Abstract
The different sialic acid (serogroups B, C, Y, and W-135) and nonsialic acid (serogroup A) capsular polysaccharides expressed by Neisseria meningitidis are major virulence factors and are used as epidemiologic markers and vaccine targets. However, the identification of meningococcal isolates with similar genetic markers but expressing different capsular polysaccharides suggests that meningococcal clones can switch the type of capsule they express. We identified, except for capsule, isogenic serogroups B [(α2→8)-linked polysialic acid] and C [(α2→9)-linked polysialic acid] meningococcal isolates from an outbreak of meningococcal disease in the U. S. Pacific Northwest. We used these isolates and prototype serogroup A, B, C, Y, and W-135 strains to define the capsular biosynthetic and transport operons of the major meningococcal serogroups and to show that switching from the B to C capsule in the outbreak strain was the result of allelic exchange of the polysialyltransferase. Capsule switching was probably the result of transformation and horizontal DNA exchange in vivo of a serogroup C capsule biosynthetic operon. These findings indicate that closely related virulent meningococcal clones may not be recognized by traditional serogroup-based surveillance and can escape vaccine-induced or natural protective immunity by capsule switching. Capsule switching may be an important virulence mechanism of meningococci and other encapsulated bacterial pathogens. As vaccine development progresses and broader immunization with capsular polysaccharide conjugate vaccines becomes a reality, the ability to switch capsular types may have important implications for the impact of these vaccines.
Neisseria meningitidis is a leading worldwide cause of meningitis and rapidly fatal sepsis in otherwise healthy individuals (1); over 350,000 cases of meningococcal disease were estimated to have occurred in 1995 (2). The problem of meningococcal disease is emphasized by the recurrence of major epidemics due to serogroups A, B, and C N. meningitidis over the last 20 years (3–9) [such as the devastating serogroup A outbreak in sub-Saharan Africa this year (W.H.O., 1996, unpublished bulletin); the recent dramatic increases in the incidence of serogroups B and C meningococcal disease in parts of North America (6–8); and the emergence in Europe and elsewhere of meningococci with decreased susceptibility to antibiotics (10)].
Differences in capsular polysaccharide chemical structure determine the meningococcal serogroups (11, 12). Meningococci of serogroups B, C, Y, and W-135 express capsules composed entirely of polysialic acid or sialic acid linked to glucose or galactose (12, 13), while the capsule of group A N. meningitidis is composed of N-acetyl mannosamine-1-phosphate (11). The currently available capsular polysaccharide vaccines for serogroups A, C, Y, or W-135 N. meningitidis are effective for control of meningococcal outbreaks in older children and adults. However, because of poor immunogenicity in young children and short-lived immunity (14), these vaccines are not routinely used for long-term prevention of meningococcal disease. In the case of group B N. meningitidis, whose (α2→8)-linked polysialic capsule is an immunotolerized self-antigen, a reliable polysaccharide vaccine is not yet available. However, rapid progress is being made in development of polysaccharide–protein conjugate vaccines, and it is hoped that, following the example of newly licensed Haemophilus influenzae type b vaccines, widespread introduction of the polysaccharide conjugates will lead to elimination of disease.
In some epidemic settings, simultaneous or closely linked meningococcal outbreaks have occurred in the same population due to different serogroups (5, 6, 15). Further, Caugant et al. (3, 16) have noted that meningococcal isolates of different serogroups may be members of the same enzyme type (ET)-5, ET-37, or ET-4 clonal complexes.
Since 1993 the number of cases of serogroup B meningococcal disease in Oregon and adjacent counties in Washington has doubled, and the overall incidence has been fivefold higher than rates observed in other parts of the United States (6). This increase was due to the first appearance in the United States of serogroup B meningococcal strains belonging to the ET-5 complex. ET-5 complex strains have been responsible for major epidemics in Norway, Iceland, Cuba, and South America over the last 20 years (3, 17, 18). Since 1994, cases of serogroup C meningococcal disease due to ET-5 complex strains were also noted in Oregon and Washington. Recent advances (19–27) in understanding the genetic basis for meningococcal capsule expression allowed us to carefully analyze the serogroup B and C ET-5 meningococcal strains responsible for the outbreak in the Pacific Northwest.
MATERIALS AND METHODS
Bacterial Strains.
Forty serogroup B and C ET-5 complex meningococcal isolates recovered from Oregon, Washington, and California in 1994 and 1995 were used in these studies. In addition, meningococcal strains GA0078 (serogroup C), GA0290 (C), NMB (B), C114 (B), M986 (B), 2996 (B), KB (B), 269B (B), FAM18 (C), 6083 (W-135), GA0929 (Y), F8229 (A), F8239 (A), NM-44/76 (B), GA1002 (W-135), N. gonorrhoeae strain FA19, and Neisseria lactamica and other commensal Neisseria spp. (refs. 21 and 28; this study) were also used.
Molecular Epidemiology.
Multiple enzyme electrophoretic (ET) typing and pulsed-field gel electrophoresis (PFGE) were performed as described (29, 30). Specific enzyme types (e.g., ET-301) were designated by the Centers for Disease Control Meningococcal Reference Laboratory (Atlanta). Serotyping of meningococcal strains was done as described (31), with the following modifications. The serotyping procedure was modified to grow meningococci on brain heart infusion agar (Difco) supplemented with 1% horse serum (GIBCO), and to use a higher concentration of cells (cell density, OD600 1.0), different blocking buffer (PBS + 0.1% Tween 20), and shorter primary antibody incubation (2.5 h).
Molecular Biology.
PCR, Southern DNA hybridization, and DNA sequencing techniques were performed as described (32). Automated sequencing using an Applied Biosystems model 377 automated DNA sequencing system was performed on some PCR templates. Oligonucleotide primers used for PCR, sequencing, and construction of Southern probes were as follows: 5′ ctrA, 5′-GTGTGGAAGTTTAATTGTAGGATG-3′; 3′ ctrA, 5′-CCACCACCAAACAATACTGCCG-3′; 5′ synX, 5′-GCAATACCATTACGTTTATCTCTC-3′; 3′ synX, 5′-GTTTCAGGATTGTTGATTACTTCAGC-3′; 5′ synB, 5′-GTCCTACGCCCTGCAGAGCTGG-3′; 3′ synB, 5′-CATTAGGCCTAAATGCCTAGG-3′; 5′ synC, 5′-GCTGAAGTTGTTAAACATCAAACAC-3′; 3′ synC, 5′-GCTACGACAGATGCAAAGGCG-3′; 5′ synD, 5′-AGAGGATTGGCTATTACATATAGC-3′; 3′ synD, 5′-AGCTCTGTTGTCGATTACTCTCC-3′; 5′ FKBP, 5′-CATTACACAGGTTGGCTGGAAGACGG-3′; 3′ FKBP, 5′-GCAGCTCGACTTCAAATATCAAAGTGGC-3′; 5′ recA, 5′-GCCAGCAGGAAGAAAACCTCG-3′; 3′ recA, 5′-GCCGTTGTAGCTGTACCACGC-3′; 5′ ctrA-synX, 5′-CACCACCAAACAATACTGCC-3′; 3′ ctrA-synX, 5′-GCTTGTTCATTTGCTACCAAGTGG-3′; 5′ galE, 5′-CCAGCATCAATATCCTGCCACG-3′; 3′ galE, 5′-CCATCATTTGTGCAAGGCTGCG-3′. Nucleotide sequences were analyzed using either the DNAstar (Madison, WI) sequence analysis software or the Genetics Computer Group (Madison, WI) Sequence Analysis Software Package, Version 7.3.1 UNIX (33). Plate transformations of meningococcal strains were performed as described (32).
RESULTS
Molecular Epidemiology.
Serogroup B and C meningococcal strains from the Pacific Northwest outbreak were studied by ET typing (29), serotyping (31), and PFGE (30). These included 35 ET-5 complex strains (29 were serogroup B and 6 were serogroup C) consecutively isolated during 1994 in Oregon and five serogroup B ET-5 complex strains recovered in 1994–1995 from Washington and California. In Oregon in 1994, ≈88% of serogroup B isolates studied were ET-5 complex strains and 17% of C isolates were ET-5 complex strains. None of the strains were from case-clusters or epidemiologically linked patients. All were ET-301 (a member of the ET-5 complex). All, except one, were serotype 4 or 15, all were subtype 1.7,16 and all except one expressed the L3,7,9 LOS immunotype. One predominant PFGE pattern (A) was seen in these isolates. None of the isolates differed from the predominant PFGE A pattern by more than three bands, indicating the isolates were closely related (34). These data, as well as similar data on other strains of this outbreak isolated in 1993–1996 showed identity or close-relatedness of the ET-5 serogroup B strains causing epidemic disease in the Pacific Northwest. In addition, the serogroup C strains isolated were identical to the dominant serogroup B strains by these molecular epidemiologic markers. These data suggested that the epidemic meningococcal clone causing the outbreak in the Pacific Northwest may express either serogroup B [(α2→8)-linked polysialic acid] or serogroup C [(α2→9)-linked polysialic acid] capsular polysaccharide. The outbreak strains were distinct by ET typing, serotyping, subtyping, and PFGE from serogroup B and C meningococcal disease isolates recovered from other parts of the country during this time.
Genetic Structure of Capsule Biosynthesis and Transport Regions in Serogroups A, B, C, Y, and W-135 N. meningitidis.
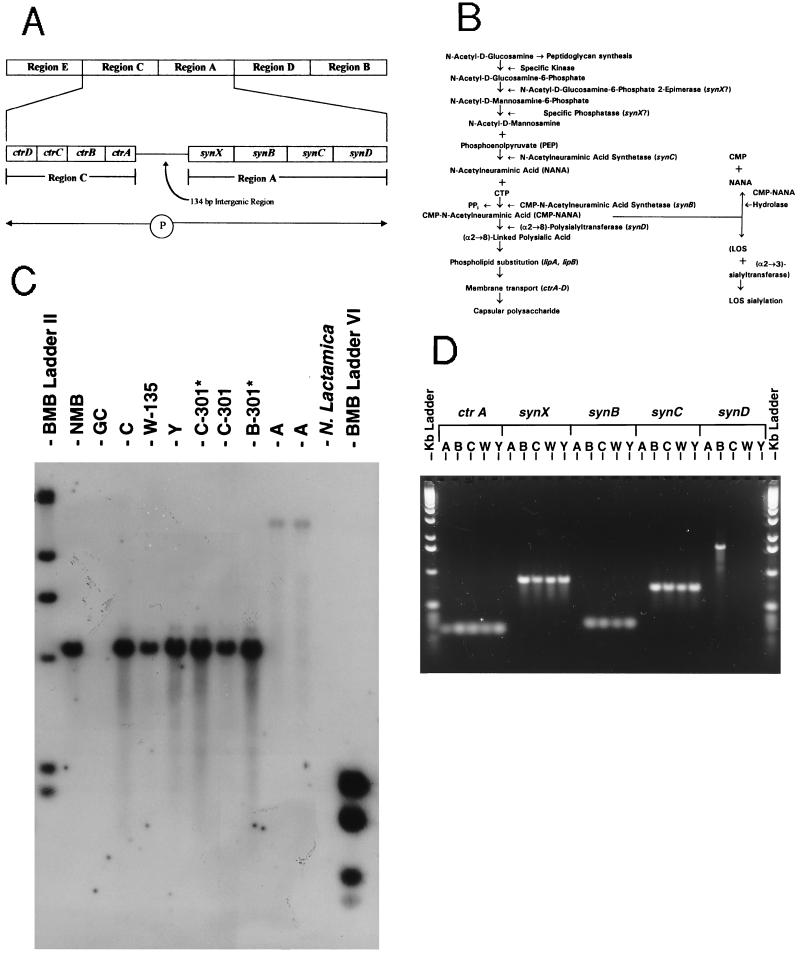
The genetic basis for serogroup B meningococcal capsule biosynthesis and membrane translocation has been under active study (19–27) and is summarized in Fig. 1A. The role of these genes in the capsule synthesis pathway is shown in Fig. 1B. Using Southern blot analysis, PCR, and DNA sequencing (21, 26), we assessed the genetic structure of the capsule transport and biosynthetic regions in strains from each of the other major meningococcal serogroups (Fig. 1 C and D). The strains of the sialic acid capsule-expressing serogroups (B, C, Y, and W-135) were found to have a similar genetic organization consisting of the ctrA capsule transport gene linked by a short intergenic region to the oppositely transcribed biosynthetic genes synX-synC. Based on identical Southern hybridization patterns for ctrA (Fig. 1C), synX, synB, and synC (data not shown); identical PCR amplification products (Fig. 1D) of ctrA, synX, synB, and synC; and similar nucleotide sequences of the ctrA–synX intergenic region (26), we concluded that ctrA and synX–synC in serogroups C, Y, and W-135 N. meningitidis were homologues of the same genes in serogroup B meningococci. In contrast, synD [which encodes the serogroup B (α2→8)-linked capsule polysialyltransferase (24)] was not detected in the serogroup C, Y, and W-135 strains by Southern hybridization or PCR amplification using probes specific for synD (Fig. 1D).
Figure 1.
Molecular analysis of capsule biosynthesis and membrane transport genes in prototype isolates of serogroup A, B, C, Y, and W-135 N. meningitidis. (A) Genetic basis for serogroup B meningococcal capsular polysaccharide. Meningococcal capsules are produced by genes encoded by the 24-kb cps gene complex (19), which is composed of five regions: E, C, A, D, and B. In serogroup B, four capsular biosynthetic genes (synX–D) are found in region A and are transcribed as an operon. Region C, adjacent to region A, contains four polycistronic genes, ctrA–D, encoding proteins that transport the phospholipid-substituted polysialic acid across the inner and outer membranes. The ctr genes are transcribed in the opposite orientation from the syn biosynthetic genes of region A, but use the same 134 bp promoter region (26). (B) The biosynthetic pathway for production of serogroup B capsule; SynX is either the N-acetyl-d-glucosamine-6-phosphate 2-epimerase that produces N-acetyl-d-mannosamine 6-phosphate or a specific phosphatase that converts N-acetyl-d-mannosamine 6-phosphate into N-acetyl-d-mannosamine (21); SynB is the CMP-N-acetylneuraminic acid (NANA) synthetase (23); SynC is the NANA synthetase (22); and SynD is the polysialyltransferase responsible for (α2→8)-linked polysialic acid chain polymerization and elongation (24). (C) Southern DNA hybridization showing ctrA homology in serogroups A (strains F8229, F8239), B (strains NMB, 1070 [B-301*]), C (strains FAM18, 1205 [C-301*], 1843 [C-301]), Y (strain GA0929), and W-135 (strain 6083) of N. meningitidis. Chromosomal DNA from each of the strains was prepared, digested with ClaI, electrophoresed through a 1.2% agarose gel, and transferred to a nylon membrane. The membrane was then probed with a 150 bp digoxigenin-labeled PCR product derived from the 5′-end of the serogroup B ctrA gene. N. lactamica and N. gonorrhoeae (GC) showed no hybridization. Molecular weight size standards (Boehringer Mannheim) flank the chromosomal digests. (D) PCR amplification of ctrA and synX–synD from serogroups A (strain F8239), B (strain NMB), C (strain FAM18), W-135 (strain 6083), and Y (strain GA0929) N. meningitidis using oligonucleotide primers derived from the individual gene sequences of serogroup B prototype strain NMB. Kb DNA ladders (BRL) flank the gel. [Fig. 1 A and B are modified with permission from ref. 26 (Copyright 1996, American Society for Microbiology).]
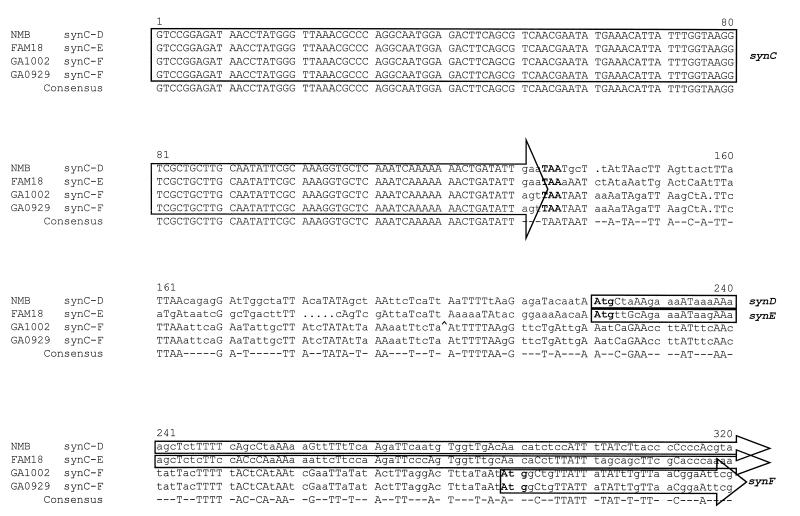
To investigate these observations further, the nucleotide sequence of the 3′ end of synC and the sequence downstream of synC were determined in serogroups C, Y, and W-135. The sequences of the 3′ end of synC from serogroups B, C, Y, and W-135 were identical up to the last codon where the sequences then diverged (Fig. 2). The 5′ ends of the downstream ORFs, which encode the putative sialic acid capsule polymerases (designated in serogroup B as synD, in serogroup C as synE, and in serogroups Y and W-135 as synF), were distinct (Fig. 2). In the serogroup Y and W-135 strains the codon for the last amino acid in synC had been replaced by a different codon (creating a change from glutamine to serine) (Fig. 2). The nucleotide sequences downstream of synC were almost identical in serogroups Y and W-135 both in the intergenic region and in the first 800 bases of the 5′ end of the predicted capsule polymerase, but were distinct from serogroups B and C.
Figure 2.
Multiple nucleotide sequence alignment of the 3′ end of synC and downstream sequence in serogroups B (strain NMB), C (strain FAM18), W-135 (strain GA1002), and Y (strain GA0929) N. meningitidis [Pretty multiple sequence comparison program of the Genetics Computer Group sequence analysis package version 7.3.1 UNIX (33)]. Consensus nucleotide matches (three or more identical) at each position are indicated in uppercase type, while differences from consensus are indicated by lowercase type. Dots indicate gaps introduced by the analysis program to facilitate alignment. The synC termination codon (TAA) and the synD/E/F start codons (ATG) are shown in boldface type. The location of an IS1301 element located downstream of the synC gene in the otherwise identical sequence of a second serogroup W-135 strain, 6083, is shown in the GA1002 sequence by a ^. The complete sequence of synE derived from serogroup C strain FAM18 is available through the GenBank data base under accession no. U75650U75650.
Thus, meningococci expressing serogroup B, C, Y, or W-135 sialic acid capsules have similar synX–synC biosynthetic genes that are linked to ctrA of the capsule membrane transport operon. However, the genes encoding the sialic acid capsule polymerases in serogroups B, C, and Y/W-135 are different. Interestingly, meningococci of serogroups Y and W-135 are almost identical in the 5′ end of this gene. These are known to be closely related serogroups, and simultaneous elaboration of both serogroup W-135 and Y capsular polysaccharides by a single strain of N. meningitidis has been reported (35).
In contrast to the sialic acid producing serogroups, serogroup A meningococci contain a ctrA homologue but did not have a ctrA–synX intergenic region or the sialic acid biosynthetic homologues synX–synD homologues (Fig. 1D). The serogroup A ctrA differed in nucleotide sequence and Southern ClaI fragment size from the sequence and location of ctrA in the sialic acid capsule-expressing serogroups (Fig. 1C). Southern blot and PCR analyses revealed that for a specific serogroup, the genes (e.g., synD, synE, synF) involved in alternative capsule polymerization were not present elsewhere in the chromosome [e.g., serogroup B strains contained synD but not synE or synF homologues (data not shown)].
Transformation of Meningococcal Capsule Biosynthesis Operon in Vitro.
Because meningococci are naturally competent for transformation, we postulated that conversion from one sialic acid-expressing capsule serogroup to another could be accomplished by homologous recombination of the sequences encoding the serogroup-specific capsule polymerase. To test this hypothesis, chromosomal DNA containing a class I Tn916 insertion interrupting synD of the serogroup B strain NMB (26) was prepared and used to transform (32) the prototype serogroup C, Y, and W-135 meningococcal strains. Tetracycline-resistant transformants were obtained at a frequency of between 1 × 10−5 and 1 × 10−7 per recipient. Acquisition of the Tn916 mutation and the adjacent synD sequence was confirmed by PCR and nucleotide sequence analysis of selected tetracycline-resistant transformants of these strains.
Analysis of Capsule Biosynthesis and Other Genes in Serogroup B and Serogroup C ET-5 Complex Strains from the Pacific Northwest Outbreak.
We hypothesized that a transformation event involving the capsule biosynthesis genes might have produced the closely related serogroup B and C meningococcal strains recovered in the Oregon and Washington outbreak. Thus, we analyzed the capsule biosynthetic and transport operons as well as unlinked genes in two serogroup B and three serogroup C ET-5 complex strains (Table 1) recovered from this outbreak. These strains by ET-type (301), serotype (15), subtype (1.7,16), immunotype (L3,7,9), and PFGE type were identical, except for the type of capsule they produced.
Table 1.
N. meningitidis isolates of the ET-5 complex recovered from patients with invasive meningococcal disease in Oregon in 1994
| ID no. | Date of onset of illness | Serogroup | Serotype | Subtype | Immunotype | ET type | PFGE type |
|---|---|---|---|---|---|---|---|
| B301#1 1070 | 06/21/94 | B | 15 | 1.7,16 | L3,7,9 | 301 | A |
| B301#2 1069 | 06/13/94 | B | 15 | 1.7,16 | L3,7,9 | 301 | A |
| C301#1 1205 | 11/19/94 | C | 15 | 1.7,16 | L3,7,9 | 301 | A |
| C301#2 1198 | 08/08/94 | C | 15 | 1.7,16 | L3,7,9 | 301 | A |
| C301#3 1204 | 10/29/94 | C | 15 | 1.7,16 | L3,7,9 | 301 | A |
(i) Capsule biosynthesis operon.
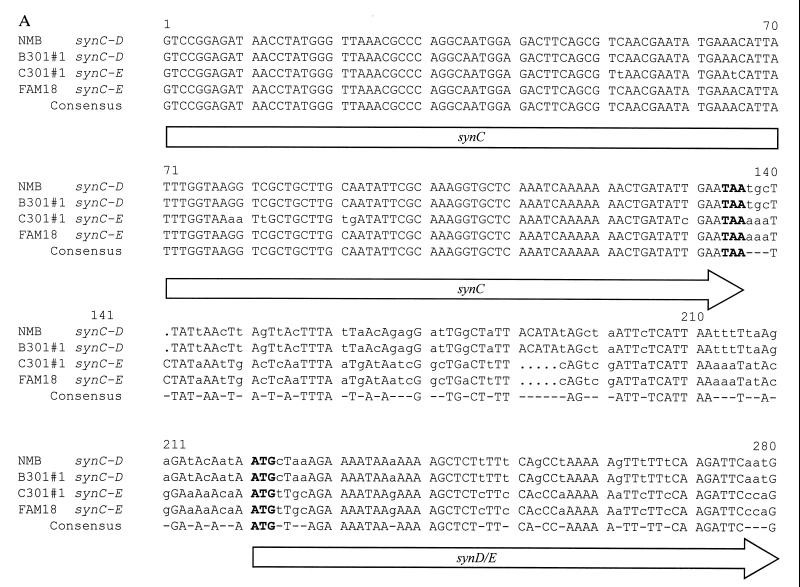
By PCR and Southern hybridization profile, the strains had similar ctrA and synX–synC homologues, but the serogroup B ET-301 strains contained a synD homologue and the serogroup C ET-301 strains contained a synE homologue. This observation was confirmed by determination of the nucleotide sequences of the intergenic region following synC and the sequences of the 5′ end of the downstream gene encoding the predicted polysialyltransferase. As shown in Fig. 3A, these regions were distinct in strain 1070 (serogroup B, ET-301) and 1205 (serogroup C, ET-301) isolates (only 63% nucleotide identity). The nucleotide sequence of synD in the B301 strain was 99% identical to synD of the prototype serogroup B strain NMB; and in the C301 strain, synE was 99% identical to synE of the prototype serogroup C strain FAM18. Nucleotide sequences of synX and synC from strains 1070 and 1205 demonstrated 1% (synX) and 5% (synC) diversity (Fig. 3 A and B1), suggesting that in addition to the polysialyltransferase, the entire synX–synD biosynthetic operon had exchanged.
Figure 3.
Genetic analysis of serogroup B301 (strains 1070 and 1069) and C301 (strains 1205, 1198, and 1204) N. meningitidis recovered from Oregon/Washington outbreak. (A) Nucleotide sequence alignment of the 3′ end of synC and downstream sequence in serogroup B strains NMB and 1070 (B-301#1), and serogroup C strains FAM18 and 1205 (C-301#1) [Pretty multiple sequence comparison program of the Genetics Computer Group sequence analysis package version 7.3.1 UNIX (33)]. The synC termination codon (TAA) and the synD/E start codons (ATG) are indicated in boldface type. (B) Nucleotide polymorphisms of the B301, C301, and other meningococcal strains within (i) a 909-bp PCR product containing the 5′ ends of both ctrA and synX and the 134-bp intergenic region separating these two genes (bp 1–319 are the 5′ end of ctrA, bp 320–453 are the 134-bp intergenic region, and bp 454–909 are the 5′ end of synX) (26), (ii) a 238-bp PCR product amplified from the 330-bp FKBP gene (28), and (iii) an 803-bp PCR product amplified from the 1128-bp recA gene (36). Regions were sequenced from strains 1070 (B301#1) (B), 1069 (B301#2) (B), FAM18 (C), 1205 (C301#1) (C), 1198 (C301#2) (C), 1204 (C301#3) (C), NMB (B), GA1002 (W-135), F8239 (A), GA0929 (Y), and GA1002 (W-135) and compared with the sequence of other neisserial strains (28, 36). The sequence of strain 1070 (B301#1) was used as the master sequence. Differences from the master sequence are indicated at the nucleotide positions within FKBP, recA, or the ctrA–synX PCR product (shown above), identity at a given position is indicated by a dash and deleted nucleotides are shown by dots.
(ii) Recombinational event.
In contrast to the biosynthesis operon, the 5′ nucleotide sequence of ctrA and the ctrA–synX intergenic region were identical in B-301 strains 1070 and 1069 and C-301 strains 1205, 1198, and 1204, but differed from other B, C, Y, and W-135 strains (Fig. 3B1). For example, the two B-301 and three C-301 strains contained the same ctrA–synX intergenic nucleotide sequence including an 8 bp deletion. In addition, the nucleotide sequence of two genes [recA (36) and fkbp (28)] not linked to capsule expression were also identical in the B-301 and C-301 strains, but the sequence differed by up to 5% from other meningococcal strains (Fig. 3 B2 and B3).
Thus, capsule switching of the epidemic serogroup B/C isolates was the result of substitution of the serogroup B synD polysialyltransferase with the serogroup C synE polysialyltransferase. Upstream of the polysialyltransferases, the recombinational event also appeared to have included the conserved CMP-NANA biosynthesis genes, synX–synC, but did not extend to ctrA or the intergenic region separating ctrA–synX, and did not involve unlinked genes. The downstream recombinational exchange is not clearly established but may have occurred upstream or within galE. PCR studies using primers specific for the 3′ end of synC and the 5′ end of galE (37) indicated that synD/synE were downstream from galE by ≈2 kb in the prototype serogroup B strain, NMB, in the prototype serogroup C strain, FAM18, and in each of the B-301 and C-301 strains. PCR amplification of chromosomal DNA from B-301 and C-301 strains using internal galE-specific primers derived from the NMB galE sequence (37) yielded a 900-bp product. However, this product was not obtained with the serogroup C prototype strains FAM18 and two other non-ET-301 serogroup C strains (GA0078-ET-17 and GA0290-ET-27).
DISCUSSION
The data indicate that capsule switching in N. meningitidis can occur by gene conversion of the capsule polymerase and that this event occurs in vivo. Presumably, co-colonization of serogroup B and C strains in the human nasopharynx and genetic exchange of capsule biosynthesis genes by transformation and allelic-exchange are the events responsible for capsule switching. The high frequency (5–10%) of meningococcal carriage in the human nasopharynx of adults (38), which appears to increase in epidemic settings, may facilitate the chances of capsule switching. These data are supported by molecular analysis of meningococcal strain collections that contain isolates with otherwise identical genetic markers (e.g., ET-type) that express different capsular polysaccharides (3, 18).
In addition to the meningococcal epidemic in the Pacific Northwest, recent cases of meningococcal disease caused by B and C strains with identical serotypes and ET types in the Czech Republic and Canada (39, 40) suggest capsule switching may be common. Indeed, the ability to switch capsules provides a selective advantage to meningococci, inasmuch as they are thereby able to evade killing, opsonization, or neutralization by preexisting anticapsular antibody. Other experimental studies suggest capsule switching may not be an isolated event in meningococci, but appears to occur in encapsulated Streptococcus pneumoniae and Haemophilus influenzae (41, 42).
These data have important implications for understanding meningococcal pathogenesis, for developing vaccine prevention strategies based on capsular polysaccharide, and for epidemiologic investigations of disease due to encapsulated bacteria. For example, meningococci of different serogroups recovered during epidemic outbreaks or from cases of endemic disease can be identical in their expression of other virulence factors (e.g., outer membrane proteins). Because they do not express (α2→8)-linked polysialic acid, the serogroup C ET-5 strains noted in this study may have significance in the current development of ET-5 outer membrane protein vaccines. Meningococcal capsule switching appears to occur among sialic acid-expressing strains by allelic replacement of the sialic acid capsule polymerase. Since the genetic structure responsible for capsule expression in group A meningococci is different (see above), homologous recombination with conversion of serogroup A strains to sialic acid capsule-expressing strains (by introduction of a sialic acid capsule biosynthetic operon) may be less likely.
These data suggest a general strategy by which meningococci and other encapsulated bacteria capable of causing epidemic outbreaks or endemic disease escape vaccine-induced or natural protective immunity. Thus, multivalent vaccines effective against all major capsular serogroups may be needed to control epidemics and possibly endemic disease. Evidence of capsule switching in encapsulated pathogens isolated in future outbreaks should be sought.
Acknowledgments
We thank Dr. Nigel Raymond for help with the PFGE studies and Lane Pucko for manuscript preparation. This work was supported by an interagency agreement and by the Emerging Infections Program of the Centers for Disease Control and Prevention, by Public Health Service Grant A140247 from the National Institutes of Allergy and Infectious Diseases, and by the Research Service of the Department of Veterans Affairs
Footnotes
References
- 1.Apicella M A. In: Principles and Practice of Infectious Diseases. Mandell G L, Douglas R G, Bennett J E, editors. New York: Churchill Livingstone; 1995. pp. 1896–1909. [Google Scholar]
- 2.W.H.O. Fighting Disease, Fostering Development. Geneva: W.H.O.; 1996. [Google Scholar]
- 3.Caugant D A, Frøholm L O, Bøvre K, Holten E, Frasch C E, Mocca L F, Zollinger W D, Selander R K. Proc Natl Acad Sci USA. 1986;83:4927–4931. doi: 10.1073/pnas.83.13.4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang J F, Caugant D A, Li X, Hu X, Pooman J T, Crowe B A, Achtman M. Infect Immun. 1992;60:5267–5282. doi: 10.1128/iai.60.12.5267-5282.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sacchi C T, Tondella M L, de Lemos A P, Gorla M C, Berto D B, Kumiochi N H, Melles C E. J Clin Microbiol. 1994;32:1783–1787. doi: 10.1128/jcm.32.7.1783-1787.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention. Morbid Mortal Week Rep. 1995;44:121–134. [Google Scholar]
- 7.Jackson L A, Schuchat A, Reeves M W, Wenger J D. J Am Med Assoc. 1995;273:383–389. [PubMed] [Google Scholar]
- 8.Wahlen C M, Hockin J C, Ryan A, Ashton F. J Am Med Assoc. 1995;273:390–394. [PubMed] [Google Scholar]
- 9.Riou J Y, Djibo S, Sangare L, Lombart J P, Fagot P, Chippaux J P, Guibourdenche M. Bull WHO. 1996;74:181–187. [PMC free article] [PubMed] [Google Scholar]
- 10.Campos J, Fuste M C, Trujillo G, Saez-Nieto J, Vazquez J, Loren J G, Vinas M, Spratt B G. J Infect Dis. 1992;166:173–177. doi: 10.1093/infdis/166.1.173. [DOI] [PubMed] [Google Scholar]
- 11.Liu T Y, Gotschlich E C, Jonssen E K, Wysocki R J. J Biol Chem. 1971;246:2849–2858. [PubMed] [Google Scholar]
- 12.Liu T Y, Gotschlich E C, Dunne F T, Jonssen E K. J Biol Chem. 1971;246:4703–4712. [PubMed] [Google Scholar]
- 13.Bhattacharjee A K, Jennings J H, Kenny C P, Martin A, Smith I C. Can J Biochem. 1976;54:1–8. doi: 10.1139/o76-001. [DOI] [PubMed] [Google Scholar]
- 14.Zollinger W D, Moran E. Trans R Soc Trop Med Hyg. 1991;85:37–43. doi: 10.1016/0035-9203(91)90339-z. [DOI] [PubMed] [Google Scholar]
- 15.Křížová P, Musílek M. Centr Eur J Public Health. 1994;3:189–194. [PubMed] [Google Scholar]
- 16.Caugant D A, Mocca L F, Frasch C E, Froholm L O, Zollinger W D, Selander R K. J Bacteriol. 1987;169:2781–2792. doi: 10.1128/jb.169.6.2781-2792.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sierra G V, Campa H C, Varcacel N M, Garcia I L, Izquierdo P L, Sotolongo P F, Casanueva G V, Rico C O, Rodriguez C R, Terry M H. NIPH Ann. 1991;14:195–207. [PubMed] [Google Scholar]
- 18.Sacchi C T, Pessoa L L, Ramos S R, Milagres L G, Camargo M C, Hidalgo N T, Melles C E, Caugant D A, Frasch C E. J Clin Microbiol. 1992;30:1734–1738. doi: 10.1128/jcm.30.7.1734-1738.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frosch M, Weisgerber C, Meyer T F. Proc Natl Acad Sci USA. 1989;86:1669–1673. doi: 10.1073/pnas.86.5.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Edwards U, Müller A, Hammerschmidt S, Gerardy-Schahn R, Frosch M. Mol Microbiol. 1994;14:141–149. doi: 10.1111/j.1365-2958.1994.tb01274.x. [DOI] [PubMed] [Google Scholar]
- 21.Swartley J S, Stephens D S. J Bacteriol. 1994;176:1530–1534. doi: 10.1128/jb.176.5.1530-1534.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ganguli S, Zapata G, Wallis T, Reid C, Boulnois G, Vann W F, Roberts I S. J Bacteriol. 1994;176:4583–4589. doi: 10.1128/jb.176.15.4583-4589.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Edwards U, Frosch M. FEMS Microbiol Lett. 1992;96:161–166. doi: 10.1016/0378-1097(92)90397-7. [DOI] [PubMed] [Google Scholar]
- 24.Frosch M, Edwards U, Bousset K, Krausse B, Weisgerber C. Mol Microbiol. 1991;5:1251–1260. doi: 10.1111/j.1365-2958.1991.tb01899.x. [DOI] [PubMed] [Google Scholar]
- 25.Frosch M, Müller D, Bousset K, Müller A. Infect Immun. 1992;60:798–803. doi: 10.1128/iai.60.3.798-803.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Swartley J S, Ahn J H, Liu L-J, Kahler C M, Stephens D S. J Bacteriol. 1996;178:4052–4059. doi: 10.1128/jb.178.14.4052-4059.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hammerschmidt S, Hilse R, van Putten J P M, Gerardy-Schahn R, Unkmeir A, Frosch M. EMBO J. 1996;15:192–198. [PMC free article] [PubMed] [Google Scholar]
- 28.McAllister C F, Stephens D S. Mol Microbiol. 1993;10:13–23. doi: 10.1111/j.1365-2958.1993.tb00899.x. [DOI] [PubMed] [Google Scholar]
- 29.Reeves M W, Perkins B A, Diermayer M, Wenger J D. Emerging Infect Dis. 1995;2:53–54. doi: 10.3201/eid0102.950203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bygraves J A, Marden M C. J Gen Microbiol. 1992;138:523–531. doi: 10.1099/00221287-138-3-523. [DOI] [PubMed] [Google Scholar]
- 31.Wedege E, Hoiby E A, Rosenqvist E, Froholm L O. J Med Microbiol. 1990;31:195–201. doi: 10.1099/00222615-31-3-195. [DOI] [PubMed] [Google Scholar]
- 32.Swartley J S, McAllister C F, Hajjeh R A, Heinrich D W, Stephens D S. Mol Microbiol. 1993;10:361–369. doi: 10.1111/j.1365-2958.1993.tb01956.x. [DOI] [PubMed] [Google Scholar]
- 33.Devereux J, Haeberli P, Smithies O. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tenover F C, Arbeit R D, Goering R V, Mickelsen P A, Murray B E, Persing D H, Swaminathan B. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brandt B L, Pier G B, Goroff D K, Altieri P L, Griffiss J M. J Gen Microbiol. 1980;118:39–43. doi: 10.1099/00221287-118-1-39. [DOI] [PubMed] [Google Scholar]
- 36.Zhou J, Spratt B G. Mol Microbiol. 1992;6:2135–2146. doi: 10.1111/j.1365-2958.1992.tb01387.x. [DOI] [PubMed] [Google Scholar]
- 37.Zhou D, Stephens D S, Gibson B W, Engstrom J, McAllister C F, Lee F K N, Apicella M A. J Biol Chem. 1994;269:11162–11169. [PubMed] [Google Scholar]
- 38.Greenfield S, Sheehe P R, Feldman H A. J Infect Dis. 1971;123:67–73. doi: 10.1093/infdis/123.1.67. [DOI] [PubMed] [Google Scholar]
- 39.Kriz P, Musilek M. In: Abstracts of the Tenth International Pathogenic Neisseria Conference. Zollinger W D, Frasch C E, Deal C D, editors. Baltimore: Conference Organizing Committee; 1996. poster 174482. [Google Scholar]
- 40.Ashton F E, Ryan A, Mancino L, Johnson W, Coulthart M, Kertesz D. In: Abstracts of the Tenth International Pathogenic Neisseria Conference. Zollinger W D, Frasch C E, Deal C D, editors. Baltimore: Conference Organizing Committee; 1996. poster 148431. [Google Scholar]
- 41.Coffey T J, Dowson C G, Daniels M, Zhou J, Martin C, Spratt B G, Musser J M. Mol Microbiol. 1991;5:2255–2260. doi: 10.1111/j.1365-2958.1991.tb02155.x. [DOI] [PubMed] [Google Scholar]
- 42.Kroll J S, Moxon E R. J Bacteriol. 1990;172:1374–1379. doi: 10.1128/jb.172.3.1374-1379.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]