Abstract
The human glioma pathogenesis-related protein (GliPR) is highly expressed in the brain tumor glioblastoma multiforme and exhibits 35% amino acid sequence identity with the tomato pathogenesis-related (PR) protein P14a, which has an important role for the plant defense system. A molecular model of GliPR was computed with the distance geometry program diana on the basis of a P14a–GliPR sequence alignment and a set of 1,200 experimental NMR conformational constraints collected with P14a. The GliPR structure is represented by a group of 20 conformers with small residual diana target function values, low amber-energies after restrained energy-minimization with the program opal, and an average rms deviation relative to the mean of 1.6 Å for the backbone heavy atoms. Comparison of the GliPR model with the P14a structure lead to the identification of a common partially solvent-exposed spatial cluster of four amino acid residues, His-69, Glu-88, Glu-110, and His-127 in the GliPR numeration. This cluster is conserved in all known plant PR proteins of class 1, indicating a common putative active site for GliPR and PR-1 proteins and thus a functional link between the human immune system and a plant defense system.
The glioma pathogenesis-related protein (GliPR) is highly expressed in the tumor glioblastoma multiforme (1), which arises from brain immune cells and accounts for over 65% of all human primary brain tumors (2). GliPR was found in all glioma cell lines and tumors studied, but was not detectable in any normal fetal or adult tissues, including normal brain, suggesting that GliPR plays an important role for tumor growth. High levels of GliPR expression can also be induced with phorbol ester in macrophages (1), which are active at the front line of the human immune system. RTVP-1, another protein that was recently found in glioblastoma multiforme (3), is almost identical to GliPR. Compared with GliPR, RTVP-1 possesses both an additional N-terminal signal sequence and a C-terminal putative transmembrane segment. Although RTVP-1 was found to be expressed also in various normal tissues, it cannot be excluded that its high expression in the tumor cells is also related to their malignant properties. GliPR and RTVP-1 exhibit high sequence homology with the plant pathogenesis-related proteins of group 1 (PR-1 proteins), which play a central role for the defense system of plants (4), for example, during the manifestation of systemic acquired resistance (5). Because proteins with a sequence identity larger than 30% are commonly believed to adopt the same fold (6), this homology is suggestive of a structural link between the human immune system and the defense system of plants (1). To follow up on this suggestion and in view of the possible role of GliPR as a potential target for drugs interfering with tumor growth, we further investigated structural similarities between GliPR and the PR-1 proteins on the level of the three-dimensional structure.
METHODS
The three-dimensional structure of GliPR was predicted on the basis of the high-quality NMR structure of P14a (7). Therefore, a subset of the input used for the P14a structure calculation (Table 1) was selected as follows: First, we identified those upper distance constraints that connect corresponding proton pairs in P14a and GliPR. For conserved residues, stereospecific assignments of constraints to isopropyl methyls (Val-38, Leu-54, Val-131, Val-132 in GliPR) and β-methylene protons (in total 18) were retained. Second, the constraints for the backbone dihedral angles φ and ψ measured for P14a were retained, except where l-amino acid residues in P14a are either deleted or replaced by Pro or Gly in GliPR. Third, the constraints for the dihedral angles χ1 obtained for P14a were kept for sites with identical amino acids in both proteins. Fourth, we introduced constraints for eight central hydrogen bonds (8) that were identified in the four-stranded β-sheet of P14a. The resulting data set, which consists of 56% of the constraints obtained with P14a (Table 1), was used as input for structure calculations of GliPR by using the program diana (9). Subsequent restrained energy-minimization in a water bath with the amber force field (10) was performed with the program opal (11). The same protocol was followed as described in ref. 7 for P14a. Clefts on the protein surface and their volumes were identified by using the program surfnet (12).
Table 1.
Structure calculation for human GliPR, using an input of conformational constraints obtained with the NMR- and homology-based approach described in the text and comparison with the NMR structure of P14a (7)
| GliPR | P14a | |
|---|---|---|
| Conformational constraints | ||
| Total number of constraints | 1,216 | 2,021 |
| Upper distance limits | 932 | 1,692 |
| Disulfide bond constraints* | 18 | 18 |
| β-Sheet hydrogen bond constraints* | 32 | — |
| ψ,φ-dihedral angle constraints | 202 | 228 |
| χ1-dihedral angle constraints | 32 | 83 |
| 20 diana conformers used to represent the structure, after energy minimization (average value ± SD) | ||
| diana target function, Å2† | 6.5 ± 0.7 | 3.1 ± 0.7 |
| NOE constraint violations, Å: Sum | 7.86 ± 0.49 | 9.20 ± 0.64 |
| Max | 0.11 ± 0.14 | 0.10 ± 0.01 |
| Dihedral angle violations, °: Sum | 25.6 ± 4.3 | 45.0 ± 5.1 |
| Max | 2.4 ± 0.3 | 2.3 ± 0.2 |
| amber energies, kcal⋅mol−1: Van der Waals | −358 ± 21 | −570 ± 12 |
| Electrostatic | −4,203 ± 182 | −4,576 ± 74 |
| Average rms deviations for different atom selections (average value ± SD) | ||
| Backbone N, Cα, C′ of constrained segments‡ | 1.66 ± 0.26 | 0.88 ± 0.08 |
| All heavy atoms of constrained segments‡ | 2.37 ± 0.24 | 1.30 ± 0.09 |
| Backbone of the regular secondary structures§ | 0.98 ± 0.19 | 0.45 ± 0.08 |
| Backbone of constrained segments‡ + core side chains¶ | 1.60 ± 0.21 | 0.91 ± 0.07 |
In GliPR, the disulfide bonds between C65–C146, C121–C125, and C141–C163, and the hydrogen bonds W91⋅⋅⋅F161, A142⋅⋅⋅I162, Q144⋅⋅⋅H160, H160⋅⋅⋅Q144, F161⋅⋅⋅W91, I162⋅⋅⋅A142, C163⋅⋅⋅N89, and N164⋅⋅⋅G140 in the β-sheet were constrained following ref. 8.
Before energy minimization.
“Constrained segments” of GliPR are those parts of the polypeptide chain for which conformational constraints were available from the NMR measurements with P14a, which includes the residues 21–38, 44–72, 87–146, 148–151, and 159–176 (see also Fig. 1).
The secondary structure elements in GliPR are as follows: α-helix I, residues 24–37; α-II, 52–64; α-III, 100–111; α-IV, 127–132; β-strand A, 49–50; β-B, 87–92; β-C, 138–145; β-D, 159–166 (see Fig. 1).
The “molecular core” of GliPR includes the residues 27, 28, 30, 31, 34, 35, 38, 45, 48, 50, 54, 55, 58, 61, 62, 65, 67, 89–91, 98, 103, 104, 106–108, 114, 116, 121, 125, 128–131, 133, 136, 139–142, 144, 146, 160, 161, 163–165, 170, 175, 176, which are indicated with “1” or “2” in Fig. 1. The displacements calculated for all these residues except C65, F67, C121, and C146, after superimposing the backbone heavy atoms N, Cα, and C′ of the “constrained segments” and the side chain heavy atoms of the core residues for minimal rms deviation, are smaller than 2.3 Å. The corresponding displacements in the NMR solution structure of P14a (7) were all smaller than 1.9 Å.
Amino acid sequence alignments were performed with the program clustalw (13). To identify sequence homologues of GliPR, the databases Swiss-Prot, Protein Identification Resource (PIR), and GenBank were screened by using the program blast (14) with default parameters (National Center for Biotechnology Information’s blast WWW server; http://www.dot.imgen.bom.tmc.edu:9331/seq-search/protein-search.html). The cutoff for the score was set to 70, and protein fragments and nearly identical sequences were eliminated. A genealogical tree was subsequently constructed for the sequence homologues with the program allall (15). For secondary structure prediction we employed the program phdsec (16), and Fig. 2 was generated by using the molecular graphics program molmol (17).
Figure 2.
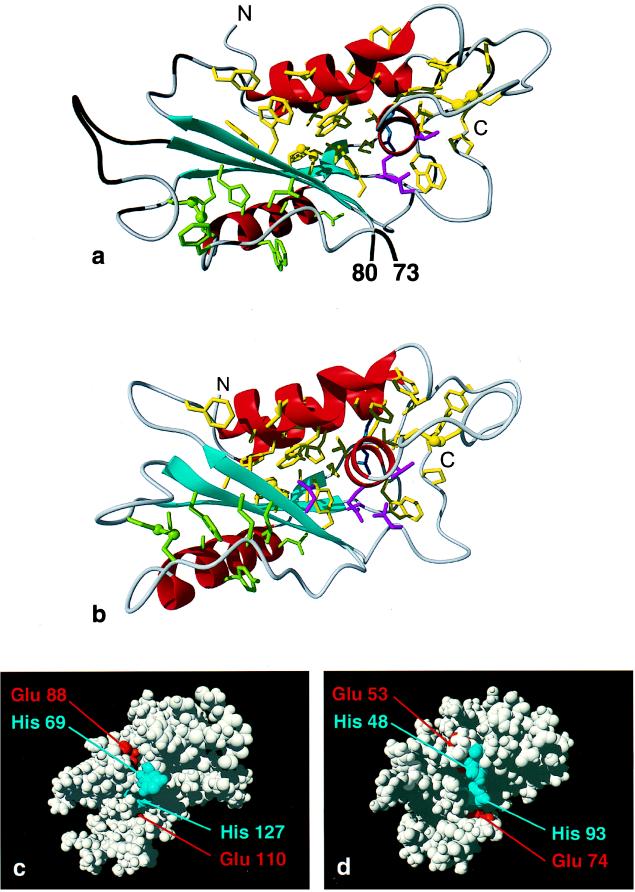
(a) Ribbon drawing of the diana conformer of GliPR with the lowest residual target function after energy minimization. The input was obtained from a NMR- and homology-based approach (see text). The helices I to IV are shown in red, the β-strands A to D in cyan, and other polypeptide segments in gray, and the polypeptide chain ends are indicated with N and C. Polypeptide segments that represent insertions into the amino acid sequence of P14a (see Fig. 1) are depicted in black. Amino acid side chains that form a large hydrophobic cluster associated with the β-sheet and the α-helices I, III, and IV are shown in yellow, with the two disulfide bonds C121–125 and C141–C163 drawn as ball and stick models, and side chains that belong to a smaller hydrophobic cluster associated with the β-sheet, α-helix II, and the disulfide bond C65–C146 are green. Polar side chains in the hydrophobic core are identified by drawing His-31, Thr-129, Gln-130, and Asn-170 in magenta and Arg-35 in blue. (b) Best diana conformer from the NMR structure determination of P14a (7) in the same orientation and with the same color code as used for GliPR in a. The magenta-colored polar side chains are His-11, Thr-95, Gln-96, and Asn-128, and the blue side chain is Arg-15. (c and d) Space-filling representations of GliPR and P14a, respectively, showing the putative active site (see text), with histidinyl and glutamyl residues depicted in blue and red, respectively. For clarity, in GliPR the insertions relative to the sequence of P14a are omitted (Fig. 1).
RESULTS AND DISCUSSION
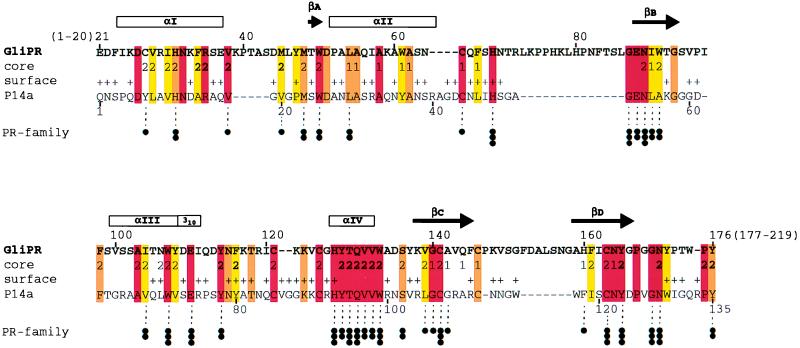
The sequence alignment of the 219-residue protein GliPR and the 135-residue protein P14a of Fig. 1 shows that GliPR possesses a 20-residue N-terminal extension and a 43-residue C-terminal extension of the polypeptide chain. Moreover, there are four peptide segments in GliPR, comprising residues 39–43, 73–86, 147, and 152–158, that are inserted into the polypeptide chain of P14a, whereas the segments 40–43, 86–87, and 133 of P14a are not present in GliPR. P14a is related with other PR-1 proteins by numerous highly conserved residues that have essential structural roles in the architecture of the P14a core (7). Although the sequence homology of P14a with GliPR is significantly lower (35% identity) than with other PR-1 proteins (>54% identity), similar conservation is readily apparent in Fig. 1, and the additions to the sequence of P14a are all located either as insertions between regular secondary structures or as extensions at the chain ends (Fig. 1). Hence, the sequence alignment of GliPR and P14a in Fig. 1 indicates similarity also on the level of the three-dimensional structures of the two proteins.
Figure 1.
Sequence alignment of the proteins GliPR and P14a. The one-letter amino acid code is used, and the numbering for GliPR is indicated above its sequence. The locations of the regular secondary structure elements identified for P14a (7) are depicted at the top, with the four α-helices and the 310-helix represented by boxes marked αI, αII, αIII, αIV, and 310, and the four β-strands, by the arrows βA, βB, βC, and βD. In the row “core” the residues forming a small hydrophobic cluster associated with the β-sheet and helix II (Fig. 2 a and b) are indicated 1, those constituting the larger core associated with the β-sheet and helices I, III, and IV (Fig. 2 a and b) with 2, and residues of helix IV and those directly contacting helix IV in P14a (Fig. 2 a and b) with 2. In the row “surface,” + identifies residues that exhibit a solvent accessibility larger than 25% in at least one of the 20 energy-minimized diana conformers of P14a. Color code: residues conserved between GliPR and P14a are orange, and those conserved also within the whole group of known plant PR-1 proteins (see figure 10 in ref. 7) are red. Conserved hydrophobicity of Ala, Val, Leu, Ile, Tyr, Phe, uncharged His, Trp, Pro, Met, or Cys in GliPR and all PR-1 proteins is highlighted in yellow. Conservation within the “PR-protein superfamily” (see text and Fig. 3), derived from multiple alignment of all 26 representative members, is indicated at the bottom: three dots label residues that are strictly conserved within the superfamily, two dots denote conservation of side chain character (hydrophobic: Ala, Val, Leu, Ile, Tyr, Phe, Trp, Met, Cys, and Pro; hydroxyl-bearing: Ser and Thr; or hydrogen bond acceptors: uncharged His, Asp, Asn, Glu, and Gln), and one dot indicates mutations from hydrophobic amino acids to Gly, Thr, or His.
Direct three-dimensional structure comparison of GliPR and P14a was based on the high-quality NMR structure of P14a (7). The structure calculations performed for GliPR with the input adapted from the experiments with P14a (see Methods) converged with low residual target function values and constraint violations (Table 1), showing that the distance geometry algorithm found sterically allowed three-dimensional folds as well as good side chain packing in the molecular core of GliPR. This is further evidenced by low van der Waals and electrostatic amber energies (10), which are comparable to those obtained for the high-quality NMR solution structure of P14a. The average rms deviations among the conformers selected to represent the structure of GliPR (Table 1) correspond to those of a good-quality NMR structure determination. The presently employed approach for homology modeling based on a high-quality NMR solution structure has the obvious advantage that the sampling of conformation space performed for the NMR structure is transferred to the target sequence, which avoids steric clashes that might arise if a single conformer of the template molecule were used to derive a set of distance restraints (for a recent review of sequence homology-based structure prediction, see ref. 18).
In the molecular model of GliPR (Fig. 2a) obtained with this NMR- and homology-based approach the regular secondary structure elements form the same, so far unique, α–β–α sandwich as P14a (Fig. 2b), where the intermediary layer is formed by a four-stranded mixed β-sheet of topology +3x, −2x, +1. The visual impression of near-identity of the polypeptide folds of GliPR and P14a is substantiated by a rms deviation of 0.88 Å calculated between the mean coordinates of the two sets of 20 diana conformers for the backbone heavy atoms of the 128 residues that are aligned in identical positions in Fig. 1. The spatial arrangement of regular secondary structure elements leads to a bipartite hydrophobic core associated with the two layer interfaces of the sandwich, which is further stabilized by the three disulfide bonds. The α-helix IV is completely buried between the β-sheet and the two-helix bundle of helices I and III, and it is fully integrated into the larger of the two clusters. For the insertions and extensions of the polypeptide chain relative to P14a (Fig. 1), for which the presently used approach does not yield conformational constraints, secondary structure prediction did not indicate any regular secondary structures.
Inspection of the three-dimensional structures shows that only His-69 and His-127 are solvent exposed and strictly conserved in GliPR and the family of plant PR-1 proteins (Fig. 1), which makes these residues prime candidates for a role in functionally active sites (19). These histidyls are in close spatial proximity to the highly conserved glutamates 88 and 110, which also possess no obvious structural role. Hence, the cluster comprising these two histidines and two glutamates is a likely candidate for the so-far-unknown active site of both GliPR (Fig. 2c) and the plant PR-1 proteins represented here by P14a (Fig. 2d). There are no other conserved hydrophobic, polar, or charged amino acid combinations on the protein surface that could account for a preserved interaction of the functionally related plant PR-1 proteins (4, 5) with their biomolecular targets. To further evaluate the likelihood that the clusters identified in Fig. 2 c and d are related to a functional site, we identified the largest surface clefts of P14a and GliPR. The calculation showed that the cleft containing the putative active site residues is the largest cleft on both protein surfaces (Fig. 2 c and d). Because statistically enzyme active sites are typically located in the largest surface cleft of a protein (20, 21) this suggests that GliPR and plant PR-1 proteins might function as enzymes with the putative active sites shown in Fig. 2 c and d. In these, His-127 is the C-terminal residue of α-helix IV, which is completely buried within the molecular core (Fig. 2 a and b). All residues of this helix and the directly succeeding Trp are strictly conserved in GliPR and the plant PR-1 proteins and the residues that contact helix IV are also highly conserved (Fig. 1), so that the distinct hydrogen bonding network identified within the core of P14a (see figure 8 of ref. 7) is maintained in GliPR. The embedding of helix IV within the core and its positional fine adjustment with a network of hydrogen bonds (22) could thus be the structural basis of the function of PR proteins. The dipole of helix IV may even serve for proper tuning of the pKa value of His-127 (23). In P14a, His-48, Glu-53, and His-93 (Fig. 2d) are flexibly disordered in solution (7), but they could adopt the conformation found for the active site histidyl and glutamyl residues in several Zn proteases. However, no Zn2+ ion could be unambiguously identified in an x-ray crystal structure of P14a obtained with crystals that were grown in the presence of Zn2+ (V. Mikol, personal communication). The two histidines could also represent the active site of a ribonuclease, but biochemical assays performed to detect ribonuclease activity were negative (E. Mösinger, personal communication).
Overall, the strict conservation of the putative active site residues (Fig. 2 c and d) strongly suggests that human GliPR and plant PR-1 proteins operate according to the same molecular mechanism, which establishes a possible functional link between the human immune system and a plant defense system. In this context the evolutionary origin of these PR proteins is of keen interest. The close structural similarity of the molecular cores of P14a and GliPR (Fig. 2 a and b) speaks against the convergent evolution of two independent ancestors, because even if this had led to the same fold and active site geometry of the two proteins it would very likely have resulted in different architectures for the molecular cores. The alternative assumption that P14a and GliPR arose from a common ancestor raises questions concerning the role of the ancestor protein, which presumably functioned at a very early stage in evolution when living organisms separated into different kingdoms and the presently known defense systems probably did not yet exist. A possible solution would be that these proteins have been horizontally transferred (24) at a much later stage of evolution—e.g., PR-1 proteins might originally have evolved in plants and subsequently been recruited by mammals.
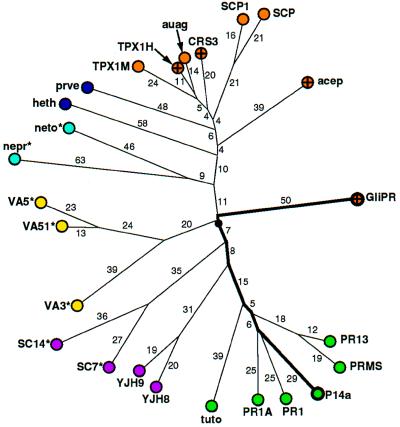
To follow up this possibility we constructed a genealogical tree (Fig. 3) for a set of 26 sequence homologues of GliPR. The accepted point mutation (PAM) distances between the sequences and the resulting topology of the tree suggest that GliPR and P14a arose from a common ancestor that has evolved into a large “PR-protein superfamily.” Its members include human GliPR, mammalian sperm coating proteins (25), plant PR-1 proteins, allergens of insect venoms (26), and snake or lizard toxins, and are thus found in the three kingdoms of animals, plants, and fungi. The underlying molecular mechanism for the action of these proteins is unknown. The alignment of their amino acid sequences with GliPR revealed a striking conservation of amino acid residues corresponding to the buried helix IV of GliPR and P14a (Figs. 1 and 2 a and b) within the superfamily (see also refs. 3, 25, and 26). In addition, numerous residues aligned with residues that form the molecular core show significant conservation (Fig. 1), in particular those that participate in the hydrogen bonding network in the core of GliPR (His-33, Asn-89, Tyr-165; see also figure 8 in ref. 7). The evolutionary distances between GliPR and P14a (PAM = 120), and between GliPR and the other members of the superfamily are comparable (Fig. 3). It may thus well be that the α–β–α sandwich fold of P14a and GliPR is characteristic for the entire superfamily. Moreover, the putative active site residues His-69, Glu-110, and His-127 of GliPR (Fig. 2 c and d) are conserved in all but one sequence, and Glu-88 of GliPR is conserved in 19 of the 26 sequences, while the other 7 possess a Gln in the corresponding position (Fig. 3), indicating that all proteins of the PR-protein superfamily could, on the basis of present knowledge of the molecular structure, operate according to the same molecular mechanism.
Figure 3.
Genealogical tree of the “PR-protein superfamily,” where the distances are given in PAM units. For clarity, distances with PAM = 1 or 2 are not indicated. Color code: mammals, orange (human proteins are further marked with a cross); plants, green; fungi, magenta; insects, yellow; nematodes, light blue; and other vertebrates, dark blue. The branches are labeled either with the first four letters of the Swiss-Prot database code in capital letters, or with an abbreviation derived from the GenBank and PIR database codes. Proteins for which the putative active site (Fig. 2 c and d and text) is not strictly conserved are identified as follows: where Glu-88 of GliPR is exchanged with Gln there is an asterisk. For “heth” the residue corresponding to His-69 of GliPR is exchanged with Glu. Abbreviations: from GenBank and PIR, acep, acidic epididymal glycoprotein, S80310; auag, autoantigen 1 of the guinea pig sperm acrosome, U35712; heth, helothermine precursor (Mexican beaded lizard), U13619; nemt, gene sequence from chromosome III of nematode Caenorhabditis elegans, Z68108; nepr, nematode protective antigen, A28112; prvn, Prepro-Cys-rich venom protein, U59447; tuto, tumor-related protein from tobacco; from Swiss-Prot, CRS3_HUMAN, PR13_HORVU, PR1A_TOBAC, PR1_ARATH, PRMS_MAIZE, SC14_SCHCO, SC7_SCHCO, SCP1_MOUSE, SCP_RAT, SGP28_X94323, TPX1_HUMAN, TPX1_MOUSE, VA3_SOLIN, VA51_VESCR, VA5_VESPE, VJH8_YEAST, and YJH9_YEAST.
Acknowledgments
We thank Drs. V. Mikol and E. Mösinger for communicating unpublished results. Financial support was obtained from the Schweizerischer Nationalfonds (project 31.49047.96) and a fellowship to C.F. from the Schweizerische Bundesstipendienkommssion.
ABBREVIATIONS
- PR
pathogenesis-related
- GliPR
glioma PR protein
- PAM
accepted point mutation
- RTVP-1
related to testes-specific, vespid, and pathogenesis proteins
- PIR
Protein Identification Resource
References
- 1.Murphy E V, Zhang Y, Zhu W, Biggs J. Gene. 1995;159:131–135. doi: 10.1016/0378-1119(95)00061-a. [DOI] [PubMed] [Google Scholar]
- 2.Morris J H, Schoene W C. In: The Pathological Basis of Disease. Robbins S L, Cotran R F, Kumar V, editors. Philadelphia: Saunders; 1984. pp. 1401–1456. [Google Scholar]
- 3.Rich T, Chen P, Furman F, Huynh N, Israel M A. Gene. 1996;180:125–130. doi: 10.1016/s0378-1119(96)00431-3. [DOI] [PubMed] [Google Scholar]
- 4.Bol J F, Linthorst H J M, Cornelissen B J C. Annu Rev Phytopathol. 1990;28:113–138. [Google Scholar]
- 5.Ryals J, Uknes S, Ward E. Plant Physiol. 1994;104:1109–1112. doi: 10.1104/pp.104.4.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Orengo C A, Jones D T, Thornton J M. Nature (London) 1994;372:631–634. doi: 10.1038/372631a0. [DOI] [PubMed] [Google Scholar]
- 7.Fernández C, Szyperski T, Bruyère T, Ramage P, Mösinger E, Wüthrich K. J Mol Biol. 1997;266:576–593. doi: 10.1006/jmbi.1996.0772. [DOI] [PubMed] [Google Scholar]
- 8.Williamson M P, Havel T F, Wüthrich K. J Mol Biol. 1985;182:295–315. doi: 10.1016/0022-2836(85)90347-x. [DOI] [PubMed] [Google Scholar]
- 9.Güntert P, Braun W, Wüthrich K. J Mol Biol. 1991;217:517–530. doi: 10.1016/0022-2836(91)90754-t. [DOI] [PubMed] [Google Scholar]
- 10.Weiner P K, Kollman P A, Nguyen D, Case D A. J Comp Chem. 1986;7:230–252. doi: 10.1002/jcc.540070216. [DOI] [PubMed] [Google Scholar]
- 11.Luginbühl P, Güntert P, Billeter M, Wüthrich K. J Biomol NMR. 1996;8:136–146. doi: 10.1007/BF00211160. [DOI] [PubMed] [Google Scholar]
- 12.Laskowski R A. J Mol Graph. 1995;13:323–330. doi: 10.1016/0263-7855(95)00073-9. [DOI] [PubMed] [Google Scholar]
- 13.Thompson J D, Higgins D G, Gibson T J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 15.Gonnet G H, Cohen M A, Benner S A. Science. 1992;256:1443–1445. doi: 10.1126/science.1604319. [DOI] [PubMed] [Google Scholar]
- 16.Rost B, Sander C. Proteins. 1994;19:55–72. doi: 10.1002/prot.340190108. [DOI] [PubMed] [Google Scholar]
- 17.Koradi R, Billeter M, Wüthrich K. J Mol Graph. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 18.Sali A. Curr Opin Biotechnol. 1995;6:437–451. doi: 10.1016/0958-1669(95)80074-3. [DOI] [PubMed] [Google Scholar]
- 19.Schulz G E, Schirmer R H. Principles of Protein Structure. Berlin: Springer; 1979. [Google Scholar]
- 20.Laskowski R A, Luscombe N M, Swindells M B, Thornton J M. Protein Science. 1996;5:2438–2452. doi: 10.1002/pro.5560051206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peters K P, Fauck J, Frömmel C. J Mol Biol. 1996;256:201–213. doi: 10.1006/jmbi.1996.0077. [DOI] [PubMed] [Google Scholar]
- 22.Sauer R T. Folding Design. 1996;1:R27–R30. doi: 10.1016/S1359-0278(96)00015-6. [DOI] [PubMed] [Google Scholar]
- 23.Sancho J, Serrano L, Fersht A R. Biochemistry. 1992;31:2253–2258. doi: 10.1021/bi00123a006. [DOI] [PubMed] [Google Scholar]
- 24.Syvanen M. Annu Rev Genet. 1994;28:237–261. doi: 10.1146/annurev.ge.28.120194.001321. [DOI] [PubMed] [Google Scholar]
- 25.Kjeldsen L, Cowland J B, Johnson A H, Borregaard N. FEBS Lett. 1996;380:246–250. doi: 10.1016/0014-5793(96)00030-0. [DOI] [PubMed] [Google Scholar]
- 26.Lu G, Villalba M, Coscia M R, Hoffman D R, King T P. J Immunol. 1993;150:2823–2830. [PubMed] [Google Scholar]