Abstract
The initiator RNAs of mammalian Okazaki fragments are thought to be removed by RNase HI and the 5′-3′ flap endonuclease (FEN1). Earlier evidence indicated that the cleavage site of RNase HI is 5′ of the last ribonucleotide at the RNA-DNA junction on an Okazaki substrate. In current work, highly purified calf RNase HI makes this exact cleavage in Okazaki fragments containing mismatches that distort the hybrid structure of the heteroduplex. Furthermore, even fully unannealed Okazaki fragments were cleaved. Clearly, the enzyme recognizes the transition from RNA to DNA on a single-stranded substrate and not the RNA/DNA heteroduplex structure. We have named this junction RNase activity. This activity exactly comigrates with RNase HI activity during purification strongly suggesting that both activities reside in the same enzyme. After junction cleavage, FEN1 removes the remaining ribonucleotide. Because FEN1 prefers a substrate with a single-stranded 5′-flap structure, the single-stranded activity of junction RNase suggests that Okazaki fragments are displaced to form a 5′-tail prior to cleavage by both nucleases.
Two classes of RNase H, RNase HI and II, have been identified in a variety of eukaryotes, purified and characterized (1–8). Class I enzymes have a native molecular mass of ≈68–90 kDa and are able to use both Mn2+ and Mg2+ as cofactors. In contrast, class II RNase H is approximately 30 kDa and can use only Mg2+ as a cofactor. Both forms of RNase H randomly degrade RNA in long RNA/DNA hybrid structures (2, 9).
RNase HI is involved in the removal of the initiator RNA primers of Okazaki fragments, an essential step during chromosomal DNA replication of the lagging strand (10). These RNAs, 7–14 nucleotides in length, are generated by the primase activity of DNA polymerase α (pol α) and prime DNA synthesis of both the leading and lagging strands at the DNA replication fork. The pol activity of this same enzyme then extends each primer with deoxynucleotides. Pol α is thought to be replaced by pol δ that continues synthesis to the next downstream fragment. There is considerable evidence that the RNA is removed by the combined action of RNase HI and a 5′ to 3′ exo/endonuclease (11–13). This latter enzyme has been called flap endonuclease (FEN1) (14) in mammals and is known as RAD two homolog (RTH1) and RAD27 in Saccharomyces cerevisiae (15). Removal of the RNA is required before DNA synthesis and ligation can complete the replication process.
The exact functions of RNase HI in DNA replication have been the subject of several studies. Drosophila RNase HI was found to stimulate synthesis by pol α (4). In addition, RNases H from yeast and calf thymus (3, 6) associate with and stimulate their cognate pol α. These observations suggest that RNase HI and pol α form a complex in vivo capable of synthesis and removal of RNA primers. Reconstitution assays with both mouse cell and SV40 replication proteins require RNase HI for the maturation of lagging strands in vitro (12, 13).
Although cleavage of long RNA/DNA hybrids is random, cleavage of certain substrates is clearly structure specific. Eder et al. (16) found that when a series of ribonucleotides were followed by a 3′ DNA segment and annealed to DNA to form a duplex, human RNase HI preferentially cleaved to leave a DNA segment with a 5′ monoribonucleotide. In reactions reconstituting Okazaki fragment processing, RNase HI from calf thymus similarly cleaved the RNA-DNA strand 5′ to the junctional ribonucleotide. At the enzyme concentrations used, the RNA primer was released by a unique cleavage with no further degradation (11). RNase HI is capable of this structure-specific cleavage regardless of the length of the RNA or the sequence context of the RNA-DNA junction (17). Therefore, RNase HI has been described as both a random and structure-specific endonuclease.
Here we show that cleavage at the RNA-DNA junction does not require the presence of any duplex structure. Highly purified RNase HI preparations contain a junction RNase activity that recognizes distinctly different aspects of substrate structure than conventional RNase H activity. The nature of the substrate, either an RNA-DNA junction or an RNA/DNA hybrid, determines that cleavage mechanism is employed.
EXPERIMENTAL PROCEDURES
Materials.
Synthetic DNA oligonucleotides were synthesized by Genosys Biotechnologies (The Woodlands, TX). [α-32P]-radiolabeled CTP and UTP (800 Ci/mmol; 1 Ci = 37 GBq) and [γ-32P] ATP (3000 Ci/mmol) were purchased from Dupont/NEN. Poly[8H]adenylic acid (≈15 Ci/mmol) was obtained from Amersham Life Sciences. Ribonucleotides and deoxyribonucleotides were purchased from Pharmacia LKB. Sequenase (version 2.0), T3 RNA polymerase, and T4 polynucleotide kinase were from Amersham Life Sciences. Restriction endonucleases were purchased from New England Biolabs. The plasmid, pBS+, was obtained from Stratagene. All other reagents were from Sigma.
Purification of RNase HI.
Two separate approaches were undertaken to purify RNase HI both starting with 100 g of fetal calf tissue. The first method, as described by Busen (18), was followed through hydroxylapatite chromatography with the exception that the (NH4)SO4 precipitation step was omitted. All columns were equilibrated with buffer A (30 mM Tris⋅HCl, pH 7.8/0.1% 2-mercaptoethanol/0.1 mM phenylmethylsulfonyl fluoride/15% glycerol) prior to use. Pooled fractions of peak RNase HI activity from the hydroxylapatite column were dialyzed into buffer A and applied to a 40 ml Affi-gel Blue column previously equilibrated with buffer A. After the column was washed with buffer A containing 250 mM MgCl2, RNase HI activity was eluted with buffer A containing 1.0 M MgCl2. Peak fractions were combined and dialyzed into buffer B (buffer A with 50 mM KCl) and applied to a 6 ml Affi-gel Blue column. The column was washed sequentially with buffer A containing 0.1, 0.3, 0.5, and 1.0 M KCl. The peak activity eluted with 300 mM KCl and was dialyzed against buffer B. The dialysate was loaded onto a 1 ml Mono-Q column and eluted with a linear salt gradient of 50–500 mM KCl. RNase HI eluted at ≈200 mM KCl and stored at 4°C. The specific activity of the peak Mono-Q fraction was 134,000 U/mg. One unit of RNase HI produces 1 nmol of trichloroacetic acid soluble ribonucleotide monomer from [3H]poly(rA)/oligo(dT) in 15 min at 37°C.
The second method for purification of RNase HI was adapted from Kawan et al. (3). After a calf thymus homogenate was prepared (11), DNA was removed by polyethylene glycol precipitation (3). Affinity chromatography over native DNA-cellulose and hydroxylapatite chromatography also were performed according to this procedure (3). Fractions containing the peak RNase H activity from the hydroxylapatite column were dialyzed into buffer A (pH 7.8) with 10 mM NaCl and applied to a 1 ml Q-Sepharose column. Activity was eluted with a 10 ml linear salt gradient from 10 to 500 mM NaCl. The active fractions were pooled, dialyzed against buffer A (pH 8.3) with 10 mM NaCl, and loaded onto a 1 ml SP-Sepharose column. A 10 to 500 mM linear salt gradient was used to elute RNase HI with a similar specific activity as the above preparation.
Glycerol Gradient Sedimentation.
Approximately 20 μg of RNase HI from the peak Mono-Q fraction was diluted to 15% glycerol in final volume of 50 μl and applied to a 3.95 ml glycerol gradient (15–35%) in buffer A with 100 mM NaCl. Protein standards consisting of catalase (232 kDa), BSA (68 kDa), and RNase (13.7 kDa) were applied to parallel gradients. Density gradient sedimentation was performed by centrifugation in a Beckman SW60.1 rotor for 72 hr at 45,000 rpm. Gradients were fractionated from the top by careful removal of 100 μl aliquots from the meniscus.
Preparation of Okazaki Fragment Substrates.
Internally labeled RNA primers were prepared by T3 RNA pol directed run-off transcription on pBS(+) plasmids digested with HindIII (19) in the presence of 625 pmol [α-32P]CTP and [α-32P]UTP, 1 mM ATP and GTP. For 5′-end-radiolabeled primers, the RNA13nt was incubated with T4 polynucleotide kinase in the presence of [γ-32P]ATP as described (30). Following separation on a denaturing 20% polyacrylamide gel, RNA primers were purified and annealed to the DNA template T1 (5′-TACCCGGGGATCCTCTAGAGTCGACCTGCAGGCATGCAAGCTTTTGTTCCCTTTAGTGAGGGTTAATTTCGAGCT-3′). Then, the RNA primers were extended with 10 units of SequenaseTM and 1.3 mM of each of the four dNTPs. These RNA-DNA primers (5′-GGGAACAAAAGCUTGCATGCCTGCAGGTCGACTCTAGAGGATCCCCGGGTA-3′) were isolated after separation through a denaturing 15% polyacrylamide gel and annealed to the appropriate template oligomer as specified in the figure legends. The RNA portion of RNA-DNA primer sequence is underlined and in italics.
DNA sequences used to create mismatched Okazaki fragments were: DNA mismatch (5′-TACCCGGGGATCCTCTAGAGTCGACCTGCAGGCATGCTAGCTTTTGTTCCCTTTAGTGAGGGTT-3′), upstream RNA mismatch (5′-TACCCGGGGATCCTCTAGAGTCGACCTGCAGGCATCCAAGCTTTTGTTCCCTTTAGTGAGGGTT-3′), and downstream RNA mismatch (5′-TACCCGGGGATCCTCTAGAGTCGACCTGCAGGCATGGAAGCTTTTGTTCCCTTTAGTGAGGG-3′). The underlined nucleotide indicates the site of mismatch.
RNase HI Assays.
Assays using Okazaki fragment substrates contained 60 mM bis⋅Tris (pH 7.0), 10 mM MgCl2, 5 mM 2-mercaptoethanol, 5% glycerol, 100 mg/ml BSA, and ≈2 fmol of substrate per 12 μl. Reactions were initiated by the addition of 1 μl of enzyme preparation that was diluted as noted in the figure legends, incubated at 37°C for times indicated and terminated with 12 μl formamide loading buffer [90% formamide (vol/vol)/10 mM EDTA/0.01% with bromophenol blue and xylene cyanole]. Cleavage products were separated by electrophoresis through a denaturing 12% polyacrylamide gel (20) and analyzed using a Molecular Dynamics PhosphorImager and imagequant software (Molecular Dynamics).
RNase H assays with [3H]poly(rA)/oligo(dT) were performed as described by Eder et al. (5) at 37°C for 10 min.
RESULTS
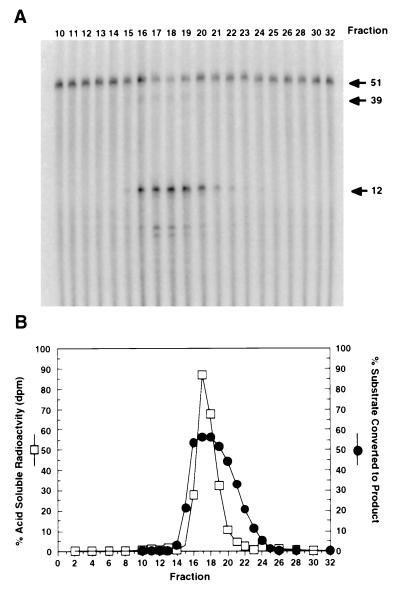
We initially began to explore the substrate specificity of RNase HI to define its role in initiator RNA removal. We wanted to verify that RNase HI, known to be a nearly random endonuclease on RNA/DNA hybrids, is responsible for the RNA-DNA junction specific cleavage observed with model double-stranded Okazaki substrates. Therefore, we purified RNase HI to a greater extent than we reported (11). Throughout purification over ion and affinity resins, we monitored both structure-specific and random endonucleolytic activity. Structure-specific cleavage was examined using a model Okazaki fragment (Fig. 1). An internally radiolabeled oligonucleotide containing a region of 13 nucleotides of RNA (RNA13nt) immediately followed by 38 nucleotides of DNA (DNA38nt) was annealed to a DNA template. After the substrate was incubated with an aliquot of each protein fraction, the reaction products were analyzed using a Molecular Dynamics PhosphorImager. Elution of the structure-specific cleavage activity from the final column, Mono-Q, is shown in Fig. 1A. Cleavage results in the formation of an RNA12nt and a 39 nucleotide segment corresponding to the remaining ribonucleotide and DNA. Smaller fragments represent additional nonspecific cleavage by RNase HI. Random endonucleolytic activity was measured on a standard RNase HI substrate [3H]poly(rA)/oligo(dT) and detected as acid soluble radioactivity. The amount of random cleavage from the Mono-Q fractions was quantitated by scintillation counting, and the data plotted as the percentage of 3H counts found in the acid soluble fraction (Fig. 1B). Additionally, the data from the structure-specific assay is presented as the percent of substrate converted to the RNA12nt product (Fig. 1B). Clearly both structure-specific and random endonucleolytic activities copurified. This provided substantial evidence that RNase HI is responsible for specific cleavage at RNA-DNA junctions.
Figure 1.
RNase HI contains both a structure-specific and random endonucleolytic activity. Aliquots (1 μl) of each fraction from the final column of the first preparation (Mono-Q) were diluted 1:4000 and tested for both RNase HI structure-specific and random cleavage activity. (A) To generate a model Okazaki substrate, an internally radiolabeled (32P)RNA13nt was extended with DNA38nt and annealed to a DNA template. Aliquots of the diluted fractions were assayed at 37°C for 10 min. Cleavage results in a specific RNA12nt fragment and an oligonucleotide39nt containing the remaining ribonucleotide and deoxynucleotides (indicated by arrows). (B) Random RNase HI cleavage was assayed on a [3H]poly(rA)/oligo(dT) substrate. RNase HI activity degrades the substrate releasing small oligonucleotides into the acid soluble fraction that were quantitated by scintillation counting. The percent of acid soluble radioactivity (□) was determined for each fraction. Additionally, the percent of the Okazaki fragment substrate converted to the RNA12nt product in A was determined and also plotted (•).
pol α/primase lacks the ability to perform 3′-5′ exonucleolytic proofreading. Consequently, it frequently misincorporates both ribo- and deoxyribonucleotides during priming and DNA synthesis (21, 22). This creates mismatched nucleotides in the 5′-end region of Okazaki fragments. We anticipated that a mismatch at or near the RNA-DNA junction would alter the structure of the RNA/DNA heteroduplex and prevent RNase HI from recognizing and cleaving the substrate. Failure of RNase HI to initiate removal of RNA primers would prevent completion of DNA replication.
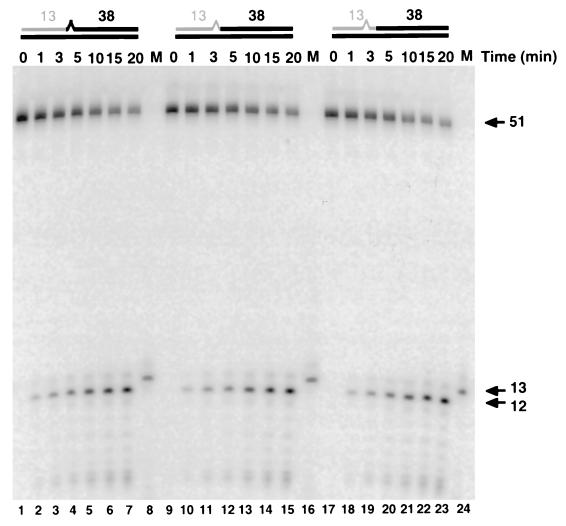
Substrates were generated to examine the ability of the RNase HI preparation to cleave the RNA-DNA junction in the presence of a mismatched base pair. A 5′-radiolabeled oligonucleotide of RNA13nt-DNA38nt was annealed to a DNA template. The template strand was designed to generate a specific mismatch with the RNA-DNA strand. Each substrate was incubated with the peak fraction of RNase HI activity from the Mono-Q column. During the reaction, aliquots were taken at selected time points and analyzed as described in the experimental procedures. Following their separation by PAGE, products were quantitated by a Molecular Dynamics PhosphorImager. When the site of mismatch was the deoxynucleotide at the RNA-DNA junction, RNase HI efficiently cleaved the substrate (Fig. 2, lanes 1–7) to generate the 12 nucleotide RNA product. While this substrate had distorted helical structure, the actual cleavage site, 5′ of the last RNA, was still able to form an RNA/DNA hybrid structure. Therefore, we tested cleavage on a substrate with the mismatch at the ribonucleotide just upstream (Fig. 2, lanes 17–23) and just downstream (lanes 9–15) of the cleavage site. Again, RNase HI specifically and efficiently cleaved 5′ of the junctional ribonucleotide. These data show that the nuclease does not require complete hybridization at the cleavage site.
Figure 2.
Mismatches near the Okazaki fragment RNA-DNA junction do not lower cleavage efficiency. A 5′-radiolabeled (32P)RNA13nt⋅DNA38nt oligonucleotide was annealed to a DNA template designed to create a specific mismatch. Above the appropriate lanes, substrate structure representations are given. Gray lines indicate RNA and solid lines depict DNA. A sawtooth segment marks the position of the mismatch relative to the template strand. The peak fraction of Mono-Q diluted 1:4,000 was assayed with the indicated substrates as described. A time course of endonucleolytic cleavage reveals that RNase HI efficiently cleaves in spite of a mismatch at the first deoxynucleotide (lanes 1–7) or a mismatch at the ribonucleotide downstream (lanes 9–15) or upstream (lanes 17–23) of the site of cleavage. For all substrates, an RNA12nt fragment was released from the substrate (arrow). The size marker (M) containing an RNA13nt transcript is also shown (lanes 8, 16, 24). Reaction times (min) are as indicated.
It was surprising that structure-specific cleavage occurred in the presence of a mismatch because the random nuclease activity of RNase HI depends on an RNA/DNA hybrid structure. In these structures, RNA/DNA hybrids form A-type helices, whereas DNA/DNA duplexes form B-type helices. Presumably, the junction in RNA-DNA/DNA substrate adopts an intermediate structure (23). We had anticipated that this unique structure could signal junction-specific cleavage. However, the mismatched substrates are still cleaved efficiently, even though they must have considerable distortion in the junction structure. Results of the mismatch experiment show that recognition of the junction cleavage site by RNase HI does not require the helical structure present at a normal RNA-DNA junction in a heteroduplex. While surprising, this conclusion is consistent with the observation that RNase HI cleaves 5′ of a single ribonucleotide embedded in a DNA-RNA-DNA/DNA heteroduplex (16, 24), anticipated to have a helical structure different from that of a duplex Okazaki fragment substrate. This observation also explains why initiator RNA mismatches do not disrupt mammalian DNA replication.
We next considered that the surrounding duplex of either RNA/DNA or DNA/DNA may be needed for RNase HI cleavage. To examine the significance of the RNA/DNA heteroduplex upstream of the site of cleavage, we utilized model Okazaki substrates lacking any RNA/DNA heteroduplex. A 5′-radiolabeled oligonucleotide of (32P)RNA13nt-DNA38nt was annealed to a DNA template complementary to only the DNA sequence. Over time, RNase HI activity released an RNA12nt (Fig. 3A). This shows that specific cleavage can occur in a substrate in which RNA/DNA hybrid cannot form. We also tested RNase HI activity on the RNA13nt-DNA38nt oligonucleotide in the absence of any DNA template. Remarkably, a specific RNA12nt segment was again released (Fig. 3B) Specific cleavage was also seen with two single-stranded Okazaki fragments having different sequences surrounding the junction (not shown). Furthermore, in comparison to a standard Okazaki substrate, the activity of RNase HI with the unannealed substrates was similar to that of a fully annealed substrate (Fig. 3C). The highly purified RNase HI displayed a cleavage activity that is independent of any heteroduplex structure.
Figure 3.
Junction-specific cleavage by RNase HI is independent of any RNA/DNA heteroduplex. Above the appropriate lanes, substrate structure representations, and reaction times (min) are given. Gray lines indicate RNA and solid lines depict DNA. Reactions were performed as described in Fig. 2. (A) A 5′-radiolabeled oligonucleotide of (32P)RNA13nt⋅DNA38nt was annealed to a DNA template complementary to only the DNA sequence (5′-TACCCGGGGATCCTCTAGAGTCGACCTGCAGGCATGCA-3′). Over time, RNase HI activity released an RNA12nt product. (B) RNase HI activity also was examined on the RNA13nt⋅DNA38nt oligonucleotide in the absence of any DNA template. Again, a specific RNA12nt segment was released. (C) RNase HI activity on a fully annealed substrate generated the RNA12nt product at the same rate as in A and B. The size marker (M) containing an RNA13nt transcript is indicated.
The ability of RNase HI to cleave a single-stranded junction substrate was surprising given that activity on single-stranded RNA had not been previously reported. Though our enzyme preparation did not measurably degrade any of a single-stranded [3H]poly(rA) substrate, the junction-specific cleavage of single-stranded RNA-DNA substrates might still represent a contaminating activity in the RNase HI preparation. Therefore, we sought to verify that junction activity on both double- and single-stranded Okazaki fragment substrates and RNA/DNA hybrids were contained within RNase HI.
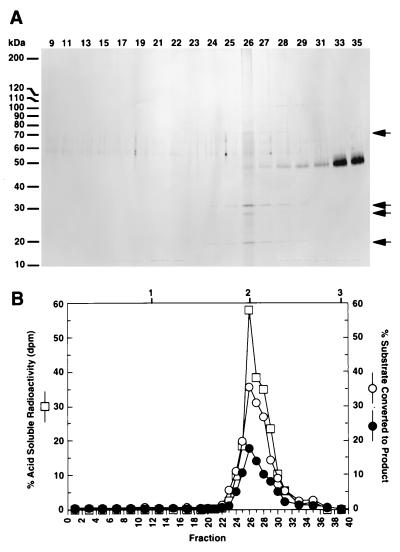
Mono-Q fractions with the highest RNase H and Okazaki fragment junction cleavage activity were subjected to glycerol gradient sedimentation. Fractions from the glycerol gradient were analyzed on an 8–14% SDS-polyacrylamide gel stained with silver (25) (Fig. 4A). Assays using either the standard RNase H [3H]poly(dA)/oligo(dT) substrate or double- and single-stranded Okazaki fragment substrates were performed (Fig. 4B). Results demonstrate that the peak of enzymatic activity on all three substrates cosediment. Cosedimentation is so exact that the ratio of activities is essentially the same for every fraction over the peak. They are also coincident with protein bands at 70, 30, 27, and 18 kDa. This represents nearly homogeneous RNase HI where the lower molecular weight species are degradation products of the full-length 70-kDa protein (7). RNase HI was also purified by a second method that involved different types of fractionation from the first procedure. Junction-specific cleavage activity on an RNA-DNA substrate again coeluted with the random RNase HI activity on the [3H]poly(rA)/oligo(dT) substrate. Fractions from the final SP-Sepharose column were analyzed by SDS/PAGE and showed the same 70-, 30-, 27-, and 18-kDa protein bands as obtained following glycerol sedimentation (data not shown). These results provide compelling evidence that RNase HI contains multiple nucleolytic activities.
Figure 4.
Junction RNase activity is tightly associated with RNase HI. (A) An aliquot of purified RNase HI was subjected to sedimentation on a 15–35% glycerol gradient. After fractionation, aliquots were analyzed by electrophoresis on an 8–14% SDS-polyacrylamide gel and stained with silver. Gel bands corresponding to RNase HI are indicated (arrows). The peak of RNase HI is fraction 26 that corresponds to a sedimentation value of ≈4.3 and a native molecular mass of ≈68 kDa as expected for RNase HI. (B) Glycerol gradient fractions (diluted 1:400) were tested for activity on [3H]RNA/DNA hybrids and Okazaki substrates as previously described. The percent of acid soluble radioactivity (□) and the percent of double-stranded (•) and single-stranded (○) Okazaki substrate to converted to product were plotted vs. fraction number. Above the graph, positions of protein standards for the glycerol gradient are: (1) RNase (13.7 kDa), (2) BSA (68 kDa), and (3) catalase (232 kDa).
The susceptibility of RNase HI activity to increasing salt concentration was measured on the [3H]poly(rA)/oligo(dT) and junction specific substrates. Overall, both activities showed similar sensitivity to titration with KCl (0–500 mM). Total activity on the two different substrates was inhibited 50% at ≈200 mM KCl. This result further supports the conclusion that both activities reside in the same protein. On the RNA-DNA substrate, total activity consisted of both the specific junction cleavage and several additional nonspecific cleavages. Inhibition of the nonspecific cleavages generally occurred at a lower salt concentration than at the specific junction site. In effect, the specificity for the junction can be increased at appropriate salt concentration.
DISCUSSION
The original model for initiator RNA removal from mammalian Okazaki fragments predicts a specific cleavage of the duplex RNA by RNase HI. This is followed by exonucleolytic removal of the remaining junction ribonucleotide by the FEN1 nuclease. Both enzymes are required because FEN1 nuclease cannot exonucleolytically cleave a triphosphorylated 5′-end, and RNase HI cannot remove the junction ribonucleotide (11). However, FEN aptly stands for “flap endonuclease” because its preferred substrate is a primer-template in which the primer has an unannealed 5′-region (14, 26). The nuclease enters the 5′-end of the unannealed tail, or flap, and slides to the point of annealing. There it performs a structure-specific endonucleolytic cleavage. The exonucleolytic activity is much less active and has been proposed to be a manifestation of endonucleolytic cleavage of the transiently unannealed 5′-nucleotide of a primer (27). Furthermore, yeast FEN1 nuclease functionally interacts with the Dna2 helicase (28, 29). This supports the interpretation that the nuclease acts in vivo after formation of a partially unannealed 5′-strand. It has puzzled us that RNase HI appears to act on fully annealed Okazaki fragments, whereas the FEN1 nuclease prefers a substrate with an unannealed tail.
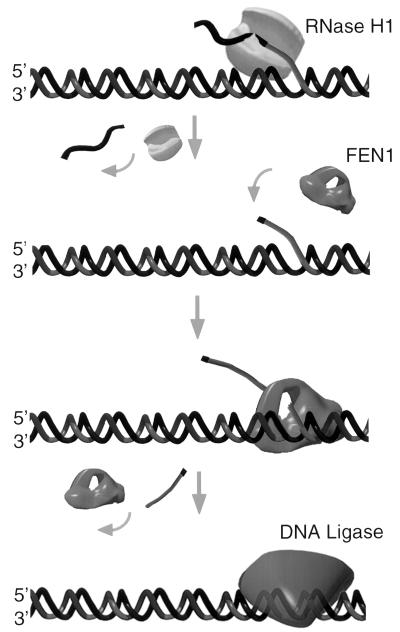
The demonstration that the calf RNase HI is also a single-strand specific junction RNase suggests an alternative pathway for the removal of initiator RNA, as illustrated in Fig. 5. In this case, strand displacement by DNA synthesis or DNA helicase activity creates an unannealed tail at the 5′-end of every Okazaki fragment. This is an efficient substrate for the junction RNase activity of RNase HI. Next, cleavage by FEN1 removes a segment of unannealed tail containing the 5′ monoribonucleotide. This fully endonucleolytic mechanism not only takes advantage of preferred substrates for both enzymes but could explain why pol α does not display high fidelity synthesis. Base substitution errors made by pol α would disappear from the chromosome if all of the nucleotides added by that polymerase were removed by the strand displacement mechanism. pol δ directed synthesis from upstream primers would replace this region with accurately base-paired nucleotides. Unlike an exonucleolytic mechanism, cleavage of the unannealed tail to remove RNA and a substantial region of DNA requires that each nuclease make only a single cleavage.
Figure 5.
Displacement model of Okazaki fragment processing. During replication, the initiator primer (black segment) is displaced by an upstream helicase or pol. RNase HI then cleaves the single-stranded RNA-DNA junction substrate, releasing the RNA primer intact. The remaining displaced RNA nucleotide and DNA forms an unannealed tail. The FEN1 nuclease binds the tail, tracks to the point of annealing and cleaves endonucleolytically removing the remaining ribonucleotide and some DNA. DNA ligase then seals the resulting nick.
When we created an Okazaki fragment that had a mismatched 5′-region including the initiator RNA and an adjacent region of DNA, FEN1 nuclease efficiently removed the entire unannealed region (19). If so, what is the need of RNase HI? We propose that RNase HI will cleave all initiator RNAs regardless of secondary structure. FEN1 is inhibited by a primer annealed to the tail flap (27, 30). Therefore, if the initiator RNA forms a stable secondary structure with another sequence on the displaced region of the Okazaki fragment, the RNA may not be removable except by RNase HI.
In light of our current work, we can interpret results obtained by Eder et al.(16) in more general terms. They found that human RNase HI can cleave just 5′ to a single ribonucleotide in double-stranded DNA, even if that ribonucleotide were mismatched. It is now evident that RNase HI recognizes the RNA-DNA junction itself and not a unique helical structure. This allows the single ribonucleotide to be recognized even though it is likely to have minimal impact on the surrounding helical configuration. Furthermore, the ability to cleave a mismatch derives from the complete lack of a requirement for RNA/DNA or DNA/DNA hybrid structure.
The cleavage of single-stranded RNA-DNA substrates by RNase HI further distinguishes junction-specific activity of RNase HI from its standard random degradation of RNA/DNA hybrids. We have emphasized this activity, which depends on recognition of a transition from RNA to DNA in the same strand, and not hybrid structure, by use of the term “junction RNase.” The presence of this activity in mammalian RNase HI has allowed us to propose a new mechanism for processing of mammalian Okazaki fragments in which both RNase HI/junction RNase and the FEN1 nuclease cleave a displaced 5′-region of these nascent DNA fragments.
Acknowledgments
The authors would like to thank Dr. C. Palaniappan, Jeffrey A. Rumbaugh, and Carole J. Barnes for their helpful discussions and critical reading of this manuscript. This work was supported by National Institutes of Health Research Grant GM24441 and Fellowship Grant GM18961 (to L.A.H.), and in part by Cancer Center Core Grant CA11198.
ABBREVIATIONS
- FEN1
flap endonuclease 1/RAD two homolog 1
- pol
DNA polymerase
- nt
nucleotide
References
- 1.Busen W, Hausen P. Eur J Biochem. 1975;52:179–190. doi: 10.1111/j.1432-1033.1975.tb03985.x. [DOI] [PubMed] [Google Scholar]
- 2.Goulian M, Heard C J. Anal Biochem. 1991;192:398–402. doi: 10.1016/0003-2697(91)90555-8. [DOI] [PubMed] [Google Scholar]
- 3.Karwan R, Blutsch H, Wintersberger U. Biochemistry. 1983;22:5500–5507. [Google Scholar]
- 4.DiFrancesco R A, Lehman I R. J Biol Chem. 1985;260:14764–14770. [PubMed] [Google Scholar]
- 5.Eder P S, Walder J A. J Biol Chem. 1991;266:6472–6479. [PubMed] [Google Scholar]
- 6.Hagemeier A, Grosse F. Eur J Biochem. 1989;185:621–628. doi: 10.1111/j.1432-1033.1989.tb15158.x. [DOI] [PubMed] [Google Scholar]
- 7.Rong Y W, Carl P L. Biochemistry. 1990;29:383–389. doi: 10.1021/bi00454a012. [DOI] [PubMed] [Google Scholar]
- 8.Frank P, Albert S, Cazenave C, Toulme J J. Nucleic Acids Res. 1994;22:5247–5254. doi: 10.1093/nar/22.24.5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crouch R J, Dirksen M L. In: Nucleases. Linn S M, Roberts R J, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1982. pp. 211–241. [Google Scholar]
- 10.Bambara R A, Murante R S, Henricksen L A. J Biol Chem. 1997;272:4647–4650. doi: 10.1074/jbc.272.8.4647. [DOI] [PubMed] [Google Scholar]
- 11.Turchi J J, Huang L, Murante R S, Kim Y, Bambara R A. Proc Natl Acad Sci USA. 1994;91:9803–9807. doi: 10.1073/pnas.91.21.9803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Waga S, Bauer G, Stillman B. J Biol Chem. 1994;269:10923–10934. [PubMed] [Google Scholar]
- 13.Ishimi Y, Claude A, Bullock P, Hurwitz J. J Biol Chem. 1988;263:19723–19733. [PubMed] [Google Scholar]
- 14.Harrington J J, Lieber M R. EMBO J. 1994;13:1235–1246. doi: 10.1002/j.1460-2075.1994.tb06373.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson R E, Kovvali G K, Prakash L, Prakash S. Science. 1995;269:238–240. doi: 10.1126/science.7618086. [DOI] [PubMed] [Google Scholar]
- 16.Eder P S, Walder R Y, Walder J A. Biochimie. 1993;75:123–126. doi: 10.1016/0300-9084(93)90033-o. [DOI] [PubMed] [Google Scholar]
- 17.Huang L, Kim Y, Turchi J J, Bambara R A. J Biol Chem. 1994;269:25922–25927. [PubMed] [Google Scholar]
- 18.Busen W. J Biol Chem. 1980;255:9434–9443. [PubMed] [Google Scholar]
- 19.Murante R S, Rumbaugh J A, Barnes C J, Norton J R, Bambara R A. J Biol Chem. 1996;271:25888–25897. doi: 10.1074/jbc.271.42.25888. [DOI] [PubMed] [Google Scholar]
- 20.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1991. [Google Scholar]
- 21.Sheaff R J, Kuchta R D. J Biol Chem. 1994;269:19225–19231. [PubMed] [Google Scholar]
- 22.Thomas D C, Roberts J D, Sabatino R D, Myers T W, Tan C K, Downey K M, So A G, Bambara R A, Kunkel T A. Biochemistry. 1991;30:11751–11759. doi: 10.1021/bi00115a003. [DOI] [PubMed] [Google Scholar]
- 23.Salazar M, Fedoroff O Y, Reid B R. Biochemistry. 1996;35:8126–8135. doi: 10.1021/bi9528917. [DOI] [PubMed] [Google Scholar]
- 24.Rumbaugh J A, Murante R S, Shi S, Bambara R A. J Biol Chem. 1997;272:22591–22599. doi: 10.1074/jbc.272.36.22591. [DOI] [PubMed] [Google Scholar]
- 25.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 26.Murante R S, Huang L, Turchi J J, Bambara R A. J Biol Chem. 1994;269:1191–1196. [PubMed] [Google Scholar]
- 27.Wu X, Li J, Li X, Hsieh C L, Burgers P M, Lieber M R. Nucleic Acids Res. 1996;24:2036–2043. doi: 10.1093/nar/24.11.2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Budd M E, Choe W C, Campbell J L. J Biol Chem. 1995;270:26766–26769. doi: 10.1074/jbc.270.45.26766. [DOI] [PubMed] [Google Scholar]
- 29.Budd M E, Campbell J L. Mol Cell Biol. 1997;17:2136–2142. doi: 10.1128/mcb.17.4.2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murante R S, Rust L, Bambara R A. J Biol Chem. 1995;270:30377–30383. doi: 10.1074/jbc.270.51.30377. [DOI] [PubMed] [Google Scholar]