of SRC-1 increased hormone-dependent activation by the WT receptor. However, activation by the L454V mutant receptor was also progressively augmented by SRC-1, restoring its function to levels attained by WT receptor (Fig. 5b). Again, coexpressed SRC-1 had negligible effects on basal promoter activity, except at the highest level of added SRC-1.
Figure 5.
Effect of coexpressed SRC-1 on the transcriptional activity of WT and L454V mutant receptors. (a) 24-well plates of JEG-3 cells were transfected with 500 ng of MALTKLUC, 50 ng of WT or mutant vectors, 100 ng Bos–β-gal and varying amounts (0–100 ng) of SRC-1 expression vector, including empty vector where necessary to keep total vector DNA constant. Following incubation in the absence or presence of 100 nM T3, reporter activity was calculated and expressed as a percentage relative to the maximum corrected luciferase activity of WT receptor with 100 ng of SRC-1. Basal (open) versus stimulated (dark shaded) levels for WT and basal (light shaded) versus stimulated (solid) for L454V mutant receptor are shown. (b) JEG-3 cells were transfected with 500 ng of PALTKLUC reporter together with other plasmids as in a. Reporter activity with or without 100 nM T3 was calculated as above. Note that the scale of the vertical axes have been interrupted to show both basal and stimulated reporter activity.
DISCUSSION
The three affected individuals in this kindred showed biochemical abnormalities which are characteristic of RTH, with markedly elevated serum FT4 and FT3 levels and a failure to suppress TSH secretion. The proband’s sister (ES) had been virtually asymptomatic and was therefore classified as a case of generalized resistance. In contrast, the proband (AJ) and her daughter (CJ) had experienced thyrotoxic symptoms, suggesting predominant pituitary resistance. The coexistence of generalized resistance and pituitary resistance phenotypes in a single family with the same receptor mutation highlights the variability of clinical features in RTH and supports previous observations that indicate that generalized resistance and pituitary resistance are differing manifestations of a single genetic disorder (7, 31).
When the transcriptional and hormone-binding properties of natural mutant receptors associated with RTH are considered, three types of mutants have been described (32): type I mutants exhibit reduced transactivation in keeping with their altered ligand binding, but higher T3 concentrations elicit full activation; type II mutants have disproportionately reduced transactivation relative to their altered ligand binding at both low and high T3 concentrations; type III mutants neither bind ligand nor transactivate. The activation profiles of the L454V mutant (Fig. 2) are most consistent with type II properties. A possible cause of impaired transactivation is an alteration in receptor DNA binding or dimerization functions. For example, another natural transactivation mutant (R429Q) exhibits impaired homodimer formation on some TREs (9, 11). However, normal homo- and heterodimer formation by the L454V mutant (Fig. 1a) does not favor this hypothesis. These assays also demonstrate that L454V mutant homodimer complexes are attenuated in the presence of T3 (Fig. 1a) and that heterodimers retain 125I-T3 binding (Fig. 1b). The latter suggests that allosteric changes in the mutant receptor following DNA binding, which might preclude ligand occupancy (29, 30) are also an unlikely explanation for its altered transactivation.
Significantly, we and others (9, 33) have identified a number of other natural RTH mutations (P453A, P453H, P453S, P453T) with impaired transactivation, involving the proline residue that immediately precedes the leucine at codon 454, as well as seven additional residues at the TRβ carboxyl terminus. This sequence motif can be delineated in a number of other nuclear receptors (15), with striking conservation of hydrophobic and negatively charged residues. Studies have shown that mutation of a negatively charged glutamic acid residue in the ER (15), retinoic acid receptor (RARα) (19), or TR (16–18, 34) impairs transactivation with little effect on ligand binding. A double mutation (L543A/L544A) of hydrophobic residues in mouse ER (mER) (15) also disrupts transactivation and the L454V mutation we have described is homologous to the proximal leucine residue in mER. Together, these findings underscore the importance of this proximal hydrophobic residue, which is conserved in a number of nuclear receptors (15), in transactivation.
Crystal structures of the ligand-binding domains of hRARγ and rTRα bound to ligand have recently been elucidated (21, 35) and confirm that this C-terminal sequence does indeed form an amphipathic α-helix. In the rTRα ligand-binding domain, some hydrophobic residues in the carboxyl-terminal α-helix (e.g., Phe-401; Fig. 3a) are oriented toward the hormone-binding cavity and participate in ligand binding, in keeping with the detrimental effects of mutations at these positions on ligand-binding affinity (16, 18). In contrast, the conserved negatively charged residue (Glu-403) is solvent exposed and is not involved in hormone binding. A similar presentation of Leu-400 (the residue homologous to Leu-454 in TRβ and Leu-543 in mER) on the surface of the receptor raised the possibility that both of these residues might be involved in receptor interaction with heterologous proteins.
A number of proteins that exhibit hormone-dependent interaction with the carboxyl-terminal domains of nuclear receptors have been described. ERAP160 (36) and RIP140 (22, 23) were identified using Far Western blot assays with ER. TIF1 (37), Trip1/Sug1 (38), and SRC-1 (24) were isolated following interaction in a yeast two-hybrid assay system with RAR/RXR, TR, or progesterone receptors, respectively. Most recently, the coactivator CBP has also been shown to interact with nuclear receptors (39). Although the precise role of these proteins remains to be elucidated, their recruitment by liganded receptors probably reflects a realignment of the carboxyl-terminal activation helix in the presence of hormone.
Our data indicate that mutation of either leucine 454 or glutamic acid 457 in the carboxyl-terminal α-helix of TRβ impairs receptor interaction with RIP140 and SRC-1. Furthermore, the most functionally deleterious mutations (L454A, E457A) at these positions are associated with the greatest reduction in RIP140 and SRC-1 binding. More conservative substitutions (L454V, E457D) result in retention of transcriptional activity in some contexts [L454V on a TREpal containing reporter, Fig. 2d; a GAL4–E457D fusion on a UASg containing reporter (18)], correlating with retention of some binding to RIP140 or SRC-1. Furthermore, we have shown that coexpressed SRC-1 not only enhances WT receptor activity (Fig. 5b), but also augments the function of the L454V mutant, restoring it to WT levels in some TRE contexts. The differing responses to cotransfected SRC-1 exhibited by WT receptor on MAL versus PAL TREs suggests that the transcriptional requirement for this factor may be modulated by TRE configuration. Similarly, the higher transcriptional activity of L454V relative to WT on PAL versus MAL also suggests that the nature of the TRE may influence the ability of the mutant receptor to recruit SRC-1.
Overall, these findings support the notion that hormone-dependent transcriptional activation by TRβ involves receptor interactions with intermediary proteins via critical residues in the C-terminal amphipathic α-helix. Whether other residues from neighboring helices are also involved in such receptor–protein interactions, to constitute an activation surface [as has been suggested for RARγ (35)], remains to be elucidated. Lastly, the association of a transactivation mutation (L454V) with RTH may also have important functional implications for hormone-dependent transcriptional inhibition by TR. Individuals harboring this mutation all exhibit raised serum thyroid hormones with a failure to suppress pituitary TSH secretion, and hormone-dependent negative regulation of a pituitary target gene (the human TSHα promoter) by the L454V mutant receptor is impaired. This suggests that residues in the amphipathic α-helix are also important for hormone-dependent transcriptional inhibition by TR, perhaps via interaction with similar accessory factors.
Finally, the properties of the L454V receptor mutant highlight the utility of analyzing naturally occurring TRβ mutations in RTH. Although the majority of natural receptor mutants are dysfunctional as a consequence of altered ligand binding, some are transcriptionally impaired despite normal hormone binding. Providing the mutant receptor also retains dominant negative activity, as in this case, an RTH phenotype should result. Thus, it is likely that other receptor mutants with unexpected properties will be identified in RTH and their analysis may identify further domains involved in transcriptional regulation and protein interaction.
Acknowledgments
This work was supported by the Wellcome Trust and the Medical Research Council (United Kingdom). V.K.K.C. is a Wellcome Senior Clinical Research Fellow. E.K. was funded by a grant from The Netherlands Organization for Scientific Research (N.W.O.).
Footnotes
Abbreviations: RTH, resistance to thyroid hormone; TSH, thyroid-stimulating hormone; TR, thyroid hormone receptor; TRβ, thyroid hormone receptor β; RXR, retinoid X receptor; WT, wild type; β-gal, β-galactosidase; ER, estrogen receptor; mER, mouse estrogen receptor; RAR, retinoic acid receptor; TRE, thyroid response element.
References
- 1.Usala S J, Bale A E, Gesundheit N, Weinberger C, Lash R W, Wondisford F E, McBride O W, Weintraub B D. Mol Endocrinol. 1988;2:1217–1220. doi: 10.1210/mend-2-12-1217. [DOI] [PubMed] [Google Scholar]
- 2.Refetoff S, Weiss R E, Usala S J. Endocr Rev. 1993;14:348–399. doi: 10.1210/edrv-14-3-348. [DOI] [PubMed] [Google Scholar]
- 3.Refetoff S, Weiss R E, Usala S J, Hayashi Y. Endocr Rev Monogr. 1994;3:336–343. [Google Scholar]
- 4.Evans R M. Science. 1988;240:889–895. doi: 10.1126/science.3283939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Forman B M, Yang C R, Au M, Casanova J, Ghysdael J, Samuels H H. Mol Endocrinol. 1989;3:1610–1626. doi: 10.1210/mend-3-10-1610. [DOI] [PubMed] [Google Scholar]
- 6.Parrilla R, Mixson A J, McPherson J A, McClaskey J H, Weintraub B D. J Clin Invest. 1991;88:2123–2130. doi: 10.1172/JCI115542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Adams M, Matthews C, Collingwood T N, Tone Y, Beck-Peccoz P, Chatterjee V K K. J Clin Invest. 1994;94:506–515. doi: 10.1172/JCI117362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meier C A, Dickstein B M, Ashizawa K, McClaskey J H, Muchmore P, Ransom S C, Menke J B, Hao E H, Usala S J, Bercu B B, Cheng S-Y, Weintraub B D. Mol Endocrinol. 1992;6:248–258. doi: 10.1210/mend.6.2.1569968. [DOI] [PubMed] [Google Scholar]
- 9.Collingwood T N, Adams M, Tone Y, Chatterjee V K K. Mol Endocrinol. 1994;8:1262–1277. doi: 10.1210/mend.8.9.7838159. [DOI] [PubMed] [Google Scholar]
- 10.Hao E, Menke J B, Smith A M, Jones C, Geffner M E, Hershman J M, Wuerth J-P, Samuels H H, Ways D K, Usala S J. Mol Endocrinol. 1994;8:841–851. doi: 10.1210/mend.8.7.7984146. [DOI] [PubMed] [Google Scholar]
- 11.Flynn T R, Hollenberg A N, Cohen O, Menke J B, Usala S J, Tollin S, Hegarty M K, Wondisford F E. J Biol Chem. 1994;269:32713–32716. [PubMed] [Google Scholar]
- 12.Nagaya T, Jameson J L. J Biol Chem. 1993;268:15766–15771. [PubMed] [Google Scholar]
- 13.Hayashi Y, Sunthornthepvarakul T, Refetoff S. J Clin Invest. 1994;94:607–615. doi: 10.1172/JCI117376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zenke M, Munoz A, Sap J, Vennstrom B, Beug H. Cell. 1990;61:1035–1049. doi: 10.1016/0092-8674(90)90068-p. [DOI] [PubMed] [Google Scholar]
- 15.Danielian P S, White R, Lees J A, Parker M G. EMBO J. 1992;11:1025–1033. doi: 10.1002/j.1460-2075.1992.tb05141.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barettino D, Vivanco-Ruiz M M, Stunnenberg H. EMBO J. 1994;13:3039–3049. doi: 10.1002/j.1460-2075.1994.tb06603.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baniahmad A, Leng X, Burris T P, Tsai S Y, Tsai M-J, O’Malley B W. Mol Cell Biol. 1995;15:76–86. doi: 10.1128/mcb.15.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tone Y, Collingwood T N, Adams M, Chatterjee V K K. J Biol Chem. 1994;269:31157–31161. [PubMed] [Google Scholar]
- 19.Durand B, Saunders M, Gausdon C, Roy B, Losson R, Chambon P. EMBO J. 1994;13:5370–5382. doi: 10.1002/j.1460-2075.1994.tb06872.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leng X, Blanco J, Tsai S Y, Ozato K, O’Malley B W, Tsai M-J. Mol Cell Biol. 1995;15:255–263. doi: 10.1128/mcb.15.1.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wagner R L, Apriletti J W, McGrath M E, West B L, Baxter J D, Fletterick R J. Nature (London) 1995;378:690–697. doi: 10.1038/378690a0. [DOI] [PubMed] [Google Scholar]
- 22.Cavaillés V, Dauvois S, Danielian P S, Parker M G. Proc Natl Acad Sci USA. 1994;91:10009–10013. doi: 10.1073/pnas.91.21.10009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cavaillés V, Dauvois S, L’Horset F, Lopez G, Hoare S, Kushner P J, Parker M G. EMBO J. 1995;14:3741–3751. doi: 10.1002/j.1460-2075.1995.tb00044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Onate S A, Tsai S Y, Tsai M-J, O’Malley B W. Science. 1995;270:1354–1357. doi: 10.1126/science.270.5240.1354. [DOI] [PubMed] [Google Scholar]
- 25.Beck-Peccoz P, Chatterjee V K K, Chin W W, DeGroot L J, Jameson J L, Nakamura H, Refetoff S, Usala S J, Weintraub B D. J Clin Endocrinol Metab. 1994;78:990–993. doi: 10.1210/jcem.78.4.8157732. [DOI] [PubMed] [Google Scholar]
- 26.Forman B M, Casanova J, Raaka B M, Ghysdael J, Samuels H H. Mol Endocrinol. 1992;6:429–442. doi: 10.1210/mend.6.3.1316541. [DOI] [PubMed] [Google Scholar]
- 27.Wahlstrom G, Sjoberg M, Andersson M, Nordstrom K, Vennstrom B. Mol Endocrinol. 1992;6:1013–1022. doi: 10.1210/mend.6.7.1324417. [DOI] [PubMed] [Google Scholar]
- 28.Yen P M, Darling D S, Carter R L, Forgione M, Umeda P K, Chin W W. J Biol Chem. 1992;267:3565–3568. [PubMed] [Google Scholar]
- 29.Schrader M, Muller K M, Nayeri S, Kahlen J-P, Carlberg C. Nature (London) 1994;370:382–386. doi: 10.1038/370382a0. [DOI] [PubMed] [Google Scholar]
- 30.Kurokawa R, DiRenzo J, Boehm M, Sugarman J, Gloss B, Rosenfeld M G, Heyman R A, Glass C K. Nature (London) 1994;371:528–531. doi: 10.1038/371528a0. [DOI] [PubMed] [Google Scholar]
- 31.Mixson A J, Renault J C, Ransom S, Bodenner D L, Weintraub B D. Clin Endocrinol (Oxford) 1993;38:227–234. doi: 10.1111/j.1365-2265.1993.tb00999.x. [DOI] [PubMed] [Google Scholar]
- 32.Meier C A, Parkison C, Chen A, Ashizawa K, Meier-Hausler S C, Muchmore P, Cheng S-Y, Weintraub B D. J Clin Invest. 1993;92:1986–1993. doi: 10.1172/JCI116793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zavacki A M, Harney J W, Brent G A, Larsen P R. Mol Endocrinol. 1993;7:1319–1330. doi: 10.1210/mend.7.10.8264663. [DOI] [PubMed] [Google Scholar]
- 34.Saatcioglu F, Bartunek P, Deng T, Zenke M, Karin M. Mol Cell Biol. 1993;13:3675–3685. doi: 10.1128/mcb.13.6.3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Renaud J-P, Rochel N, Ruff M, Vivat V, Chambon P, Gronemeyer H, Moras D. Nature (London) 1995;378:681–689. doi: 10.1038/378681a0. [DOI] [PubMed] [Google Scholar]
- 36.Halachmi S, Marden E, Martin G, MacKay H, Abbondanza C, Brown M. Science. 1994;264:1455–1458. doi: 10.1126/science.8197458. [DOI] [PubMed] [Google Scholar]
- 37.Le Douarin B, Zechel C, Garnier J M, Lutz Y, Tora L, Pierrat B, Heery D, Gronemeyer H, Chambon P, Losson R. EMBO J. 1995;14:2020–2033. doi: 10.1002/j.1460-2075.1995.tb07194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee J W, Ryan F, Swaffield J C, Johnston S A, Moore D D. Nature (London) 1995;374:91–94. doi: 10.1038/374091a0. [DOI] [PubMed] [Google Scholar]
- 39.Kamei Y, Xu L, Heinzel T, Torchia J, Kurokawa R, Gloss B, Lin S-C, Heyman R A, Rose D W, Glass C K, Rosenfeld M G. Cell. 1996;85:403–414. doi: 10.1016/s0092-8674(00)81118-6. [DOI] [PubMed] [Google Scholar]