Abstract
The yeast nicotinamidase Pnc1p acts in transcriptional silencing by reducing levels of nicotinamide, an inhibitor of the histone deacetylase Sir2p. The Pnc1p structure was determined at 2.9 Å resolution using MAD and MIRAS phasing methods after inadvertent crystallization during the pursuit of the structure of histidine-tagged yeast isocitrate dehydrogenase (IDH). Pnc1p displays a cluster of surface histidine residues likely responsible for its co-fractionation with IDH from Ni2+-coupled chromatography resins. Researchers expressing histidine-tagged proteins in yeast should be aware of the propensity of Pnc1p to crystallize, even when overwhelmed in concentration by the protein of interest. The protein assembles into extended helical arrays interwoven to form an unusually robust, yet porous superstructure. Comparison of the Pnc1p structure with those of three homologous bacterial proteins reveals a common core fold punctuated by amino acid insertions unique to each protein. These insertions mediate the self-interactions that define the distinct higher order oligomeric states attained by these molecules. Pnc1p also acts on pyrazinamide, a substrate analog converted by the nicotinamidase from Mycobacterium tuberculosis into a product toxic to that organism. However, we find no evidence for detrimental effects of the drug on yeast cell growth.
Keywords: nicotinamidase, Sir2p, X-ray crystallography, kinetic analyses, NAD+, multiple isomorphous replacement, multiwavelength anomalous diffraction
Pnc1p from Saccharomyces cerevisiae is a zinc-dependent nicotinamidase (EC 3.5.1.19) that catalyzes the deamidation of nicotinamide to produce ammonia and nicotinic acid [1; 2] (Figure 1Ai). The latter compound is converted back to NAD+ in a series of reactions catalyzed by other enzymes in the NAD+ salvage pathway. Pnc1p has received considerable attention [3; 4; 5] because nicotinamide is an inhibitor of Sir2p [6; 7; 8], an NAD+-dependent histone deacetylase required for transcriptional silencing in yeast cells subjected to calorie restriction [9]. Deletion of the PNC1 gene correlates with an increase in nicotinamide levels and a decrease in transcriptional silencing [7; 10], whereas overexpression of Pnc1p correlates with an extension of replicative life span [11].
Figure 1.
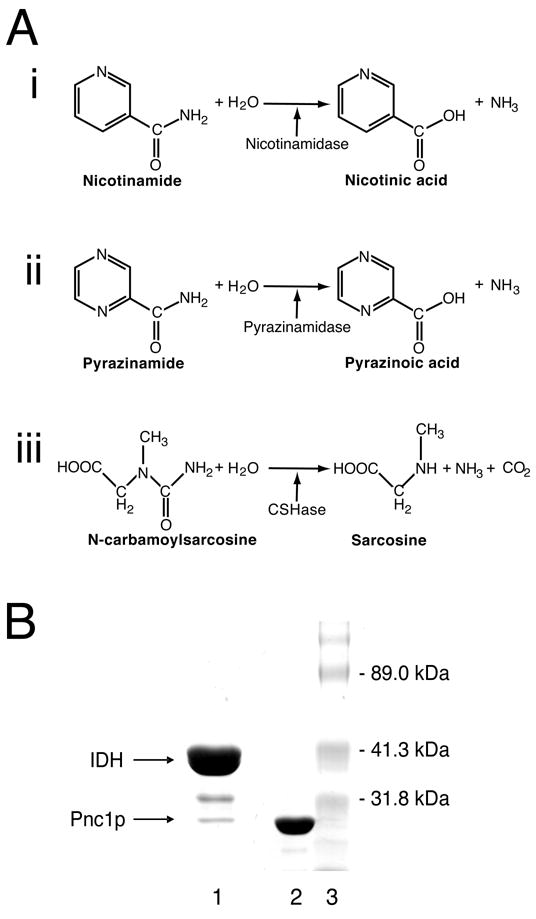
Three classes of hydrolase enzymes, the reactions they catalyze, and the purification of the Pnc1p nicotinamidase from yeast. A) Hydrolase reactions catalyzed by nicotinamidase, pyrazinamidase, and N-carbamoylsarcosine hydrolase. Nicotinamidases catalyze the conversion of nicotinamide (pyridine-3-carboxamide) to nicotinic acid (pyridine-3-carboxylic acid) and ammonia. Pyrazinamidases catalyze the conversion of pyrazinamide (pyrazine-2-carboxamide) to pyrazinoic acid (pyrazine-2-carboxylic acid) and ammonia. N-carbamoylsarcosine aminohydrolases catalyze the conversion of N-carbamoylsarcosine (2-(carbamoyl-methyl-amino)acetic acid to sarcosine 2-(Methylamino)acetic acid), ammonia and carbon dioxide. B) Isolation of Pnc1p. Coomassie stained denaturing gel of protein samples obtained following Ni2+-NTA column chromatography. Lane 1 was loaded with 30 μg of protein isolated from yeast expressing histidine-tagged IDH. The IDH1 and IDH2 subunits that assemble to form the functional IDH octamer [44] are not resolved because they have similar molecular masses [36; 37]. Lane 2 was loaded with 10 μg of protein isolated from E. coli expressing recombinant Pnc1p. Lane 3 was loaded with molecular mass standards. Mass spectrometry revealed that the band migrating between IDH and Pnc1p in lane 1 contains an IDH degradation product.
Pnc1p demonstrates limited sequence similarity to the products of the Escherichia coli and Mycobacterium tuberculosis pncA genes [2; 12; 13], and to the product of the Pyrococcus. horikoshii gene with locus tag PH0999, which until now represented the only nicotinamidase for which structural information was known [14]. The E. coli and M. tuberculosis nicotinamidases can also act on pyrazinamide [2; 12; 15], converting it to ammonia and pyrazinoic acid (Figure 1Aii). Pyrazinoic acid is active against M. tuberculosis, and when pyrazinamide is administered in combination with isoniazid and rifampin, the conventional tuberculosis treatment can be substantially shortened [16]. Today, pyrazinamide remains a front-line treatment of M. tuberculosis infection, although resistant strains have been identified that contain alterations in the pncA gene [13; 17; 18]. Collectively, these prokaryotic nicotinamidases demonstrate distant sequence similarity to the N-carbamoylsarcosine amidohydrolase (CSHase, Figure 1Aiii) from Arthrobacter sp. [19; 20], one of three enzymes found in a pathway for the degradation of creatinine to glycine [21], and to a putative hydrolase (YcaC) of unknown function from E. coli [22].
Pnc1p was purified initially over 30 years ago using conventional column chromatography [1], but more recently due to its unexplained ability to bind Ni2+-coupled affinity matrices [2]. In the present study, Pnc1p was crystallized unexpectedly as a minor contaminant in the purification of a histidine-tagged form of yeast isocitrate dehydrogenase (IDH). Crystals believed to be of IDH were later identified as Pnc1p during manual tracing of an experimental electron density map. The structure determination and refinement was completed because of the general interest in the role of Pnc1p in the modulation of Sir2 function and its link to aging, and because it represents the first nicotinamidase structure to be determined from a eukaryote.
The results of the structural analysis reveal a cluster of solvent-exposed residues that may account for the co-fractionation of Pnc1p with histidine-tagged IDH from Ni2+-coupled chromatography resins and show that Pnc1p can assemble into a robust superstructure consisting of interconnecting “hubs” and “spokes” interspersed with large solvent channels. Comparison of Pnc1p with the structures of PH0999, YcaC, and CSHase reveals a common core fold broken by amino acid insertions that are unique to each protein. These amino acid insertions appear to mediate the assembly of these proteins into distinct higher order oligomeric states.
Materials and Methods
Expression and purification of Pnc1p
When isolated from yeast, Pnc1p elutes as a contaminant in the purification of histidine-tagged yeast IDH as described [23; 24]. For heterologous expression in E. coli, the PNC1 gene was amplified from yeast genomic DNA using the polymerase chain reaction (PCR) and cloned into pET-17b (Novagen), which was subsequently transformed into E. coli strain BL21(DE3). A single transformant colony was grown in 1.0 L LB medium at 37°C to OD600nm = 0.5. Isopropyl-β-D-galactoside was added to 1.0 mM, and cultivation was continued for 6 h at 30°C. Cells were harvested by centrifugation and lysed by sonication. Recombinant Pnc1p was purified using immobilized metal affinity chromatography with a Ni2+-nitrilotriacetic (NTA) column as described [23]. Protein concentrations were determined using the Bradford method [25]. The yield was ~35 mg Pnc1p/L culture. Protein samples from yeast and E. coli were electrophoresed using 10% polyacrylamide sodium dodecyl sulfate gels and stained with Coomassie blue. The relative amounts of Coomassie blue-stained polypeptides were estimated using densitometry. Individual bands were excised and subjected to in-gel tryptic digestion prior to their identification by the Mass Spectrometry Laboratory at the UTHSCSA.
Crystallization, data collection, structure determination and refinement
Pnc1p was crystallized in space group R3 with a=b=302.0 Å, c=112.1 Å [23; 24]. Heavy atom derivatives were prepared by transferring suitable crystals to solutions containing the reservoir solution plus 10 mM K2OsCl6, 10 mM K2Pt(SCN)6 or 0.5 M NaI, and soaked for 2 – 24 h, depending on their tolerance for the heavy atom used. Native and derivative Pnc1p crystals were cryoprotected with a 1 min soak in reservoir solution made 50% (w/v) in D-sorbitol and flash-cooled by plunging into liquid nitrogen prior to data collection. Diffraction data were taken at the Advanced Light Source beamline 8.2.1 and the Advanced Photon Source beamline 19BM. All data were integrated and scaled using HKL-2000 [26].
Molecular replacement using coordinates of bacterial isocitrate dehydrogenases and isopropylmalate dehydrogenases currently available in the Protein Data Bank (http://www.rcsb.org/pdb/home/home.do) failed. The structure was subsequently determined with a combination of multiple isomorphous replacement with anomalous scattering (MIRAS) and multiwavelength anomalous diffraction (MAD) phasing methods using the data sets in Table 1. Osmium atoms were located first, and phases were calculated using SOLVE and RESOLVE [27; 28], resulting in a mean figure of merit of 0.46 to 4.3 Å resolution. This initial phase set was used to locate the platinum and iodine atoms through anomalous difference Fourier analyses. Calculations combining phases from the three heavy atom data sets were performed using SHARP [29], resulting in a mean figure of merit of 0.49 to 3.6 Å resolution. The initial Pnc1p backbone was traced, and this backbone model was fit manually into six additional regions of the experimental electron density map in the asymmetric unit. The noncrystallographic symmetry (NCS) operators obtained after the manual fitting of these chains were used to perform sevenfold noncrystallographic averaging, substantially improving the electron density for the amino acid side chains. The model was completed through iterative cycles of computational refinement using REFMAC [30] and manual model rebuilding using COOT [31]. During crystallographic refinement, “loose” non-crystallographic (NCS) restraints were applied to each of the seven Pnc1p monomers in the asymmetric unit. Translation/libration/screw (TLS) refinement [32] was invoked in the final cycles with each Pnc1p monomer as a TLS group. The stereochemistry of the final model was assessed with the program PROCHECK [33]. All figures containing molecular models were prepared with the program PyMOL (DeLano, W.L. http://www.pymol.org) and structure-based sequence alignments were made with the help of the programs ESPript [34] and MUSTANG [35].
Table 1.
X-ray Diffraction Data and Refinement Statistics for Pnc1p Determined by MIRAS.
| Native | K2OsCl6 | K2OsCl6 | K2OsCl6 | K2Pt(SCN)6 | NaI | |
|---|---|---|---|---|---|---|
| Diffraction Data | ||||||
| Beamline | APS 19BM | ALS 8.2.1 | ALS 8.2.1 | ALS 8.2.1 | APS 19BM | ALS 8.2.1 |
| Resolution (Å)a | 50-2.9(2.95–2.90) | 50.0-4.0(4.14-4.0) | 50.0-4.0(4.14-4.0) | 50.0-4.0(4.14-4.0) | 50.0-4.8(4.97-4.8) | 50.0-3.6(3.73-3.6) |
| Wavelength (Å) | 1.0332 | 1.1401 | 1.1405 | 1.1390 | 1.0719 | 1.8000 |
| Completeness (%) | 100 (100) | 99.7 (99.9) | 99.0 (93.2) | 99.2 (94.9) | 99.4 (99.8) | 99.7 (98.8) |
| Redundancy | 4.4 (4.3) | 3.7 (3.5) | 3.7 (3.1) | 3.7 (3.1) | 2.8 (2.7) | 2.3 (2.1) |
| Average I/σI | 21.0 (3.4) | 11.6 (2.9) | 12.1 (2.9) | 12.1 (2.6) | 10.3 (3.7) | 7.5 (1.9) |
| Rsym (on I) (%)b | 0.070(0.559) | 0.097(0.434) | 0.096(0.432) | 0.097(0.456) | 0.094(0.311) | 0.111(0.529) |
| Sites | 31 | 31 | 31 | 23 | 92 | |
| Phase Power iso/anc | 2.6/1.1 | 2.4/0.70 | 2.9/0.52 | 0.50/0.62 | 0.85/0.72 | |
| Rcullis iso/anod | 0.26/0.83 | 0.24/0.89 | 0.25/0.93 | 0.99/0.92 | 0.88/0.92 | |
| (SHARP Figure of Merit to 3.6 Å: 0.49) | ||||||
|
| ||||||
| Refinement | ||||||
| Resolution (Å) | 2.9 | |||||
| # reflections | 80,251 | |||||
| F/σF>0 | ||||||
| Rwork/Rfree (%) | 18.9/22.0 | |||||
| Number of atoms | Ramachandran Plot (% resides each region) | |||||
| Protein | 12,341 | Most favored | 86.4 | |||
| Zinc ions | 7 | Additionally allowed | 12.9 | |||
| Water | 0 | Generously allowed | 0.7 | |||
| B- factors (Å2) | Disallowed | 0.0 | ||||
| Protein (mean) | 77.4 | |||||
| Metal ions (mean) | 89.9 | |||||
| Wilson | 71.5 | |||||
| RMS deviations | ||||||
| Bond lengths (Å) | 0.012 | |||||
Bond angles (°) 1.331
The numbers in parentheses correspond to the highest resolution shell.
Rsym = Σ|Ihkl−(Ihkl)|/Σ(Ihkl), where Ihkl is the observed intensity and (Ihkl) is the average intensity of multiple symmetry-related observations of that reflection.
Phasing Power = (|Fh|/ε, where Fh is the calculated heavy-atom structure factor amplitude and ε is the residual lack of closure error.
Rcullis = (ε)/(|Fph−Fp|) where ε is the residual lack of closure error and Fph and Fp are structure factor amplitudes from heavy-atom derivative and native data sets, respectively.
Kinetic analyses
Enzymatic activities were assayed as described [2] at 30°C. Recombinant Pnc1p (~1.0 μg/reaction) was incubated for 3 min with nicotinamide (concentrations ranging from 0–3.0 mM) or with pyrazinamide (concentrations ranging from 0–6.0 mM) in 400 μl 10 mM Tris-HCl (pH 7.4), 150 mM NaCl, and 1 mM MgCl2. The ammonia produced in the reaction was quantified using a Sigma diagnostic kit (catalogue #AA0100). A unit of activity is defined as 1.0 μmol ammonia/min. Kinetic data were analyzed using Sigma Plot (SPSS Inc.). Kinetic parameters represent averages of two independent determinations. Nicotinamide and pyrazinamide (pyrazinecarboxamide) were obtained from Sigma-Aldrich.
Phenotype analyses
A pnc1Δ gene disruption mutant (Accession No. Y14405) was obtained from the EUROSCARF collection of viable yeast gene-disruption strains (www.unifrankfurt.de/fb15/mikro/euroscarf/index.html). The disruption was confirmed by PCR using genomic DNA from the pnc1Δ strain as the template. The corresponding parental BY4742 strain (MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0) and the pnc1Δ strain were grown overnight in rich YP medium (1% yeast extract, 2% Bacto-peptone) with 2% glucose as the carbon source. Dilutions of the cultures were plated on 2% agar plates containing YP glucose medium and pyrazinamide in concentrations ranging from 0–8 mM. Colony growth was assessed after growth at 30°C for 3 days.
Results and Discussion
Pnc1p purification, crystallization, structure determination, and refinement
Figure 1B (lane 1) shows the proteins in a sample eluted from a Ni2+ affinity chromatography column loaded with a cleared lysate derived from yeast overexpressing histidine-tagged IDH, a ~302 kDa heterooctameric enzyme composed of four IDH1 (38 kDa) and four IDH2 (37.5 kDa) subunits [36; 37]. Crystals suitable for X-ray diffraction grown from this eluate were presumed to be of IDH given its overwhelming dominance over contaminating proteins, and given the large unit cell dimensions that were consistent with the expected mass of an IDH heterooctamer [23; 24]. In addition, the initial partial trace of the experimental electron density map revealed a multi-stranded β-sheet flanked by α;-helices and a prominent loop containing a two-stranded antiparallel β-sheet, features found in both the IDH and Pnc1p structures (Supplementary Figure 1). As the building of the protein model progressed, however, the backbone deviated from that expected for IDH. The coordinates for the traced main chain residues were submitted to the DALI 3D structure comparison server [38] to search the Protein Data Bank for a similar fold. The pyrazinamidase/nicotinamidase from P. horikoshii (PH0999) [Protein Data Bank entry 1IM5 [14]] was returned as a structural homolog. A subsequent search of the yeast genome with this bacterial nicotinamidase sequence returned the sequence of the 24 kDa nicotinimidase Pnc1p, which was an excellent fit with the symmetry averaged experimental election density. Mass spectrometry confirmed that Pnc1p was present in the minor contaminating band of the original protein sample used in crystallization experiments (Figure 1B, lane 1), and indicated that the band of intermediate mass was an IDH degradation product. Table 1 shows the X-ray diffraction data, MAD phasing, and protein structure refinement statistics. The high overall B-factor of the diffraction data is likely a result of a high solvent content of 78% (Vm = 5.6). The coordinates and structure factors have been deposited into the Protein Data Bank with identifier 2H0R.
The large volume of the unit cell, the overwhelming dominance of IDH over Pnc1p in the protein sample used for crystallization, the presence of seven Pnc1p molecules in the asymmetric unit, and the coincidental structural similarity shared by Pnc1p and IDH resulted in a case of mistaken identity [23; 24]. Although it was fortuitous that the work provided knowledge of a structurally uncharacterized molecule that plays a role in transcriptional silencing, a simple silver-stained denaturing gel or mass spectrometric analysis of the protein coming from a dissolved crystal would have prevented the initial misidentification of the protein. Researchers using yeast expression systems to isolate His-tagged proteins should be forewarned that Pnc1p can be expected to be a contaminant when using Ni2+-affinity chromatography resins and that contaminating Pnc1p can crystallize even though it may be a minor component of the protein sample of interest (see below). The facile crystallization of Pnc1p is underscored by the observation that recombinant Pnc1p produced crystal specimens isomorphous with those used in this study in approximately 30% of all crystallization trials spanning a wide range of pH values, with many different precipitating agents, and in the absence of any alternative crystal forms. Intriguingly, Pnc1p is not the only representative of this fold family to crystallize as a minor contaminant in the overwhelming presence of another molecule. Several crystal forms of the product of the E. coli ycaC gene (see below) were unexpectedly obtained during crystallization trials of a commercially prepared sample of E. coli L-glutamic acid decarboxylase [22].
Pnc1p topology
Figure 2 shows the sequence alignment of Pnc1 with PH0999 modified relative to that derived from a BLAST search to reflect the similarities and differences of the two proteins when they are structurally aligned. The sequences of E. coli YcaC and Arthrobacter sp. CSHase (see below) were added to this structure-based alignment with the assistance of the program Mustang [35]. Figure 3A shows that the overall Pnc1p fold consists of a six-stranded parallel β-sheet flanked by three α;-helices on one side and two on the other. Figure 3B shows that the largest insertion I1 (residues 62–90), near residues 63–67 of PH0999, forms an extended loop within which is contained a two-stranded antiparallel β-sheet. I2 (residues 105–115), inserted at residues 82–87 of PH0999, forms an α;-helix not found in the bacterial enzyme. I3 (residues 135–143), inserted near residues 107–109 of PH0999, forms a loop that interacts with I1.
Figure 2.
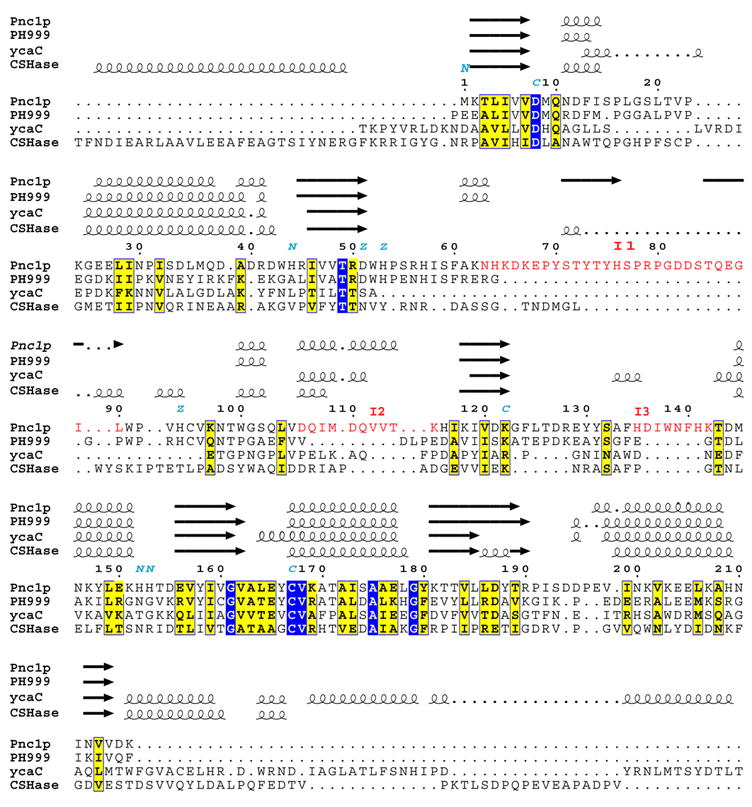
Aligned sequences of yeast Pnc1p, PH0999, YcaC, and CSHase. The alignment of these sequences obtained from a BLAST search was modified based on the structural data and prepared using the programs Mustang [35] and ESPript [34]. Identical residues are boxed and highlighted in blue. Homologous residues are boxed and highlighted in yellow. The three insertion elements I1, I2, and I3 in Pnc1p not found in PH0999 are shown in red lettering. Indicated are conserved residues that form catalytic triads (c) and Zn-binding sites (z) in Pnc1p and PH0999, and the residues (n) of Pnc1p proposed to form the surface cluster responsible for the affinity of the protein for Ni2+-coupled affinity resins.
Figure 3.
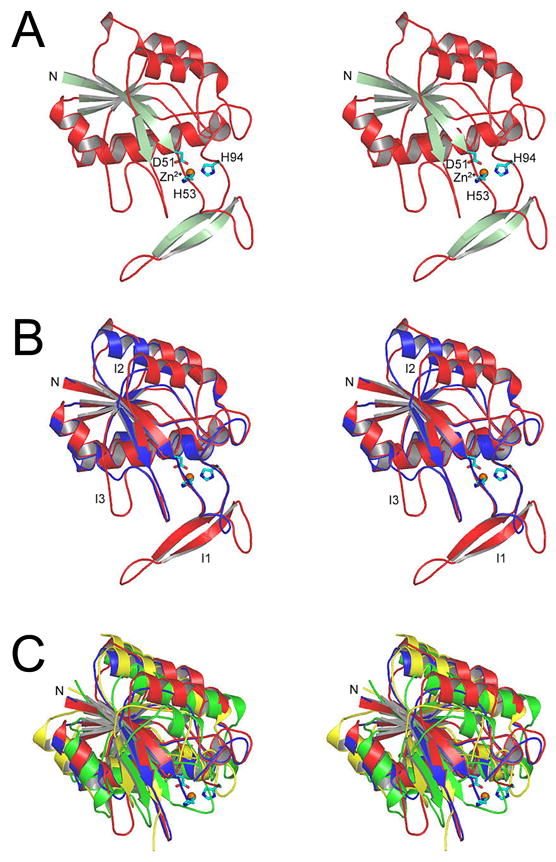
Divergent stereo views showing the overall Pnc1p fold and its similarity to three bacterial enzymes. A) The Pnc1p fold. The amino terminus is marked as N, and metal-binding residues D51, H53, and H94 are indicated. B) Structural alignment of Pnc1p and the bacterial nicotinamidase/pyrazinamidase PH0999. The backbone of Pnc1p is in shown in red and PH0999 is shown in blue. Three prominent insertions in Pnc1p as compared to PH0999 are labeled. C) Superposition of the common core folds of Pnc1p (red), PH0999 (blue), YcaC (green), and CSHase (yellow). The structural elements inserted in YcaC and CSHase relative to PH0999 and Pnc1p are shown in Figures 6 and 7.
Although Pnc1p demonstrates only weak sequence similarity with YcaC (~17% identity over 149 aligned residues) and CSHase (~16% identity over 171 aligned residues), Figure 3C shows that the common core fold of these proteins is highly conserved. The root mean square (rms) deviation between corresponding Cα; atoms of Pnc1p and the Cα; atoms of aligned residues for both YcaC and CSHase is 2.3 Å. YcaC and CSHase demonstrate ~15% identity over 165 aligned residues with a rms deviation of 2.2 Å between corresponding Cα atoms of aligned residues.
Pnc1p active site and affinity for Ni2+ resins
Pnc1p contains a metal-binding site formed by residues D51, H53, and H94 and an additional triad of residues consisting of C167, K122, and D8 (Figure 4A) that are presumed to be involved in catalysis. This first triad of amino acids is spatially conserved with respect to residues D52, H54, and H71 that participate in the binding of zinc in PH0999, while the second group is conserved spatially with respect to C133, K94, and D10 of PH0999, Cys118, R84, and D19 of YcaC, and Cys177, K144, and D51 of CSHase, all of which are presumed to act as the catalytic triads in these enzymes. A rare cis-peptide bond found in hydrolases of this family is observed between residues V162 and A163 in Pnc1p and aligns with the cis peptide bonds found between residues 128 and 129 in PH0999, between residues 113 and 114 in YcaC, and between residues 172 and 173 of CSHase. The functional importance of the catalytic cysteine residue is based on the sensitivity of CSHase to thiol-reactive agents. Structural analyses revealed a covalent bond between this cysteine and aldehyde inhibitors of that enzyme [20]. In PH0999 and Pnc1p, the thiolate moiety of the catalytic cysteine is proposed to attack the carbonyl carbon of nicotinamide (or pyrazinamide) to form a tetrahedral intermediate, and a Zn-activated water molecule is proposed to attack the resulting thioester bond to release nicotinic (or pyrazinoic) acid [14]. The cis-peptide bond orients an amide proton that, together with the amide proton of the catalytic cysteine, form an oxyanion hole to stabilize the carbonyl oxygen of the tetrahedral intermediate [14]. In the PH0999 structure, a Zn-bound water molecule sits adjacent to the position predicted to be occupied by the thioester bond, supporting the suggested role of the metal ion in catalysis [14]. The presence of structurally equivalent Zn-binding sites in Pnc1p and PH0999 and the absence of such a site in CSHase and YcaC suggest that Zn-mediated catalysis is unique to the nicotinamidase/pyrazinamidase enzymes of this family of hydrolases [14; 19; 22].
Figure 4.
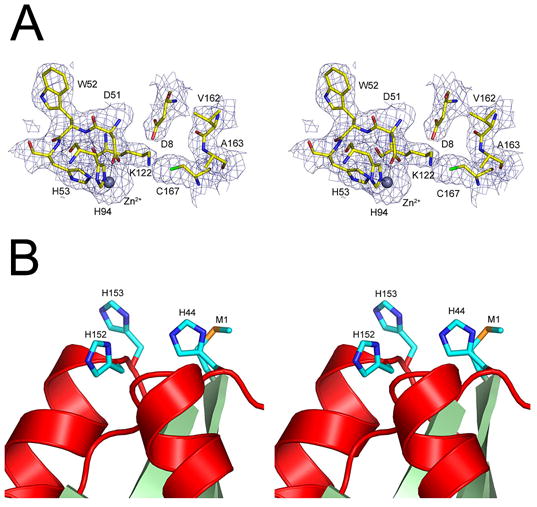
The active site and putative Ni2+-binding cluster in Pnc1p. A) 2Fo – Fc SIGMAA-weighted [45] electron density superimposed on the model of the Pnc1p metal-binding site residues (D51, H53, and H94) and catalytic triad residues (C167, K122, and D8). Also shown are residues V162 and A163 which contribute to a putative oxyanion hole as a result of their cis-peptide bond [14; 22]. B) The cluster of solvent exposed methionine and histidine residues located near the amino terminus of Pnc1p. These residues are proposed to be responsible for the observed interaction of Pnc1p with Ni2+-NTA resin. This site is occupied by metal ions in heavy atom derivative crystal forms but not in native crystal forms.
To determine whether endogenous Pnc1p co-purified with IDH due to affinity for the Ni2+-NTA column rather than an affinity for IDH, the PNC1 gene was cloned and expressed in E. coli as described in Materials and Methods. As suspected, the bacterially-expressed enzyme lacking an affinity tag could be purified to near homogeneity using only Ni2+-NTA column chromatography (Figure 1B, lane 2). A methionine (M1) and three histidine residues (H44, H152, and H153) form a cluster near the N-terminus (Figure 4B) that is likely responsible for this observed affinity of Pnc1p for nickel resins. The observation that osmium ions used for phasing bind to this cluster supports this notion. As these residues are not conserved in PH0999, CSHase, YcaC, or in related enzymes, the functional significance of this cluster is unknown.
Pnc1p superstructure
Figure 5A shows that the seven Pnc1p molecules in the asymmetric unit form a helical filamentous array, burying a total of ~1100 Å2 or 10.6% of the total solvent accessible surface area for each polypeptide. Each Pnc1p molecule is related to its two adjacent molecules by an average rotation of ~206° around the long axis of the array. The heptameric helical arrays of Pnc1p in each asymmetric unit assemble end-to-end to form an intricate, higher order framework of unbroken helical fibers running the entire length of the crystal. As shown in Figures 5B and 5C, the lynchpins of this superstructure are the blue Pnc1p molecules that reside at one end of the helical array in each asymmetric unit and are positioned on a crystallographic three-fold screw axis. In addition to binding two Pnc1p proteins along the long axis of the unbroken helical filaments, each blue Pnc1p protein binds two additional blue Pnc1p molecules along an axis approximately normal to the unbroken filament axes, forming “hubs” through which the unbroken filamentous “spokes” radiate. Figure 5C shows a view normal to the three-fold screw axis, revealing how the unbroken “spoke” filaments become interwoven to form an intricate porous mesh.
Figure 5.
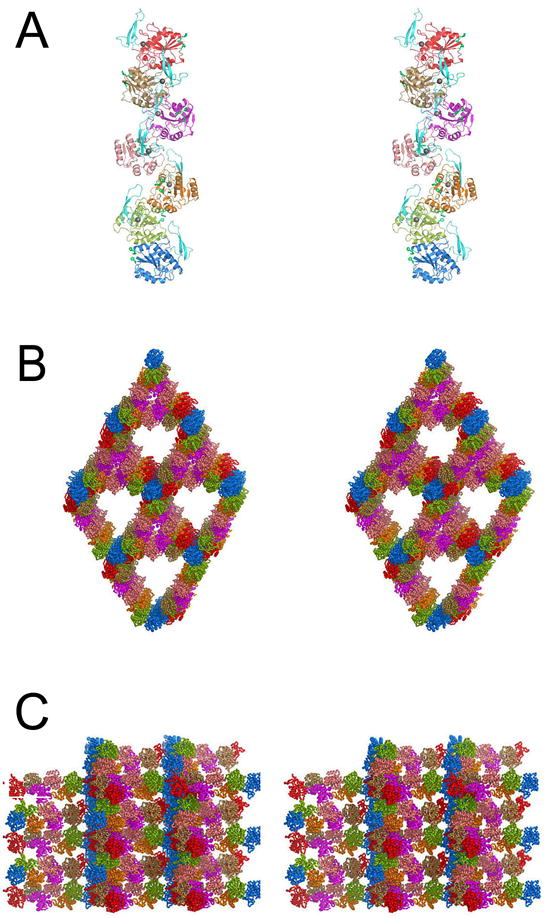
Pnc1p filamentous arrays form a porous superstructure in the crystal. A) The seven Pnc1p molecules of the asymmetric unit associate to form a helical filamentous array. Each monomer of Pnc1p is shown in a different color (see text). B) Superstructure of Pnc1p helical fibers in the crystal. The color scheme is as in A). The view is looking along the crystallographic 3-fold (center of the large solvent channels) and 3-fold screw axes (the blue “hub” Pnc1p residues and the center of the small solvent channels bordered by the salmon and magenta Pnc1p molecules). C) Superstructure of Pnc1p helical fibers in the crystal. The color scheme is as in A) and B). The view is normal to the view shown in B). Each unbroken helical array of Pnc1p is related to its neighbor along the vertical “hub” array of blue Pnc1p molecules by a 3-fold screw axis of rotation.
Because recombinant Pnc1p assembles into the higher order superstructure shown in Figure 5 over a wide range of pH and precipitating agent conditions and in the overwhelming presence of IDH, it is tempting to speculate that this assembly may have significance in vivo. When preliminary sedimentation equilibrium data are fit to various models including a single ideal species, two component non-interacting species, monomer-dimer equilibrium, or a fixed molecular weight distribution, the best fit (lowest variance) is observed for the fixed distribution model (data not shown), suggesting that Pnc1p self-associates through the sequential addition of monomers. If filamentous assembly does occur in vivo, it is possible that it might affect compartmentalization, e.g. if elevated levels of nicotinamide in the nucleus relative to the cytosol dictated a need for enhanced levels of Pnc1p in the nucleus. The Pnc1p protein in these arrays are predicted to be active, as the I1 insertion forming the two-stranded antiparallel β-sheet (lower portion of Figure 3A and 3B), the active site, and the surface His cluster near the N-terminus (Figure 4A) are all positioned on the exterior of the filament where they would be accessible to the nicotinamide substrate, metal ions, and other proteins.
The oligomeric structures of Pnc1p, YcaC and CSHase are mediated by insertion elements
Unlike Pnc1p, which associates into helical, unbroken filamentous arrays (Figure 5), YcaC and CSHase are observed as discrete globular higher order oligomers [20; 22]. Figures 6A and 7A show the YcaC and CSHase proteins superimposed with Pnc1p in the same orientation as shown in Figure 3C, revealing the regions of amino acid insertion that are distinct in each molecule. The two C-terminal helices inserted in YcaC not present in Pnc1p or CSHase provide the self-interactions that mediate the assembly of YcaC into a ring of four subunits related by a 90° rotation, which associates with another ring of YcaC molecules through a 180° rotation to generate an octamer of ~180 kDa (Figure 6B). Figure 6C shows that if the Pnc1p were packed into the YcaC octameric architecture, its I1 insertion containing the 2-stranded β-sheet would occupy the same space as the C-terminal helices of YcaC, although this structural element would be provided by the adjacent molecule in the four-membered ring. However, the nature of the resulting interactions would obviously be quite different and unfavorable for octameric assembly. Similarly, Figure 7A shows that the inserted N-terminal helix and lengthy C-terminal tail in CSHase not present in Pnc1p or YcaC provide the interactions that mediate its assembly into a tetramer with 222 symmetry (Figure 7B). Figure 7C shows that if the Pnc1p were packed into the YcaC octameric architecture, its I1 insertion would be incompatible with the interactions required around two of the two-fold axes of rotation found in the CSHase tetramer. Thus the distinct insertion elements that punctuate the overall common core fold of these enzymes, which otherwise contain superimposable active site regions, appear to modulate the assembly of these proteins into their unique higher order oligomeric states.
Figure 6.
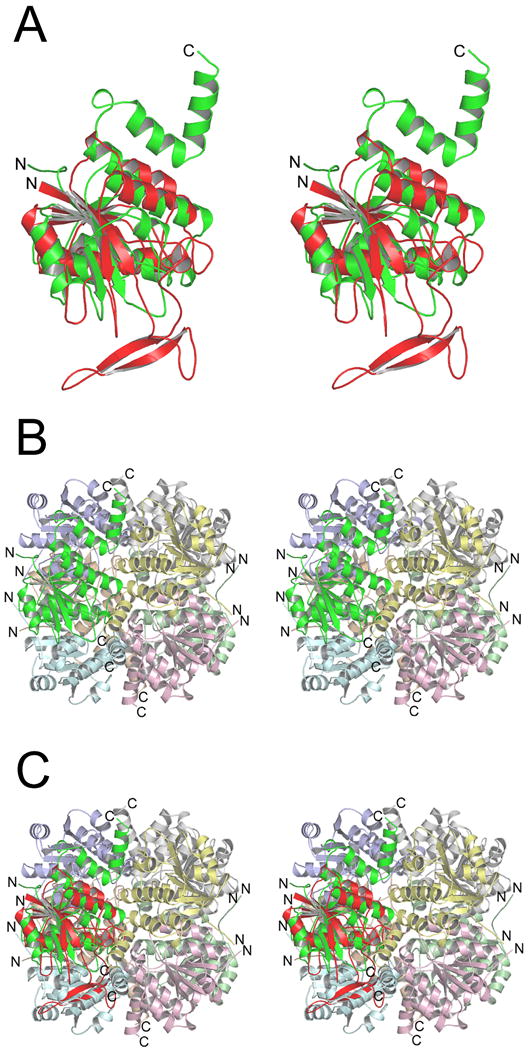
The insertion elements of Pnc1p and YcaC mediate assembly into distinct higher order oligomers (see text). The views are the same as in Figure 3. A) Superposition of Pnc1p (red) with YcaC (green) revealing the distinct amino acid insertions in each enzyme relative to the common core fold. The two green C-terminal helices in YcaC mediate its assembly into the functional octamer shown in the next panel. B) The YcaC monomer assembles into an octamer with 422 symmetry. C) Superposition of the Pnc1p molecule onto the position of each YcaC molecule in the octamer reveals that the two-stranded β-sheet insert in Pnc1p would occupy the same space as the green helices from YcaC.
Figure 7.
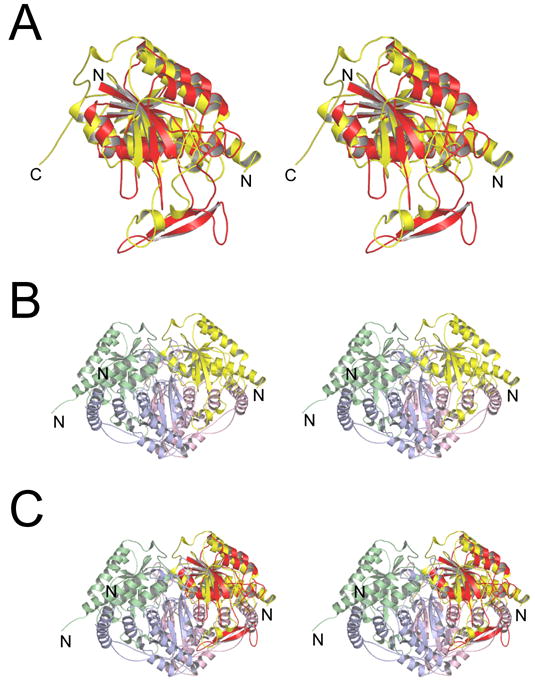
The insertion elements of Pnc1p and CSHase mediate assembly into distinct higher order oligomers (see text). A) Superposition of Pnc1p (red) with CSHase (yellow) revealing the distinct amino acid insertions in each enzyme relative to the common core fold. B) The yellow N-terminal helix and extended C-terminal tail in CSHase mediate its assembly into the functional tetramer with 222 symmetry. C) Superposition of the Pnc1p molecule onto the position of each CSHase molecule in the tetramer reveals that the two-stranded β-sheet insert in Pnc1p would disrupt the interaction across the two-fold axes in CSHase.
Pnc1p and Sir2p
Nicotinamide is both a product and a noncompetitive inhibitor of the NAD+-dependent deacetylation reaction catalyzed by yeast Sir2p [6; 7; 8]. Deletion of the yeast PNC1 gene results in a ~10-fold increase in cellular nicotinamide levels [7], suggesting that Pnc1p plays a direct role in preventing its accumulation. Normal levels of nicotinamide in yeast cells are estimated to be ~10 μM if distributed throughout the cell or ~150 μM if the metabolite is localized in the nucleus [7]. These levels are similar to those that produce inhibition of Sir2p [IC50 values of 50–120 μM [4; 8]] Furthermore, exogenous addition of nicotinamide has been shown to inhibit Sir2p-dependent silencing and to reduce life span in yeast [4; 5; 39], whereas exogenous addition of isonicotinamide, an antagonist of nicotinamide, enhances transcriptional silencing in wild-type or pnc1Δ cells [7]. Collectively, these results suggest that Sir2p-dependent processes may be sensitive to cellular concentrations of nicotinamide and that Pnc1p may play a critical role in regulating levels of this metabolite. However, there is also evidence that levels of NADH may contribute to control of Sir2p-dependent processes [40; 41]. Related to this, NADH has been shown to be a competitive inhibitor of Sir2p [40], and NAD+ has been shown to be an inhibitor of Pnc1p [1].
Pnc1p expression is elevated in response to cellular stress [2; 5; 11], and such conditions may correlate with a need for increased Sir2p activity. Pnc1p was demonstrated to be localized to the cytosol, peroxisomes, and the nucleus [11], whereas other NAD+ salvage pathway enzymes are enriched primarily in the nucleus alone [3; 10]. This pattern of localization might reflect regulation of Sir2p homologue activity in cellular compartments other than the nucleus. Although the studies described above focused primarily on Sir2p activity as related to replicative life span in yeast, there is evidence that Sir2p does not promote chronological life span extension [42]. Replicative life span is an index of the number of times a cell divides in a lifetime, whereas chronological life span is a measure of the survival time of non-replicating cells. The roles of Pnc1p and the NAD+ salvage pathway in the latter model of cellular aging remain to be clarified.
Kinetic Analyses
The kinetics of the Pnc1p enzyme were characterized using both nicotinamide and pyrazinamide as substrates. As shown in Figure 1A, the two substrates differ only at position 1 in the pyrazinamide six-membered ring. The action of Pnc1p toward these structurally related compounds was quite similar, with measured Vmax values of 50.2 ± 0.1 and 59.4 ± 1.5 units/mg for nicotinamide and pyrazinamide, respectively. The apparent Km value for nicotinamide (0.21 ± 0.02 mM) was three-fold lower than that for pyrazinamide, and was similar to the value previously reported [2]. Calculated Hill coefficients suggested cooperativity with respect to nicotinamide (2.1 ± 0.2) but little with respect to pyrazinamide (1.3 ± 0.03).
Nicotinamidases as drug targets
Mammalian cells lack a nicotinamidase analogous to Pnc1p, and the NAD+ salvage pathway involves a nicotinamide phosphoribosyltransferase that converts nicotinamide and PRPP to nicotinamide mononucleotide and pyrophosphate [43]. The absence of a nicotinamidase in humans is the basis for the efficacy of treatment of M. tuberculosis infections with pyrazinamide [17]. Since Pnc1p can also use nicotinamide and pyrazinamide as substrates as described above, we examined growth of parental and pnc1Δ yeast strains on YP glucose plates containing pyrazinamide. No differences in number or colony size were observed for either strain as a function of pyrazinamide concentrations ranging from 0 to 8 mM (data not shown). Thus, despite the robust pyrazinamidase activity of Pnc1p, yeast cells appear resistant to this drug. Several types of bacteria have nicotinamidase/pyrazinamidase activities, but are also apparently resistant to the drug [12]. For example, the nicotinamidase encoded by the E. coli pncA gene is constitutively expressed, but no effect on growth was observed in the presence of 10 mM pyrazinamide. Thus, mechanisms involved in the permeability or toxicity of this drug in M. tuberculosis may not be conserved in other organisms.
Supplementary Material
Supplementary Figure 1: Similarity of the yeast Pnc1p protein to a subdomain of bacterial isocitrate dehydrogenase (IDH) and isopropyl malate dehydrogenase (IMDH) proteins. A) Pnc1p. B) Residues 91-246 of IMDH from Sulfolobus tokodaii (Protein Data Bank entry 1WPW, unpublished) in a similar orientation.
Acknowledgments
This work was supported in part by grants from the NIH-GM051265 (to LMH) and the Robert A. Welch Foundation (to PJH). We thank Dr. Karyl I. Minard for critical comments on the manuscript, Drs. Christopher Carroll and Susan Weintraub for mass spectrometric analyses, and Drs. Borries Demeler and Virgil Schirf for preliminary analytical ultracentrifugation analyses. Support for the X-ray Crystallography Core Laboratory by the VPR and the Executive Research Committee at the University of Texas Health Science Center at San Antonio is also gratefully acknowledged.
Abbreviations and Symbols
- PH0999
Pyrococcus horikoshii nicotinamidase
- CSHase
Arthrobacter sp. N-carbamoylsarcosine amidohydrolase
- YcaC
Escherichia coli hydrolase of unknown function
- IDH
Saccharomyces cerevisiae NAD+-specific isocitrate dehydrogenase
- RMS
root mean square
- MAD
multiwavelength anomalous diffraction
- MIRAS
multiple isomorphous replacement with anomalous scattering
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Calbreath DF, Joshi JG. Inhibition of nicotinamidase by nicotinamide adenine dinucleotide. J Biol Chem. 1971;246:4334–9. [PubMed] [Google Scholar]
- 2.Ghislain M, Talla E, Francois JM. Identification and functional analysis of the Saccharomyces cerevisiae nicotinamidase gene, PNC1. Yeast. 2002;19:215–24. doi: 10.1002/yea.810. [DOI] [PubMed] [Google Scholar]
- 3.Anderson RM, Bitterman KJ, Wood JG, Medvedik O, Cohen H, Lin SS, Manchester JK, Gordon JI, Sinclair DA. Manipulation of a nuclear NAD+ salvage pathway delays aging without altering steady-state NAD+ levels. J Biol Chem. 2002;277:18881–90. doi: 10.1074/jbc.M111773200. [DOI] [PubMed] [Google Scholar]
- 4.Bitterman KJ, Anderson RM, Cohen HY, Latorre-Esteves M, Sinclair DA. Inhibition of silencing and accelerated aging by nicotinamide, a putative negative regulator of yeast sir2 and human SIRT1. J Biol Chem. 2002;277:45099–107. doi: 10.1074/jbc.M205670200. [DOI] [PubMed] [Google Scholar]
- 5.Gallo CM, Smith DL, Jr, Smith JS. Nicotinamide clearance by Pnc1 directly regulates Sir2-mediated silencing and longevity. Mol Cell Biol. 2004;24:1301–12. doi: 10.1128/MCB.24.3.1301-1312.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jackson MD, Schmidt MT, Oppenheimer NJ, Denu JM. Mechanism of nicotinamide inhibition and transglycosidation by Sir2 histone/protein deacetylases. J Biol Chem. 2003;278:50985–98. doi: 10.1074/jbc.M306552200. [DOI] [PubMed] [Google Scholar]
- 7.Sauve AA, Moir RD, Schramm VL, Willis IM. Chemical activation of Sir2-dependent silencing by relief of nicotinamide inhibition. Mol Cell. 2005;17:595–601. doi: 10.1016/j.molcel.2004.12.032. [DOI] [PubMed] [Google Scholar]
- 8.Sauve AA, Schramm VL. Sir2 regulation by nicotinamide results from switching between base exchange and deacetylation chemistry. Biochemistry. 2003;42:9249–56. doi: 10.1021/bi034959l. [DOI] [PubMed] [Google Scholar]
- 9.Lin SJ, Defossez PA, Guarente L. Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science. 2000;289:2126–8. doi: 10.1126/science.289.5487.2126. [DOI] [PubMed] [Google Scholar]
- 10.Sandmeier JJ, Celic I, Boeke JD, Smith JS. Telomeric and rDNA silencing in Saccharomyces cerevisiae are dependent on a nuclear NAD+ salvage pathway. Genetics. 2002;160:877–89. doi: 10.1093/genetics/160.3.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anderson RM, Bitterman KJ, Wood JG, Medvedik O, Sinclair DA. Nicotinamide and PNC1 govern lifespan extension by calorie restriction in Saccharomyces cerevisiae. Nature. 2003;423:181–5. doi: 10.1038/nature01578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frothingham R, Meeker-O’Connell WA, Talbot EA, George JW, Kreuzer KN. Identification, cloning, and expression of the Escherichia coli pyrazinamidase and nicotinamidase gene, pncA. Antimicrob Agents Chemother. 1996;40:1426–31. doi: 10.1128/aac.40.6.1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scorpio A, Zhang Y. Mutations in pncA, a gene encoding pyrazinamidase/nicotinamidase, cause resistance to the antituberculous drug pyrazinamide in tubercle bacillus. Nat Med. 1996;2:662–7. doi: 10.1038/nm0696-662. [DOI] [PubMed] [Google Scholar]
- 14.Du X, Wang W, Kim R, Yakota H, Nguyen H, Kim SH. Crystal structure and mechanism of catalysis of a pyrazinamidase from Pyrococcus horikoshii. Biochemistry. 2001;40:14166–72. doi: 10.1021/bi0115479. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Y, Wade MM, Scorpio A, Zhang H, Sun Z. Mode of action of pyrazinamide: disruption of Mycobacterium tuberculosis membrane transport and energetics by pyrazinoic acid. J Antimicrob Chemother. 2003;52:790–5. doi: 10.1093/jac/dkg446. [DOI] [PubMed] [Google Scholar]
- 16.Steele MA, Des Prez RM. The role of pyrazinamide in tuberculosis chemotherapy. Chest. 1988;94:845–50. doi: 10.1378/chest.94.4.845. [DOI] [PubMed] [Google Scholar]
- 17.Scorpio A, Lindholm-Levy P, Heifets L, Gilman R, Siddiqi S, Cynamon M, Zhang Y. Characterization of pncA mutations in pyrazinamide-resistant Mycobacterium tuberculosis. Antimicrob Agents Chemother. 1997;41:540–3. doi: 10.1128/aac.41.3.540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Y, Telenti A, editors. Molecular Genetics of Mycobacterium. ASM Press; Washington, D.C: 2000. [Google Scholar]
- 19.Romao MJ, Turk D, Gomis-Ruth FX, Huber R, Schumacher G, Mollering H, Russmann L. Crystal structure analysis, refinement and enzymatic reaction mechanism of N-carbamoylsarcosine amidohydrolase from Arthrobacter sp. at 2.0 A resolution. J Mol Biol. 1992;226:1111–30. doi: 10.1016/0022-2836(92)91056-u. [DOI] [PubMed] [Google Scholar]
- 20.Zajc A, Romao MJ, Turk B, Huber R. Crystallographic and fluorescence studies of ligand binding to N-carbamoylsarcosine amidohydrolase from Arthrobacter sp. J Mol Biol. 1996;263:269–83. doi: 10.1006/jmbi.1996.0574. [DOI] [PubMed] [Google Scholar]
- 21.Kim JM, Shimizu S, Yamada H. Purification and characterization of a novel enzyme, N-carbamoylsarcosine amidohydrolase, from Pseudomonas putida 77. J Biol Chem. 1986;261:11832–9. [PubMed] [Google Scholar]
- 22.Colovos C, Cascio D, Yeates TO. The 1.8 A crystal structure of the ycaC gene product from Escherichia coli reveals an octameric hydrolase of unknown specificity. Structure. 1998;6:1329–37. doi: 10.1016/s0969-2126(98)00132-4. [DOI] [PubMed] [Google Scholar]
- 23.Hu G, Taylor AB, McAlister-Henn L, Hart PJ. Crystallization and preliminary X-ray crystallographic analysis of yeast NAD+-specific isocitrate dehydrogenase. Acta Crystallograph Sect F Struct Biol Cryst Commun. 2005;61:486–8. doi: 10.1107/S1744309105010651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu G, Taylor AB, McAlister-Henn L, Hart PJ. Crystallization and preliminary X-ray crystallographic analysis of yeast NAD+-specific isocitrate dehydrogenase: Corrigendum. Acta Crystallogr F. 2006;F62:709. doi: 10.1107/S1744309105010651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–54. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 26.Otwinowski Z, Minor W. In: International Tables for Crystallography. Rossman M, Arnold E, editors. Kluwer, Kluwer; Dordrecht, The Netherlands: 2001. pp. 226–235. [Google Scholar]
- 27.Terwilliger TC. Maximum-likelihood density modification. Acta Crystallogr D Biol Crystallogr. 2000;56:965–72. doi: 10.1107/S0907444900005072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Terwilliger TC, Berendzen J. Automated MAD and MIR structure solution. Acta Crystallogr D Biol Crystallogr. 1999;55:849–61. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de La Fortelle E, Bricogne G. Methods in Enzymology. Academic Press; New York: 1997. Maximum-likelihood heavy-atom parameter refinement; pp. 472–494. [DOI] [PubMed] [Google Scholar]
- 30.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 31.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 32.Winn MD, Isupov MN, Murshudov GN. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr D Biol Crystallogr. 2001;57:122–33. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
- 33.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Cryst. 1993;26:283–291. [Google Scholar]
- 34.Gouet P, Courcelle E, Stuart DI, Metoz F. ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics. 1999;15:305–8. doi: 10.1093/bioinformatics/15.4.305. [DOI] [PubMed] [Google Scholar]
- 35.Konagurthu AS, Whisstock JC, Stuckey PJ, Lesk AM. MUSTANG: a multiple structural alignment algorithm. Proteins. 2006;64:559–74. doi: 10.1002/prot.20921. [DOI] [PubMed] [Google Scholar]
- 36.Cupp JR, McAlister-Henn L. NAD+-dependent isocitrate dehydrogenase. Cloning, nucleotide sequence, and disruption of the IDH2 gene from Saccharomyces cerevisiae. J Biol Chem. 1991;266:22199–205. [PubMed] [Google Scholar]
- 37.Cupp JR, McAlister-Henn L. Cloning and characterization of the gene encoding the IDH1 subunit of NAD+-dependent isocitrate dehydrogenase from Saccharomyces cerevisiae. J Biol Chem. 1992;267:16417–23. [PubMed] [Google Scholar]
- 38.Holm L, Sander C. Protein structure comparison by alignment of distance matrices. J Mol Biol. 1993;233:123–38. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- 39.Kaeberlein M, Hu D, Kerr EO, Tsuchiya M, Westman EA, Dang N, Fields S, Kennedy BK. Increased life span due to calorie restriction in respiratory-deficient yeast. PLoS Genet. 2005;1:e69. doi: 10.1371/journal.pgen.0010069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin SJ, Ford E, Haigis M, Liszt G, Guarente L. Calorie restriction extends yeast life span by lowering the level of NADH. Genes Dev. 2004;18:12–6. doi: 10.1101/gad.1164804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin SJ, Kaeberlein M, Andalis AA, Sturtz LA, Defossez PA, Culotta VC, Fink GR, Guarente L. Calorie restriction extends Saccharomyces cerevisiae lifespan by increasing respiration. Nature. 2002;418:344–8. doi: 10.1038/nature00829. [DOI] [PubMed] [Google Scholar]
- 42.Fabrizio P, Gattazzo C, Battistella L, Wei M, Cheng C, McGrew K, Longo VD. Sir2 blocks extreme life-span extension. Cell. 2005;123:655–67. doi: 10.1016/j.cell.2005.08.042. [DOI] [PubMed] [Google Scholar]
- 43.Magni G, Amici A, Emanuelli M, Orsomando G, Raffaelli N, Ruggieri S. Enzymology of NAD+ homeostasis in man. Cell Mol Life Sci. 2004;61:19–34. doi: 10.1007/s00018-003-3161-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Keys DA, McAlister-Henn L. Subunit structure, expression, and function of NAD(H)-specific isocitrate dehydrogenase in Saccharomyces cerevisiae. J Bacteriol. 1990;172:4280–7. doi: 10.1128/jb.172.8.4280-4287.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Read RJ. Improved Fourier coefficients for maps using phases from partial structures with errors. Acta Crystallogr. 1986;A42:140–149. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1: Similarity of the yeast Pnc1p protein to a subdomain of bacterial isocitrate dehydrogenase (IDH) and isopropyl malate dehydrogenase (IMDH) proteins. A) Pnc1p. B) Residues 91-246 of IMDH from Sulfolobus tokodaii (Protein Data Bank entry 1WPW, unpublished) in a similar orientation.


