Abstract
Cultivation methods have contributed to our present knowledge about the presence and diversity of microbes in naturally occurring communities. However, it is well established that only a small fraction of prokaryotes have been cultivated by standard methods and, therefore, the prokaryotes that are cultivated may not reflect the composition and diversity within those communities. Of the two prokaryotic phylogenetic domains, Bacteria and Archaea, members of the former have been shown to be ubiquitous in nature, with ample evidence of vast assemblages of uncultured organisms. There is also now increasingly compelling evidence that the Archaea, which were once thought to occupy a limited number of environments, are also globally widespread. Here we report the use of molecular phylogenetic techniques, which are independent of microbial cultivation, to conduct an assessment of Archaea in a soil microbial community. Small subunit ribosomal RNA genes of Archaea were amplified from soil and cloned. Phylogenetic and nucleotide signature analyses of these cloned small subunit ribosomal RNA gene sequences revealed a cluster of Archaea from a soil microbial community that diverge deeply from the crenarchaeotal line of descent and has the closest affiliation to the lineage of planktonic Archaea. The identification and phylogenetic classification of this archaeal lineage from soil contributes to our understanding of the ecological significance of Archaea as a component of microbial communities in non-extreme environments.
Keywords: small subunit ribosomal RNA, planktonic, microbial communities, microbial diversity, molecular ecology
Assessment of the microbial diversity in the environment has long challenged microbiologists and microbial ecologists. Few microbes have a sufficiently distinct cellular morphology to be identified by microscopic techniques. Cultivation of microbes as a means to characterize microbial communities has severe limitations, since the majority of microbes observed microscopically in an environmental sample cannot be cultivated by standard methods (for reviews see refs. 1 and 2). For this reason, methods that are independent of culturing are important to characterize the diversity of microorganisms in their environments. One such method is the molecular phylogenetic analysis of small subunit ribosomal RNA (SSU rRNA) gene sequences (3–15). This approach has resulted in the discovery of entirely new phylogenetic lineages, some of which are major constituents of the environmental communities that were not detected by traditional cultivation techniques (1, 16).
Many studies have focused on the identification and diversity of prokaryotes of the domain Bacteria (17) in environmental assemblages of microorganisms. Rather less attention has been given to the diversity and ecological significance of prokaryotes of the domain Archaea (17). This may be due to the fact that the majority of Archaea have been isolated from extreme environments or specialized ecological niches. All cultured members of the Crenarchaeota, one of the two kingdoms of Archaea, are thermophiles. The other kingdom of Archaea, the Euryarchaeota, is comprised of extreme halophiles, sulfur reducers, sulfate reducers, thermophilic heterotrophs, and methanogens. However, recent molecular phylogenetic studies (5, 7, 8, 11, 18, 19) have indicated that Archaea may be more diverse and widespread than what is represented by cultured members of the Archaea.
Soil is a familiar, yet poorly characterized, microbial environment in which less than 0.1% of the microscopically observed microorganisms present are cultured by standard techniques (20, 21). Recent surveys of soil microbial populations with molecular phylogenetic techniques revealed an enormous diversity of as yet uncharacterized microbes (6, 11, 15). In these studies, total DNA was isolated directly from soil samples, SSU rRNA gene sequences were amplified by PCR, and the amplified SSU rRNA gene sequences were characterized by nucleotide sequence and phylogenetic analyses. The primer sets used for the amplification were designed to selectively amplify SSU rRNA gene sequences from most members of the domain Bacteria (6) or from most members of the three primary phylogenetic domains, Archaea, Bacteria, and Eucarya (11, 15). Using the latter primer set, one group identified and cloned a partial SSU rRNA gene sequence (FIE16) that was most closely related to those of Archaea (11).
Our work was designed to assess the diversity of Archaea present in a soil microbial community and to serve as the starting point for a study of the ecology of Archaea in soil. Small subunit rRNA gene sequences of Archaea were selectively amplified from total DNA extracted directly from soil and cloned. Sequence and phylogenetic analyses of these clones identified a group of Archaea in a soil environment that diverge deeply from the crenarchaeal line of descent and has closest affiliation to the lineage of planktonic Archaea (22).
MATERIALS AND METHODS
Soil Sampling and Analyses.
A subsurface (2–10 cm) soil sample was collected in August 1995, from the West Madison Agricultural Research Station (Madison, WI), and stored on ice until processed. The soil type is a Plano silt loam containing 61% sand, 23% silt, and 16% clay, with 1.7% organic matter. The soil pH was 7.0. Soil analyses were performed by the Soil Testing Laboratory of the University of Wisconsin–Madison, as described (23).
Extraction and Purification of Total DNA from Soil Sample.
Total DNA was isolated from the soil sample by a modification of the method described by Porteus et al. (24), which is designed to isolate total DNA from a variety of cell types. Five hundred milligrams of soil were resuspended in 500 μl of Solution A (250 mM NaCl/100 mM Na2EDTA) and sonicated for 3 min in a Branson 2200 bath sonicator. Lysozyme was then added to 0.5 mg/ml and the mixture was incubated at 37°C for 30 min with occasional agitation. Proteinase K was added to a final concentration of 2.0 mg/ml and the mixture was incubated for a further 30 min. After incubation, 500 μl of Solution B [250 mM NaCl/100 mM Na2EDTA/4% (wt/vol) SDS] and 75 μl of 5 M guanidine isothiocyanate were added and the mixture was gently agitated by inversion. The mixture was then incubated at 68°C for 1 hr with occasional agitation. After incubation, the sample was mixed 1:1 with 0.1-mm diameter zirconia/silica beads and homogenized in a Mini-Beadbeater (Type BX-4; Biospec Products, Bartlesville, OK) at 3000 rpm for 45 sec. The samples were centrifuged to remove the beads and the supernatant fluid was recovered. One hundred and fifty microliters of hexadecyltrimethyl ammonium bromide (CTAB) extraction solution [2% (wt/vol) CTAB/100 mM Tris·Cl, pH 8.0/20 mM Na2EDTA/1.4 M NaCl] was added to the supernatant fluid and mixed by inversion. The mixture was incubated at 65°C for 15 min with occasional agitation and then extracted sequentially with equal volumes of chloroform:isoamyl alcohol (24:1), phenol:chloroform:isoamyl alcohol (24:24:1), and chloroform:isoamyl alcohol (24:1). An equal volume of isopropanol was added to the recovered supernatant fluid and total DNA was recovered by centrifugation. Total DNA was resuspended in 500 μl of 10 mM Tris·Cl (pH 8.0) and purified for PCR amplification by four rounds of ultrafiltration using Microcon-100 microconcentrators (Amicon).
Amplification and Cloning of Archaeal SSU rRNA Genes.
Purified total DNA was used as template for PCR amplification. The oligonucleotide primer sequences used for amplification were 23FPL (8) (5′-GCGGATCCGCGGCCGCTGCAGAYCTGGTYGATYCTGCC-3′) and 1492R (25) (5′-GGYTACCTTGTTAACGACTT-3′), where R is a purine and Y is a pyrimidine. The underlined sequence in oligonucleotide 23FPL corresponds to a NotI recognition site that was used for directional cloning of amplified products. This primer set was designed to amplify selectively nearly full-length (1.4 kb) archaeal SSU rRNA genes and could amplify >95% of the archaeal SSU rRNA gene sequences in the prokaryotic SSU data base of the Ribosomal Database Project (RDP) (release date May 18, 1995) at the University of Illinois, Urbana (22). PCR amplification was performed with a RoboCycler Gradient 96 Temperature Cycler (Stratagene). The cycling parameters were: 1 min denaturation at 94°C followed by 30 cycles of 94°C for 30 sec, 55°C for 1.5 min, and 72°C for 2.5 min. PCR amplification reaction conditions were: 10 ng of template, 40 pmol of each primer, 200 μM of each dNTP (Boehringer Mannheim), 1.25 Pfu DNA polymerase (Stratagene), and 5 μl of 10× Pfu DNA polymerase reaction buffer in a 50-μl reaction volume. Positive (containing 20 ng of Sulfolobus acidocaldarius genomic DNA) and negative (containing no exogenous template) control reactions were also performed. No amplified products were observed with the negative control reaction and an amplified product of the expected size (1.4 kb) was observed when the template was S. acidocaldarius or total DNA isolated from soil. Amplified products from four independent reactions using total DNA from soil as template were pooled, purified with QIAquick PCR purification columns (Qiagen, Chatsworth, CA), digested with the restriction enzyme NotI (Stratagene), and again purified by spin column purification. Digested products were directionally cloned by ligation to NotI–HincII digested pGEM11Zf(+) (Promega).
Nucleotide Sequencing of Cloned SSU rRNA Genes.
The nucleotide sequences of the cloned SSU rRNA genes were determined using an automated DNA sequencer (Applied Biosystems model 377). Plasmid DNA was purified with QIAprep plasmid purification kits (Qiagen) and used as template for PCR cycle sequencing with Prism Ready Reaction Dyedeoxy Terminator Sequencing Kit (Applied Biosystems). SSU rRNA-specific (8, 25) and M13 forward and reverse oligonucleotide primers (26) were used to determine the nucleotide sequences of the cloned SSU rRNA genes. An additional archaeal-based sequencing primer, 285FA (5′-AGCCCGGAGATGGGCACTGAG-3′), was designed and used for sequence analysis. Nucleotide sequences of the cloned SSU rRNA genes were obtained by sequencing both template strands at least twice.
Sequence and Phylogenetic Analyses.
Analyses of the cloned SSU rRNA gene sequences were performed using similarity_rank and align_sequence from the RDP and Basic Local Alignment Search Tool (blast) (27). These analyses were used to determine that the cloned sequences were SSU rRNA gene sequences and to estimate the degree of similarity to other SSU rRNA gene sequences. The data bases used for RDP and blast analyses were the prokaryotic SSU rRNA data base and the non-redundant nucleotide sequence data base, which contains all non-redundant GenBank, European Molecular Biology Laboratory, DNA Data Base in Japan, and Protein Data Base sequences, respectively.
Nucleotide sequences of the cloned SSU rRNA genes were manually aligned into putative secondary structures and were also submitted to the check_chimera program at the RDP to detect the presence of chimeric artifacts.
Sequences were manually aligned to SSU rRNA sequence data from the RDP based on primary and secondary structure considerations using the Genetic Data Environment (GDE) multiple sequence editor distributed by the RDP. Phylogenetic analyses were restricted to nucleotide positions that were unambiguously alignable in all sequences. Least-squares distance matrix analyses (28), based on evolutionary distances, were estimated from similarity values by using the correction of Kimura (29). Phylogenetic analyses using neighbor joining and parsimony methods were performed using the Phylogenetic Inference Package (phylip) (version 3.57c, J. Felsenstein, University of Washington, Seattle). Maximum likelihood analyses were performed using fastdnaml (version 1.1.1; distributed by RDP; ref. 22). For transversion-distance and transversion-parsimony analyses (30), the base composition of the unambiguously aligned sequences was modified to reflect only purines and pyrimidines. For the phylogenetic analyses, the order in which the sequences were added was jumbled so as to avoid potential bias introduced by the order of sequence addition. Bootstrap analysis (31) was used to provide confidence estimates for phylogenetic tree topologies.
RESULTS
To assess the presence and diversity of Archaea in soil, total DNA was isolated directly from soil and archaeal SSU rRNA gene sequences were selectively amplified by PCR and cloned. The first step in our overall strategy was to obtain partial nucleotide sequence information for 35 insert-containing plasmids. This sequence information was used to determine that the cloned inserts were SSU rRNA gene sequences from Archaea, to identify unique nucleotide sequences within the clone collection, and to select representative clones for full-length sequencing.
Approximately 400 nt of sequence from the 5′ terminal end (corresponding to positions 24–424 of the SSU rRNA gene sequence, Escherichia coli numbering) was first obtained from each of the insert-containing plasmids. Analysis of these data using blast and software provided by the RDP showed that 34 of the 35 cloned inserts were most similar to SSU rRNA gene sequences of Archaea. All of these cloned sequences had the highest similarity (78–84% nucleotide sequence identity) to Group I marine Archaea sequences (SBAR12, WHARQ, and ANTARCTIC12), which were recovered from uncultivated bacterioplankton in oceanic environments and phylogenetically placed in the planktonic Archaea subdivision of the Crenarchaeota (22). The cloned sequences from soil were more similar to each other (the least similar having 88% nucleotide sequence identity) than to any data base sequences. Alignment of the cloned sequences to SSU rRNA gene sequences from representative species of Archaea revealed secondary structure features consistent with the proposed structure of SSU rRNA of Archaea. Also, the cloned sequences had signature nucleotides or features diagnostic of Archaea (32) in 18 of the 19 relevant positions that were contained within the 400 nt of sequence information obtained for each cloned sequence (Table 1). When considered together, results of these analyses indicate that the cloned sequences are derived from SSU rRNA genes of members of the domain Archaea.
Table 1.
Interdomain nucleotide signature analysis of SCA clones
| Nucleotide position(s) of signature feature | Signature features of
|
|||
|---|---|---|---|---|
| Eucarya | Bacteria | Archaea | SCA clones | |
| 31 (bulged base) | Absent | Present | Absent | Euc, Arch |
| 33·551 | A·U | A·U | Y·R | Arch |
| 44.1·397 | −·A | −·A | U·A | Arch |
| 47.1 (extra base) | Present | Absent | Present | Euc, Arch |
| 52·359 | G·C | Y·R | G·C | Euc, Arch |
| 53·358 | C·G | A·U | C·G | Euc, Arch |
| 113·314 | C·G | G·C | C·G | Euc, Arch |
| 121 | A | Y | C | Bact, Arch |
| 292·308 | R·U | G·C | G·C | Bact, Arch |
| 307 | Y | Y | G | Arch |
| 335 | A | C | C | Bact, Arch |
| 338 | A | A | G | Arch |
| 339·350 | C·G | C·G | G·Y | Arch |
| 341·348 | U·A | C·G | C·G | Bact, Arch |
| 361 | C | R | C | Euc, Arch |
| 365 | A | U | A | Euc, Arch |
| 367 | U | U | C | Arch |
| 377·386 | Y·R | R·Y | Y·G | Bact |
| 393 | A | A | G | Arch |
Nucleotide signature features of SSU rRNA gene sequences defining the three domains, Eucarya (Euc), Bacteria (Bact), and Archaea (Arch) (32), were compared to relevant positions of the SCA clones. Numbering (nucleotide position) is based on the E. coli SSU rRNA gene sequence. · denotes a base pair. R and Y represent purines and pyrimidines, respectively.
Similar analyses determined that one cloned sequence, SCA1168, was derived from a SSU rRNA gene sequence of a member of the domain Bacteria, having 86% nucleotide sequence identity to the SSU rRNA gene sequence of Nannocystis exedens.
Of the 34 cloned archaeal SSU rRNA gene sequences, 2 nucleotide sequences occurred more than once in the collection. Clones SCA1145 and SCA1170 represent nucleotide sequences that were found in 16 and 2 cloned sequences, respectively. The other 16 clones had nucleotide sequences that were represented once in the clone collection. Of these 16 unique clones, 10 had nucleotide sequences that were highly similar to those of other clones within the collection, differing at 1–7 nt positions (out of approximately 400 nt). Clones that had nucleotide sequences similar to those of other clones were grouped and one representative clone was selected from each group. For each of these representative clones and some selected others chosen to represent the nucleotide sequence diversity within our clone collection, we determined the full-length nucleotide sequences (approximately 1400 nt).
The full-length sequences of 10 cloned SSU rRNA genes (designated as SCA clones hereafter) were checked for the presence of chimeric sequences, which can arise in PCR amplification using a mixed population of template DNA (33–35). Evaluation by the check_chimera program and inspection of putative secondary structure features did not detect the presence of chimeric sequences in the full-length cloned sequences.
Similarity searches with the full-length sequences of the SCA clones also revealed the highest similarity to SSU rRNA gene sequences of members of the planktonic Archaea in the prokaryotic SSU rRNA data base at the RDP. The highest similarity (89–98% nucleotide sequence identity) of the SCA clone sequences was to PAD19 and FIE16, which are partial SSU rRNA gene sequences previously isolated from soil (11). Similarity to these sequences was not detected using the 5′ terminal sequences of the SCA clone sequences, since the sequence information available for PAD19 and FIE16 correspond to the 3′ terminal sequence of SSU rRNA genes.
Phylogenetic relationships of the SCA clone sequences and SSU rRNA gene sequences of representative members from the domains Eucarya, Bacteria, and Archaea were determined using maximum-likelihood, neighbor-joining, and parsimony methods. Fifty taxa from the three phylogenetic domains were chosen that exemplify the phylogenetic diversity and the range of G+C content within each domain. Because the full-length SCA clone sequences had high nucleotide sequence identity (91–97%) to each other, a representative subset of SCA clone sequences (SCA1145, SCA1150, and SCA1180) was used for the phylogenetic analyses.
Initial phylogenetic analyses were performed using only full-length sequences and involved a data set of 857 nt positions. All three methods of phylogenetic analysis resulted in trees with similar topologies and consistently determined that the SCA clones branch deeply from the crenarchaeal line of descent (analysis not shown). Bootstrapping of neighbor-joining and parsimony analyses supported this topology in 86 and 66 (out of 100) trees, respectively. SSU rRNA gene sequences representing the planktonic Archaea were not included in the initial phylogenetic analyses, since the sequence information for members of this group consists of only partial SSU rRNA gene sequences.
The G+C content (55–58%) of the sequences of the SCA clones is lower than that of the SSU rRNA gene sequences of the cultured Crenarchaeota and thermophilic Euryarchaeota (63–69%) that were used in the phylogenetic analyses. Differences in base composition among sequences have been shown to promote artifacts in phylogenetic analyses when SSU rRNA gene sequences from mesophilic and thermophilic species are compared (30). The latter are considerably higher in G+C content than the former. As a result, an artificial phylogenetic clustering of the thermophilic sequences can arise.
To determine if base composition differences among the sequences used for the phylogenetic analyses affected the placement of the SCA clone lineage within the Crenarchaeota, transversion analyses were performed. The use of transversion analyses can remove some of the biases associated with phylogenetic placement of sequences with different base compositions (28). Transversion distance and transversion parsimony analyses support the placement of the SCA clone lineage as a deeply branching group within the Crenarchaeota in 94 and 76 (out of 100) trees, respectively (data not shown). The use of transversion analyses resulted in higher bootstrap support for placement of the SCA clone lineage than the substitution analyses, indicating that the base compositional differences among the sequences used in the phylogenetic analyses has a small, but detectable, effect on the placement of this lineage within the Crenarchaeota.
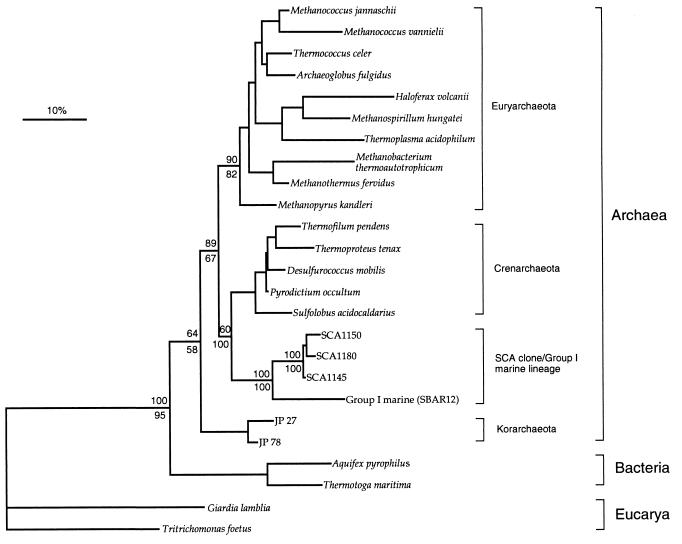
Further phylogenetic analyses were used to determine the relationship of the SCA clones to members of the planktonic Archaea. Maximum likelihood, neighbor-joining, parsimony, and transversion analyses using a data set of 637 nt positions resulted in trees with similar topologies and consistently indicated an affiliation of the SCA clones to the Group I marine Archaea (Fig. 1). Bootstrapping of neighbor-joining, parsimony, and transversion analyses revealed that the SCA clones were affiliated with the Group I marine Archaea in 100 (out of 100) trees. These analyses also supported the placement of the SCA clone/Group I marine Archaea lineage as a deeply divergent branch within the Crenarchaeota and stabilized the problematic placement of the Group I marine Archaea observed in previous studies (5, 18).
Figure 1.
Inferred unrooted phylogenetic tree of archaeal SSU rRNA gene sequences cloned from soil, illustrating close affiliation of these sequences (designated SCA clones) with those of the Group I marine sequences of the Crenarchaeota. Tree was inferred by neighbor-joining analysis of 637 homologous positions of sequence from each organism or clone. Scale bar represents 10 mutations per 100 nt of homologous sequence positions. The percentage of 100 bootstrap resamplings that support some of the major topological elements in neighbor joining (above line) and parsimony (below line) analyses is indicated.
The phylogenetic placement of the SCA clones represented in Fig. 1 was further assessed by an intradomain nucleotide signature analysis (32) (Table 2). Nine of the 10 SCA clone sequences had more signature sequence features in common with Crenarchaeota than with Euryarchaeota (10 vs. 6 features). These SCA clones also shared all 16 signature sequence features with the Group I marine Archaea. One SCA clone, SCA1180, differed from the other SCA clones at two sequence signature positions (34·550 and 1252) and had 11 and 4 sequence signature features in common with the Crenarchaeota and Euryarchaeota, respectively. Thus, both phylogenetic and intradomain nucleotide signature analyses showed an affiliation of the SCA clones with the Group I marine Archaea.
Table 2.
Intradomain nucleotide signature analysis of SCA clones
| Nucleotide position(s) of signature feature | Signature features of
|
||||
|---|---|---|---|---|---|
| Eury | Cren | Grp I marine | SCA clones | SCA 1180 | |
| 27·556 | G·C | C·G | Cren | Cren | Cren |
| 28·555 | G·Y | C·G | Cren | Cren | Cren |
| 30·553 | Y·R | G·C | Cren | Cren | Cren |
| 34·550 | U·G | C·G | Eury | Eury | U·U |
| 289·311 | C·G | G·C | Eury | Eury | Eury |
| 501·544 | R·Y | C·G | Eury | Eury | Eury |
| 503·542 | C·G | G·C | Cren | Cren | Cren |
| 504·541 | Y·R | G·Y | Eury | Eury | Eury |
| 513·538 | C·G | U·A | Cren | Cren | Cren |
| 518 | C | U | Cren | Cren | Cren |
| 658·747 | Y·R | G·C | Cren | Cren | Cren |
| 692 | U | C | Eury | Eury | Eury |
| 965 | Y | G | Cren | Cren | Cren |
| 1074·1083 | A·C | G·U | Cren | Cren | Cren |
| 1244·1293 | Y·R | R·Y | Cren | Cren | Cren |
| 1252 | U | C | Eury | Eury | Cren |
Nucleotide signature features of SSU rRNA gene sequences defining the two archaeal kingdoms, Euryarchaeota (Eury) and Crenarchaeota (Cren), and the Group I marine Archaea (Grp I marine), were compared to relevant positions of the SCA clone sequences. Sequence signatures of the two archaeal kingdoms and the Group I marine Archaea are taken from Winker and Woese (32) and Barns et al. (8), respectively. SCA clones include SCA1145, SCA1150, SCA1151, SCA1154, SCA1158, SCA1166, SCA1170, SCA1173, and SCA1175 sequences. Numbering (nucleotide position) is based on the E. coli SSU rRNA gene sequence. · denotes a base pair. R and Y represent purines and pyrimidines, respectively.
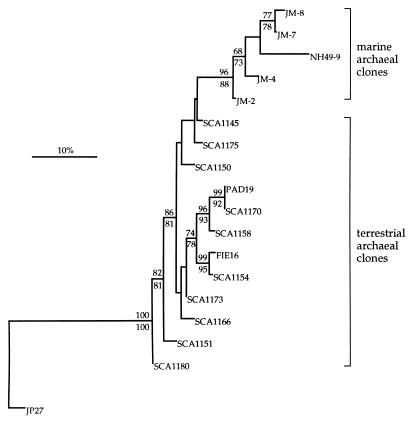
Within the planktonic Archaea are two cloned SSU rRNA gene sequences, PAD19 and FIE16 (11), which were obtained from soil. Further phylogenetic analyses were performed with the SCA clones and members of the planktonic Archaea to determine the relationship of the SCA clone sequences to those of PAD19 and FIE16, as well as to other sequences within this group. A small data set of 261 nt positions was used for these analyses, since there is a limited amount of sequence information available for PAD19 and FIE16 (286 and 287 nt, respectively). Analyses using maximum-likelihood, distance, and parsimony methods resulted in phylogenetic trees with similar topologies and consistently indicated a close affiliation of several of the SCA clone sequences (SCA1154, SCA1158, and SCA1170) to PAD19 and FIE16 (Fig. 2). These analyses also supported the resolution of the SCA clone and planktonic Archaea sequences into two discrete groups, one of which is associated with a terrestrial environment and the other with a marine environment.
Figure 2.
Inferred phylogenetic tree of archaeal SSU rRNA gene sequences cloned from soil, illustrating close affiliation of these sequences (designated SCA clones) with those within the planktonic Archaea. The tree was rooted to the korarchaeotal sequence JP27 (8). Planktonic Archaea clones used in this analysis are from marine bacterioplankton (NH49-9; ref. 7), soil (PAD19 and FIE16; ref. 11), and a marine holothurian mid-gut (JM-2, JM-4, JM-7, and JM-8; ref. 18). Tree was inferred by maximum likelihood analysis of 261 homologous positions of sequence from each clone. Scale bar represents 10 mutations per 100-nt of homologous sequence positions. The percentage of 100 bootstrap resamplings that support some of the major topological elements in maximum likelihood (above line) and parsimony (below line) analyses is indicated. Only bootstrap values >60 are shown for these analyses.
DISCUSSION
The identification of Archaea from soil is important to understanding the ecological significance of Archaea in the biosphere and for the analysis of naturally occurring microbial communities. Because many Archaea have been identified in specialized, often extreme, environmental niches, Archaea have not been considered to be important in the ecology of microbial communities in different environments. However, the identification of novel groups of Archaea in marine (3, 5, 7, 18) and terrestrial (ref. 11, this study) environments indicates that Archaea are more diverse and widespread than previously thought.
From analysis of SSU rRNA genes cloned from soil, we have identified a highly clustered group of soil Archaea. Phylogenetic analyses of the full-length soil Archaea clone sequences, designated as SCA clones, placed this lineage as a deeply branching group within the Crenarchaeota that has no close affiliation to any cultivated member of the Archaea. These analyses also determined that the SCA clone lineage has phylogenetic affinity to the Group I marine Archaea, which are members of the planktonic division in the Crenarchaeota. Nucleotide signature sequence analysis also supported the affiliation of the SCA clone sequences with the Crenarchaeota and affinity to the Group I marine Archaea. These analyses showed that 9 of the 10 SCA clone sequences had more signature sequence features in common with Crenarchaeota than with Euryarchaeota (10 vs. 6 features) and shared all 16 signature sequence features with the Group I marine Archaea. From our analyses, the phylogenetic affiliation of the SCA clone lineage to the planktonic Archaea is well supported. However, the exact phylogenetic placement of the SCA clone/planktonic Archaea lineage within the Archaea will require the acquisition of more SSU rRNA gene sequence data, as well as other genotypic and phenotypic data, from members of this lineage.
Of the 34 cloned sequences analyzed, only 2 sequences were represented more than once in the clone collection. It is unlikely that the small collection of clones examined in this study reflects the full diversity of Archaea in the soil environment sampled, since 16 of the clone sequence types appeared only once in the collection. The frequency of the sequence types within the clone collection may not represent the distribution of archaeal sequences present in the soil sampled because of potential biases arising from the small size of the soil sample, the extraction and recovery of DNA, amplification by PCR (36–38), and cloning. Of the 16 unique sequence types within the clone collection, 10 had nucleotide sequences that differed from other sequence types at 1–7 nt positions. These differences may be attributed to microheterogeneity of SSU rRNA gene sequences within an individual or between related members of the Archaea population or may be the result of base-incorporation errors during amplification, cloning artifacts, or sequencing errors. However, since such heterogeneity was not observed in the duplicates of the sequence types typified by SCA1145 and SCA1170, it is unlikely that introduced artifacts are responsible for the degree of sequence heterogeneity within the clone collection.
Because the SCA clone sequences obtained in this study showed no close phylogenetic affiliation to any cultured Archaea, it is difficult to predict the phenotypic properties and ecological role of this group of soil Archaea. However, it is interesting that the SCA clones were affiliated with PAD19 and FIE16 (11), which are SSU rRNA gene sequences cloned from a geographically distant soil. This affiliation suggests that these sequences may represent a lineage of non-thermophilic Crenarchaeota that are associated with a terrestrial environment. Molecular analyses, such as in situ hybridization with oligonucleotide probes specific for this archaeal lineage, are warranted to determine the relative abundance of Archaea in soil and whether they exist as free-living organisms or are endosymbionts of protozoan or arthropod hosts.
Methanogenesis has long been associated with the microbial communities in sediments and flooded soils. Molecular and cultivation analyses of methanogens from these environments have identified the presence of microbes affiliated with members of the Euryarchaeota (14, 39). Even though the SCA clone lineage placed in the Crenarchaeota and does not show any affiliation to known methanogens, we are unable to exclude the possibility that this lineage could contain novel groups of methanogens found in a soil environment. Our future work will be directed to microscopic and culturing studies designed to characterize the physiology and ecological roles of this lineage of soil Archaea.
Soil microbial communities appear to be far more complex than has been demonstrated by standard culturing methods. To assess the vast diversity of uncultured organisms in soil demands new biological and computational techniques that can accommodate this complexity. Our long-term goal is to develop a molecular hybridization method, which uses an array of oligonucleotides derived from SSU rRNA gene sequences, for analysis of soil microbial communities. The identification of this lineage of Archaea in soil provides a wealth of previously unknown molecular information that is useful for designing oligonucleotides for such arrays and for assessing the composition and diversity of soil microbial communities.
Acknowledgments
We gratefully acknowledge Marcelo Gravina de Moraes and Deborah Joseph for helpful discussions, Marcelo Gravina de Moraes for critical reading of the manuscript, and James S. Ireland for assistance with the computational analyses. This work was supported by a National Institutes of Health postdoctoral fellowship to S.B.B., an Environmental Protection Agency cooperative agreement (CR822902-01-0), and a grant from the McKnight Foundation. Computational analyses were made possible in part by instrumentation provided by an Industrial and Economic Development and Research Grant, University–Industry Research Program at the University of Wisconsin–Madison.
Footnotes
Abbreviations: SSU rRNA, small subunit ribosomal RNA; RDP, Ribosomal Database Project.
Data deposition: The SSU rRNA gene sequences reported in this paper have been deposited in the GenBank data base [accession nos. U62811U62811 (SCA1145), U62812U62812 (SCA1150), U62813U62813 (SCA1151), U62814U62814 (SCA1154), U62815U62815 (SCA1158), U62816U62816 (SCA1166), U62817U62817 (SCA1170), U62818U62818 (SCA1173), U62819U62819 (SCA1175), U62820U62820 (SCA1180).
References
- 1.Staley J T, Konopka A. Annu Rev Microbiol. 1985;39:321–346. doi: 10.1146/annurev.mi.39.100185.001541. [DOI] [PubMed] [Google Scholar]
- 2.Roszak D B, Colwell R R. Microbiol Rev. 1987;51:365–379. doi: 10.1128/mr.51.3.365-379.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Giovannoni S J, Britschgi T B, Moyer C L, Field K G. Nature (London) 1990;345:60–62. doi: 10.1038/345060a0. [DOI] [PubMed] [Google Scholar]
- 4.Ward D M, Weller R, Bateson M M. Nature (London) 1990;345:63–65. doi: 10.1038/345063a0. [DOI] [PubMed] [Google Scholar]
- 5.DeLong E F. Proc Natl Acad Sci USA. 1992;89:5685–5689. doi: 10.1073/pnas.89.12.5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liesack W, Stackebrandt E. J Bacteriol. 1992;174:5072–5078. doi: 10.1128/jb.174.15.5072-5078.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fuhrman J A, McCallum K, Davis A A. Appl Environ Microbiol. 1993;59:1294–1302. doi: 10.1128/aem.59.5.1294-1302.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barns S M, Fundyga R E, Jeffries M W, Pace N R. Proc Natl Acad Sci USA. 1994;91:1609–1613. doi: 10.1073/pnas.91.5.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Amann R I, Ludwig W, Schleifer K-H. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moyer C L, Dobbs F C, Karl D M. Appl Environ Microbiol. 1995;61:1555–1562. doi: 10.1128/aem.61.4.1555-1562.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ueda T, Suga Y, Matsuguchi T. Eur J Soil Sci. 1995;46:415–421. [Google Scholar]
- 12.Ohkuma M, Kudo T. Appl Environ Microbiol. 1996;62:461–468. doi: 10.1128/aem.62.2.461-468.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weidner S, Arnold W, Pühler A. Appl Environ Microbiol. 1996;62:766–771. doi: 10.1128/aem.62.3.766-771.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hales B A, Edwards C, Ritchie D A, Hall G, Pickup R W, Saunders J R. Appl Environ Microbiol. 1996;62:668–675. doi: 10.1128/aem.62.2.668-675.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Borneman J, Skroch P W, O’Sullivan K M, Palus J A, Rumjanek N G, Jansen J L, Nienhuis J, Triplett E W. Appl Environ Microbiol. 1996;62:1935–1943. doi: 10.1128/aem.62.6.1935-1943.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DeLong E F, Wu K Y, Prezelin B B, Jovine R V M. Nature (London) 1994;371:695–697. doi: 10.1038/371695a0. [DOI] [PubMed] [Google Scholar]
- 17.Woese C R, Kandler O, Wheelis M L. Proc Natl Acad Sci USA. 1990;87:4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McInerey J O, Wilkinson M, Patching J W, Embley T M, Powell R. Appl Environ Microbiol. 1995;61:1646–1648. doi: 10.1128/aem.61.4.1646-1648.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Preston C M, Wu K Y, Molinski T F, DeLong E F. Proc Natl Acad Sci USA. 1996;93:6241–6246. doi: 10.1073/pnas.93.13.6241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Torsvik V L. Soil Biol Biochem. 1980;12:15–21. [Google Scholar]
- 21.Torsvik V L, Salte K, Sørheim R, Goksøyr J. Appl Environ Microbiol. 1990;56:776–781. doi: 10.1128/aem.56.3.776-781.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maidak B L, Olsen G J, Larsen N, Overbeek R, McCaughney M J, Woese C R. Nucleic Acids Res. 1996;24:82–85. doi: 10.1093/nar/24.1.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schulte E F, Peters J B, Hodgson P R. Wisconsin Procedures for Soil Testing, Plant Analysis, and Feed and Forage Analysis. Univ. of Wisconsin–Madison: Dept. of Soil Sci.; 1987. [Google Scholar]
- 24.Porteus L A, Armstrong J L, Seidler R J, Watrud L S. Curr Microbiol. 1994;29:301–307. doi: 10.1007/BF01577445. [DOI] [PubMed] [Google Scholar]
- 25.Lane D J. In: Nucleic Acid Techniques in Bacterial Systematics. Stackebrandt E, Goodfellow M, editors. New York: Wiley; 1991. pp. 115–175. [Google Scholar]
- 26.Maniatis T, Fritsch E F, Sambrook J. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1982. [Google Scholar]
- 27.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 28.Olsen G J. Methods Enzymol. 1988;164:793–812. doi: 10.1016/s0076-6879(88)64084-5. [DOI] [PubMed] [Google Scholar]
- 29.Kimura M. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 30.Woese C R, Achenbach L, Rouviere P, Mandelco L. Syst Appl Microbiol. 1991;14:364–371. doi: 10.1016/s0723-2020(11)80311-5. [DOI] [PubMed] [Google Scholar]
- 31.Felsenstein J. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 32.Winker S, Woese C R. Syst Appl Microbiol. 1991;14:305–310. doi: 10.1016/S0723-2020(11)80303-6. [DOI] [PubMed] [Google Scholar]
- 33.Shuldiner A R, Nirula A, Roth J. Nucleic Acids Res. 1989;17:4409. doi: 10.1093/nar/17.11.4409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liesack W, Weyland H, Stackebrandt E. Microb Ecol. 1991;21:191–198. doi: 10.1007/BF02539153. [DOI] [PubMed] [Google Scholar]
- 35.Kopczynski E D, Bateson M M, Ward D M. Appl Environ Microbiol. 1994;60:746–748. doi: 10.1128/aem.60.2.746-748.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reysenbach A, Giver L J, Wickham G S, Pace N R. Appl Environ Microbiol. 1992;58:3417–3418. doi: 10.1128/aem.58.10.3417-3418.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Farrelly V, Rainey F A, Stackebrandt E. Appl Environ Microbiol. 1995;61:2798–2801. doi: 10.1128/aem.61.7.2798-2801.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suzuki M T, Giovannoni S J. Appl Environ Microbiol. 1996;62:625–630. doi: 10.1128/aem.62.2.625-630.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rajagopal B S, Belay N, Daniels L. FEMS Microbiol Ecol. 1988;53:153–158. [Google Scholar]