Abstract
Neuroadaptive changes that occur in the development of ethanol tolerance may be the result of alterations in gene expression. We have shown that PKCγ wild-type mice develop tolerance to the sedative-hypnotic effects of ethanol after chronic ethanol treatment; whereas, mutant mice do not, making these genotypes a suitable model for identifying changes in gene expression related to tolerance development. Using a two-stage process, several genes were initially identified using microarray analyses of cerebellar tissue from ethanol-treated PKCγ mutant and wild-type mice. Subsequent confirmation of a subset of these genes using qRT-PCR was done to verify gene expression changes. A total of 109 genes from different functional classifications were identified in these groups on the microarrays. Eight genes were selected for verification: three, Twik-1, Plp, and Adk2, were chosen as genes related to tolerance; another three, Hsp70.2, Bdnf, and Th, were chosen as genes related to resistance to tolerance; and two genes, JunB and Nur77, were selected as candidate genes sensitive to chronic ethanol. The results from the verification experiments indicated that Twik-1, which codes for a potassium channel, was associated with tolerance and appeared to be dependent on the presence of PKCγ. No genes were confirmed to be related to resistance to tolerance; however, expression of two of these, Hsp70.2 and Th, were found to be sensitive to chronic ethanol and were added to the transcription factors, JunB and Nur77, confirmed by qRT-PCR, as a subset of genes that respond to chronic ethanol.
Keywords: Microarray, chronic ethanol, qRT-PCR, cerebellum, mouse, PKCγ
INTRODUCTION
Chronic exposure to ethanol induces persistent behavioral changes such as tolerance, dependence, addiction, and relapse. These behavioral changes are likely the result of ethanol-induced neuroadaptation in response to repeated exposure to the drug (Hoffman et al., 2000; Nestler and Aghajanian, 1997). Evidence suggests that changes in gene expression underlie these neuroadaptive processes (Rahman and Miles, 2001). Recently, several studies have used DNA microarrays to profile gene expression changes in rodents to identify genes related to alcohol preference (Edenberg et al., 2005; McBride et al., 2005; Mulligan et al., 2006; Murphy et al., 2002) and genes that respond to acute ethanol administration (Kerns et al., 2005). Gene expression changes after chronic ethanol treatment have been observed in mouse and rat hippocampus (Daniels and Buck, 2002; Saito et al., 2002) and in striatum (Smith et al., 2006) as well as in brain regions from human alcoholics (Flatscher-Bader et al., 2005, 2006; Lewohl et al., 2000; Mayfield et al., 2002). The investigations of chronic ethanol-induced gene expression alterations have identified genes from many different functional groups that are associated with the long-lasting effects of chronic ethanol exposure that encompass tolerance, dependence, and addiction. It has been hypothesized that the development of tolerance leads to dependence, ultimately resulting in addiction (Kalant, 1996). Therefore, the identification of genes associated with tolerance would provide genetic information concerning the first step in this process. Kalivas (2005) suggests that experiments that are designed to isolate specific parts of this temporal sequence would go a long way in distinguishing neuroadaptations related to each phase.
Data from this laboratory have shown that chronic ethanol diet produces tolerance to the sedative-hypnotic effects of ethanol in protein kinase C γ (PKCγ) wild-type mice, but not in PKCγ null mutant littermates (Bowers et al., 1999). This difference in tolerance development provides a unique comparison for identifying genes associated specifically with ethanol-induced tolerance since the PKCγ genotypes are isogenic except for the lack of PKCγ in the mutant line. We have shown that ethanol potentiation of GABAA receptor function (Harris et al., 1995; Proctor et al., 2003) and 5-HT2A/C (Bowers et al., submitted) receptor function are different between the genotypes in naïve animals. In order to identify patterns of gene expression that might reveal important neural pathways as well as single gene changes related to tolerance, we used Affymetrix microarrays to compare gene expression in the cerebellum of PKCγ mutant and wild-type mice after control- and chronic ethanol-diet. Those genes whose expression changes in wild-type mice, but not mutants, after chronic ethanol might be expected to be associated with the development of tolerance. Gene expression changes seen in mutant, but not wild-type mice, might be expected to be associated with a resistance to tolerance development; and gene expression changes observed in both genotypes could be classified as ethanol-responsive genes associated with the effects of chronic treatment. A two stage strategy was used with the microarray analyses providing a list of putative genes followed by confirmation using quantitative real time reverse transcriptase polymerase chain reactions (qRT-PCR) of a subset of genes from each of these groups.
We chose to perform the analyses in cerebellum because of its role in the sedative-hypnotic effects of ethanol, specifically the genetic correlation between Purkinje cell depression and the duration of the loss of righting reflex (Sorensen et al., 1980; Spuhler et al., 1982). PKCγ is neuronal specific and it is located throughout the brain including the Purkinje cells of the cerebellum (Naik et al., 2000). The cerebellum has been shown to be a brain region particularly sensitive to the toxic effects of chronic alcohol. Specifically, Purkinje cells and granule cells are vulnerable to cell loss and may represent the cerebellar atrophy seen in alcoholics (Andersen, 2004; Karhune et al., 1994; Oberdoerster and Rabin, 1999; Phillips et al., 1987).
MATERIALS and METHODS
2.1 Animals
PKCγ null mutant mice were derived using gene-targeting and homologous recombination techniques (Abeliovich et al., 1993) and are currently bred on an F1 C57BL/6 X 129/S6 mixed genetic background at the Institute for Behavioral Genetics (Boulder, CO). The F1 generations are bred from heterozygous crosses from two congenic strains: C57BL/6.PKCγ and 129/S6.PKCγ. This breeding strategy produces homozygous mutant, heterozygous, and homozygous wild-type genotypes within a single litter, thereby providing wild-type littermate controls for each experiment. The PKCγ mutation is maintained in a heterozygous condition on C57BL/6 and 129/S6 inbred strains because homozygous null mutant mice do not survive on the C57BL/6 background (Bowers and Wehner, 2001). Genotyping was done as described by Bowers et al. (1999). Mice were housed in like-sex groups of 2–5 composed of all genotypes and were maintained on a 12-hour light/dark cycle (lights on at 0700). Food and water were available ad libitum. All animal use procedures were performed in accordance with the NIH Guide for Care and Use of Laboratory Animals and were approved by the University of Colorado IACUC.
2.2 Chronic Ethanol Treatment
The basic experimental design was similar to that used to compare striatal gene expression in PKCγ wild types with mutants (Smith et al., 2006). Wild-type and mutant littermates were derived from 28 different litters for these studies. Mice were singly housed for chronic ethanol treatment and were weighed every other day. Ethanol was administered in a protein-enriched liquid diet that had been shown previously to produce tolerance in PKCγ wild types as described by Bowers et al. (1999) according to the following schedule: Day 1: 0% ethanol derived calories (EDC), Days 2 to 4: 18.5% EDC, Days 5–11: 27% EDC. Ethanol was replaced with sucrose in the control diet to provide a caloric balance to the calories derived from ethanol. Mutants consume less ethanol diet (Bowers et al., 1999) than wild-type mice; therefore, to control for volume, wild-type ethanol diet and all control diet animals had their intake yoked to the average mutant ethanol diet intake from the previous day. Wild types and mutants do not differ in ethanol metabolism (Harris et al., 1995). The liquid diet is the sole source of nutrition; therefore, if any animal lost more than 15% of their body weight due to this regimen, single food pellets (4.0 g) were provided to all mice to facilitate weight gain.
Blood ethanol concentrations were determined in an independent experiment using heterozygous PKCγ mice (n = 8) because of the possibility that stress related to removal of blood would influence gene expression. On Day 12, immediately after diets were removed, blood was collected from the retro-orbital sinus and ethanol concentrations determined as described by Smolen et al. ( 1986). Ethanol concentrations averaged 212.73 + 81.7 mg% in this subset of mice.
2.3 Experimental Design of Gene Expression Studies
The analyses of cerebellar gene expression in PKCγ wild types and mutants were designed to be performed in two stages. In the first stage, Affymetrix arrays were used to provide a list of putative candidate genes that appeared to differ as a function of genotype and/or treatment. Because the number of samples was small in the microarray experiments, the power of the statistical analyses was limited. Therefore, changes in gene expression were verified in a subset of genes of interest using qRT-PCR techniques with RNA extracted from a group of mice treated independently from those used in the microarray experiments. An additional feature of the qRT-PCR confirmation process was that primer and probe sequences were designed from a different location on the gene than was used for the probe sequences on the arrays.
To prevent withdrawal, cerebella were removed immediately after removal of the diets at 0900 hr. For the microarray analyses, cerebellar tissues were collected from individual control- and ethanol-treated mice and RNA isolated from individual mice. Total cerebellar RNA was pooled from three mice for a total of 16 pools from 48 mice (12 mutant control-diet; 12 mutant ethanol-diet; 12 wild-type control diet; 12 wild-type ethanol diet). For the qRT-PCR confirmation studies, RNAs from individual mice from different litters and different ethanol treatment experiments were analyzed. Routinely, tissues from 9–15 mice from each group were analyzed in the qRT-PCR studies.
2.5 Tissue Preparation, RNA Isolation, cRNA Preparation
Procedures were identical to those described in Smith et al. (2006). Briefly, immediately after the diets were removed, mice were sacrificed by cervical dislocation. Cerebella were dissected on ice and frozen in liquid nitrogen, then stored at −70°C until use. Total RNA was isolated using the RNeasy Midi Kit (Qiagen, Valencia, CA) following polytron homogenization. RNA samples were stored individually in separate tubes at −70°C. RNA sample preparation and hybridization were carried out at the Gene Expression Core Facility at the University of Colorado Health Sciences Center (Denver, CO) according to the protocol outlined in the Expression Analysis Technical Manual (Affymetrix, Inc., Santa Clara, CA).
Biotin-labeled cRNA was synthesized from the ds-cDNA template by in vitro transcription (IVT) using ENZO Bioarray High Yield RNA Transcript Labeling Kit (T7) (Enzo Diagnostics, Inc. Farmingdale, NY) in the presence of biotinylated ribonucleotides and was purified using the RNeasy Mini Kit (Qiagen) for RNA cleanup. cRNA was fragmented for 35 min at 94°C in fragmentation buffer (Affymetrix, Inc.) then added to hybridization cocktail (Affymetrix, Inc.). Biotinylated cRNA samples were hybridized to MG-U74Av2 GeneChip (Affymetrix, Inc.) and were incubated for 16 hours at 45°C in a GeneChip Hybridization Oven 460 (Affymetrix, Inc.). The hybridized samples were washed and stained with Streptavidin-phycoerythrin using a GeneChip Fluidics Station 400 (Affymetrix, Inc.).
2.6 Quantitative Real-Time RT-PCR (qRT-PCR)
Quantitative RT-PCRs were performed as described in Smith et al. (2006) using the ABI PRISM™ Sequence Detection System 7000 and the ABI Gene Amp PCR System 9700 (Applied Biosystems Inc., Foster City, CA) controlled by ABI PRISM 7000 SDS Software Version 1.0 and SDS 7900HT Software Version 2.2, respectively (Applied Biosystems, Inc.). BLAST analyses were conducted for all primer and probe sequences to insure that that they were specific for their respective genes and were selected from a different region of the gene than those represented by the Affymetrix MG-U74Av2 probe sets (Liu et al., 2003). The gene used as the reference gene was hypoxanthine guanine phosphoribosyl transerase (HPRT). Cerebellar RNA was isolated as described above and stored in 1ug/ul aliquots at −70°C until used. qRT-PCR was conducted using the TaqMan® One-Step RT-PCR Master Mix Reagents Kit (Applied Biosystems, Inc.).
RNA samples were run in triplicate for the genes of interest and for the reference gene within the same experiment. Triplicate Cts were averaged for each sample. The choice of HPRT as a reference gene was supported by the observation that expression of HPRT did not differ among the experimental groups. Relative quantification of gene expression (relative amount of target RNA) was determined using an equation adapted from the equation 2−ΔΔCt outlined in Applied Biosystem User Bulletin #2.
2.7 Statistical Analysis
Four U74Av2 microarrays were analyzed for each genotype/treatment combination with each individual sample consisting of RNA pooled from three mice (see above). Individual arrays were assessed for various quality control parameters as described in the Affymetrix GeneChip Expression Analysis Technical Manual (2004). Probe sets were converted to a single expression value using the Tukey’s one-step biweight procedure (MAS5, Affymetrix). Global scaling was applied such that the average signal intensity of the array was set to a default target signal of 500. All subsequent analyses were conducted in GeneSpring GX (v. 7.2, Agilent Technologies) or MS Excel 2000. Probe sets called “absent” by the MAS5 algorithm (i.e., undetectable levels of the transcript) were removed under the condition that retained transcripts were required to be present in 3 out of 4 arrays in at least 1 of the 4 genotypes/treatment conditions. Higher level analyses are described below under Results.
Relative amounts of RNA calculated from the qRT-PCR experiments were analyzed by two-way ANOVA with diet-treatment and genotype as independent variables. Because the qRT-PCR assays were done to confirm results from the microarrays, planned comparisons were done apriori within genotype using one-tailed Student’s t-tests to determine diet effects.
RESULTS
3.1 Microarrays
Of the 12,488 probe sets on the U74Av2 microarray, 6315 reached the criterion of “present” (i.e., expressed at a detectable level) in at least 3 out 4 samples from each group (wild-type ethanol and control, mutant ethanol and control). Using this basic set of genes, a one-way ANOVA was conducted to test for differences due to ethanol treatment within each genotype. Because the experimental design required that confirmation studies would be performed, a liberal statistical criteria of p < 0.05, and a fold change of greater than + 1.3, were applied to array results. In general, changes in gene expression rarely reached over two-fold (mutant: 1.39 + 0.02 fold; wild-type: 1.38 + 0.04 fold). This is consistent with results from microarray data comparing PKCγ mutant and wild-type gene expression in striatum (Smith et al, 2006) as well as results from other gene expression studies in human and mouse (Lewohl et al, 2000; Daniels and Buck, 2002). Genes from each genotype were subjected to functional group analysis using the DAVID Bioinformatics Resources Database (http://niaid.abcc.ncifcrf.gov/home.jsp). All genes were categorized into functional groups at GO Level 2. The parent GO levels were Biological Processes, Cellular Components, and Molecular Function, which were divided further into Level 2 GO terms. Each group was enriched by the genes listed within that group at a significance level of p < 0.05. There was considerable overlap among the genes for assignment to particular groups; therefore, the tables list the genes in the order in which they appeared first in the analysis, but are also identified with additional group assignments as they appeared later by the superscript classifications. Genes that changed in both genotypes were also analyzed; however, no genes were significantly assigned to any functional group. Table 1 lists 44 genes whose expression changed by chronic ethanol treatment in mutant mice only; 50 EST’s were also significantly changed (data not shown). Of these 44 genes, 27 were up-regulated and 17 were down-regulated. Table 2 lists 43 genes whose expression changed as a result of chronic ethanol-treatment in wild-type mice only; 43 ESTs were also significantly changed (data not shown). Twenty-six genes were up-regulated and 17 were down-regulated. The functional group analyses resulted in more gene-enriched groups in the mutant mice compared to wild-type mice. In addition to the functional group analyses, genes from each genotype were subjected to KEGG pathway mapping using DAVID. In the mutant mice, no genes significantly mapped to a particular pathway; however, in the wild-type mice three genes significantly mapped to the inositol phosphate metabolism pathway at p < 0.05. These genes were dystrophia myotonica-protein kinase, cdc-like kinase, and inositol (myo)-1(or4)-monophosphatase 1. These three genes also mapped to the phosphatidylinositol signaling system pathway at p = 0.075. This suggests that in wild-type mice chronic ethanol may activate the phosphatidylinositol system; however, in mice that lack PKCγ this pathway may be disrupted.
TABLE 1.
PKCγ mutant expression changes due to chronic ethanol diet. Cellular process with fold change ≥ ±1.3; p<0.05. Genes are divided into functional groups; p < 0.05.
| Second Level GO Term | Gene Name | Gene Description | Fold Change | Sixth Level GO Term | Genbank Accession | Locuslink Accession | Affymetrix Accession |
|---|---|---|---|---|---|---|---|
| Cellular Physiological Processes | Lyn | Yamaguchi sarcoma viral (v-yes-1) oncogene homolog 1,3,13 | −1.46 | ATP binding; protein kinase activity | M57696 | 17096 | 103349_at |
| Igflr | Insulin-like growth factor I receptor1,3,12,13 | 1.39 | Insulin–like growth factor receptor signaling pathway | AF056187 | 16001 | 102224_at | |
| Arl4 | ADP-ribosylation factor-like 41,9,10,11,12 | 1.71 | GTP binding; GTPase activity | Y12577 | 11861 | 92805_s_at | |
| Tle4 | Transducin-like enhancer of split 4, homolog of Drosophila E(spl)1,2,3,9,10,11,13 | 1.81 | Wnt receptor signaling pathway | U61363 | 21888 | 102959_at | |
| Th | Tyrosine hydroxylase 1,3,8,9,11 | 2.23 | Catecholamine biosynthesis | M69200 | 21823 | 100690_at | |
| Col4a1 | Procollagen, type IV, alpha 11,10,11 | 1.51 | Extracellular matrix structural constituent | M15832 | 12826 | 101093_at | |
| Clqr1 | Complement component 1 Q subcomponent, receptor 11,4,9,10,11,12 | 1.73 | Calcium binding; defense response | AF081789 | 17064 | 93694_at | |
| Spp1 | Secreted phosphoprotein 11,2,4,5,8,12,13 | 1.71 | Cell adhesion; cytokine activity | X13986 | 20750 | 97519_at | |
| Grik2 | Glutamate receptor, ionotropic, kainate 2 (beta 2)1,12,13 | 1.57 | Glutamate-gated ion channel; ion transport | X66117 | 14806 | 98863_at | |
| Bdnf | Brain derived neurotrophic factor1,2,3,5,8,9,10,11,12,13 | −2.07 | Growth factor activity; feeding behavior | X55573 | 12064 | 102727_at | |
| Rad50 | RAD50 homolog (S. cerevisiae)1,3,4,7,9,10,11,14 | −1.35 | ATP binding; DNA repair | U66887 | 19360 | 100459_at | |
| Casp3 | Caspase3, apoptosis related cysteine protease1,3,4,5,10,14 | 1.42 | Caspase activity; apoptosis; B-cell homeostasis | U63720 | 12367 | 98437_at | |
| Bat8 | HLA-B associated transcript 81,3,9,10,11,12 | 1.43 | DNA binding; chromatin modification | AF109906 | 110147 | 100946_at | |
| Hist 1 H2ab | Histone 1, H2ab1,3,9,10,11 | 1.33 | DNA binding; nucleosome assembly | M33988 | 319172 | 94805_f_at | |
| Rbp1 | Retinol binding protein 1, cellular1,3 | 1.41 | Retinoid metabolism | X60367 | 19659 | 104716_at | |
| Gas1 | Growth arrest specific 11,2,5,13 | 1.31 | Cell cycle arrest | X65128 | 14451 | 94813_at | |
| Gadd45 a | Growth arrest and DNA-damage-inducible 45 alpha1,2,4,7,9,10,11,13 | 1.59 | Cell cycle arrest; response to DNA damage | U00937 | 13197 | 102292_at | |
| Dnmt2 | DNA methyltransferase 21,3,9,10,11 | −1.68 | DNA binding; proteolyis | AF012129 | 13434 | 93470_at | |
| Hmgn2 | High mobility group nucleosomal binding domain 21,3,9,10,11 | 1.52 | DNA binding; DNA packaging | X12944 | 15331 | 101589_at | |
| Zfp61 | Zinc finger protein 611,3,9,10,11,14 | −1.34 | Regulation of transcription; DNA-dependent | L28167 | 22719 | 102303_i_at | |
| Irx3 | Iroquois related homeobox 31,3,9,10,11 (Drosophila) | −1.41 | DNA binding; regulation of transcription | Y15001 | 16373 | 99034_at | |
| Ctsz | Cathepsin Z1,3,9,10,11,12,14 | 1.39 | Lysosome; proteolysis | AJ242663 | 64138 | 92633_at | |
| Ddc | Dopa decarboxylase1,3 | −1.75 | Catecholamine biosynthesis | AF071068 | 13195 | 160074_at | |
| Usp9x | Ubiquitin specific protease 9,X chromosome1,3,14 | −1.33 | Ubiquitin-dependent protein catabolism | U67874 | 22284 | 99102_at | |
| Bcat2 | Branched chain aminotransferase 2, mitochondrial1,3,9,10,11,12 | 1.44 | Transferase activity; metabolism | AF031467 | 12036 | 100443_at | |
| Psmb10 | Proteasome(prosome, macropain) subunit, beta type10 1,3,10,14 | −1.32 | Endopeptidase activity | Y10875 | 19171 | 101486_at | |
| Tpi | Triosephosphate isomerase1,3 | −1.31 | Fatty acid biosynthesis | L31777 | 21991 | 99566_at | |
| Hsd17b 12 | Hydroxysteroid (17-beta) dehydrogenase 121,3 | −1.45 | Oxidoreductase activity; steroid biosynthesis | AF064635 | 56348 | 94276_at | |
| Pja1 | Praja1, RING-H2 motif containing1,3,13 | 1.64 | Ligase activity; protein catabolism | U06944 | 18744 | 101461_f_at | |
| Nab2 | Ngfi-A binding protein 21,2,3,9,10,11 | −1.35 | Regulation of transcription | U47543 | 17937 | 100962_at | |
| Agt | Angiotensin1,2,3,4,6,8,12,13 | 1.33 | Regulation of blood pressure | AF045887 | 11606 | 101887_at | |
| Hspa2 | Heat shock protein 21,3,4,9,10,11,13 | 1.63 | Chaperone activity; response to heat | M20567 | 15512 | 99816_at | |
| Hspa1b | Heat shock protein 1B1,2,3,4,5,7,13 | 1.43 | Response to heat; anti-apoptosis | AF109906 | 15511 | 97809_at | |
| Per2 | Period homolog 2 (Drosophila)1,3,6,10,11,13 | −2.40 | Circadian rhythm; signal transduction | AF036893 | 18627 | 93694_at | |
| Rhythmic Processes | Cry1 | Cryptochrome 1 (photolyase-like)6 | −1.33 | Circadian rhythm; lyase activity | AB000777 | 12952 | 94420_f_at |
| Membrane Bound Organelle | Cops7a | COP9 (constitutive photo-morphogenic) homolog, subunit 7a (Arabidopsis thaliana)9,10,11 | −1.31 | Cellular and developmental processes | AF071316 | 26894 | 92874_at |
| Anxa11 | Annexin A119,10,11,13 | 1.45 | Binds to calcyclin | U65986 | 11744 | 102815_at | |
| Extracellular Space | Ogn | Osteoglycan12,13 | 1.82 | Growth factor activity | D31951 | 18295 | 99549_at |
| Intracellular | Mtcp1 | Mature T-cell proliferation 110 | 1.36 | Mitochondrion | Z35294 | 26894 | 103043_at |
| Copa | Coatomer protein complex subunit alpha1,9,10,11,12 | −1.31 | Mediates protein transport | AJ010391 | 12847 | 96854_at | |
| Ggh | Gamma-glutamyl hydrolase1,3,9,10,11,12,14 | 1.41 | Lysosome; hydrolase activity | AF051102 | 14590 | 93575_at | |
| Nefh | Neurofilament, heavy polypeptide 1,9,10,11 | −1.30 | Intermediate filament | M35131 | 18026 | 103234_at | |
| No Assignment to Functional Group at GO Level 2 at p < 0.05 | A33010-N21Rik | RIKEN cDNA A330103N2 gene | 1.39 | AF056187 | 102224_at | ||
| Myadm | Myeloid-associated differentiation marker | 1.33 | Integral to membranes | AJ001616 | 50918 | 96285_at |
Functional Groups as determined by DAVID Bioinformatics Functional Group Analysis (http://niaid.abcc.ncifcrf.gov/home.jsp):
Cellular Physiological Processes;
Negative Regulation of Biological Processes;
Metabolism;
Response to Stress;
Death;
Rhythmic Processes;
Response to Endogenous Stimuli;
Behavior;
Membrane-Bound Organelle;
Intracellular;
Intracellular Organelle;
Extracellular Space;
Protein Binding;
Hydrolase Activity.
TABLE 2.
PKCγ wild-type expression changes due to chronic ethanol diet with fold change ≥ ±1.3; p < 0.05. Genes are divided in functional groups; p < 0.05.
| Second Level GO Term | Gene Name | Gene Description | Fold Change | Sixth Level GO Term | Genbank Accession | Locuslink Accession | Affymetrix Accession |
|---|---|---|---|---|---|---|---|
| Cellular Physiological Processes | Prkcd | Protein kinase C delta1,3,7 | 1.37 | Protein serine/threonine kinase activity; amino acid phosphorylation | X60304 | 18753 | 104531_at |
| Ank1 | Ankyrin 1 erythroid 1,4,6 | 1.35 | Transcription factor activity; cytoskeleton | X69064 | 11733 | 10441_s_at | |
| Edn1 | Endothelin 11,2,3,6 | 1.89 | G-protein coupled receptor protein signaling pathway | U35233 | 13614 | 102737_at | |
| Tle1 | Transducin-like enhancer of split 1, homolog of Drosophila E(sp1)1,4,5,6 | −1.32 | Wnt receptor signaling pathway; regulation of transcription | U61362 | 21885 | 102425_at | |
| Egr1 | Early growth response 11,4 | 1.42 | Regulation of transcription | M28845 | 13653 | 98579_at | |
| Chd1 | Chromodomain helicase DNA binding protein 1 1,4,5,7 | 1.4 | Chromatin assembly/disassembly; ATP-dependent helicase activity | L10410 | 12648 | 101459_at | |
| Apaf1 | Apoptotic protease activating factor 11,2,3,4,5,6,7 | −1.41 | Apoptosis; brain development; ATP binding | AF064071 | 11783 | 103796_at | |
| Sept6 | Septin 61,7 | 1.33 | GTP binding; cytokinesis | AB023622 | 56526 | 92462_at | |
| Tfrc | Transferrin receptor1,4,5 | −3.44 | Endocytosis; iron ion transporter activity; proteolyisis/peptidolysis | X57349 | 22042 | X57349_5_at | |
| Kcnk1 | Potassium channel, subfamily K, member 11,4,5,6 | −1.73 | Potassium ion transport; integral to membrane | AF033017 | 16525 | 102335_at | |
| Dm15 | Dystrophia myotonica kinase, B151,7 | 1.42 | Protein serine/threonine kinase activity; meiosis | Z38015 | 13401 | 93431_at | |
| Mt3 | Metallothionein 31,3,4,5 | 1.37 | Metal ion binding; negative regulation of neurogenesis | M93310 | 17751 | 95340_at | |
| Alas2 | Aminolevulinic acid synthase 2, erythroid1,5 | −2.00 | 5-aminolevulinate syntahse activity; biosynthesis | M15268 | 11656 | 92768_at | |
| Dao1 | D-amino acid oxidase1,4,5,7 | 1.38 | Electron transport; tRNA aminoacylation for protein translation | D10210 | 13142 | 103602_at | |
| Lamp2 | Lysosomal membrane glycoprotein 21,4,5,7 | 1.35 | tRNA aminoacylation for protein translation; lysosome | M32017 | 16784 | 100136_at | |
| Clk | CDC-like kinase1,4,5,7 | −1.31 | Protein sesrine/threonine kinase activity | M38381 | 12747 | 93274_at | |
| Klf4 | Kruppel-like factor 4 (gut)1,4,5 | 1.53 | Regulation of transcriptiom | U20344 | 16600 | 99622_at | |
| Max | Max protein1,4,5,6 | −1.32 | Regulation of transcription | M63903 | 17187 | 99095_at | |
| Tcfap2b | Transcription factor AP-2 beta1,4,5,6 | 1.33 | Transcription factor activity; DNA binding | X78197 | 21419 | 92903_at | |
| Rad23b | RAD23b homolog (S. cerevisiae)1,3,4,5 | 1.43 | DNA repair; response to DNA damage | X92411 | 19359 | 96102_at | |
| Snrpd1 | Small nuclear ribonucleoprotein D11,4 | −1.64 | mRNA processing; spliceosome complex | M58558 | 20641 | 10057_at | |
| Bpgm | 2,3-bisphosphoglycerate mutase1 | −1.34 | Catalytic activity; phosphotransferases | X13586 | 12183 | 94815_at | |
| Sucla2 | Succinate-Coenzyme A ligase, ADP-forming, beta subunit 1,4 | −1.34 | Catalytic activity; glycolysis | AF058955 | 20916 | 93501_f_at | |
| Impa1 | Inositol (myo)-1(or4)-monophosphotase 11 | −1.33 | Myo-ionositol metabolism; magnesium ion binding | AF042730 | 55980 | 101498_at | |
| Ptp4a2 | Protein tyrosine phosphatase 4a21 | 1.38 | Protein amino acid dephosphorylation | AF064071 | 11783 | 103796_at | |
| C1qa | Complement component 1,q subcomponent, alpha polypeptide1,3,5 | 1.42 | Complement activation, classical pathway; extracellular space | X58861 | 12259 | 98562_at | |
| Response to Stress | Crhr1 | Corticotrophin releasing hormone receptor 13 | −1.41 | G-protein coupled receptor activity; membrane | X72305 | 12921 | 95290_at |
| Intracellular/Intracelular Organelle | Banp | Btg3 associated nuclear protein4 | −1.34 | Putative transcription factor | AF091234 | 53325 | 98512_at |
| Mybpc3 | Myosin binding protein C, cardiac4,6 | 1.42 | Actin binding; structural constituent of cytoskeleton | AF059576 | 17868 | 160582_at | |
| Ddx3y | DEAD (Asp-Glu-Ala-Asp) box polypeptide 3, Y-linked4,5,7 | −2.75 | ATP-dependent RNA helicase activity; nucleic acid binding | AJ007376 | 26900 | 103842_at | |
| Psme3 | Proteaseome (prosome, macropain) 28 subunit, 34 | 1.38 | Proteasome activator complex | AB007139 | 19192 | 93803_at | |
| Grb2 | Growth factor receptor bound protein 24,5,6 | 1.38 | RAS protein signal transduction | U07617 | 14784 | 10134_at | |
| Snca | Synuclein, alpha1,4,6 | −1.33 | Cytoplasm | AF044672 | 20617 | 93273_at | |
| Lmna | Lamin A1,4,6 | 1.35 | Intermediate filament | D49733 | 16905 | 98159_s_at | |
| Cacybp | Calcyclin binding protein1,4,5,6 | −1.34 | Molecular bridge | U97327 | 12301 | 92596_at | |
| Adk2 | Adenylate kinase 21,4,5,7 | 1.64 | Catalytic activity | AB020202 | 11637 | 95148_at | |
| Protein Binding | Wbp2 | WW domain binding protein 26 | 1.35 | Binding protein | U40826 | 22378 | 92683_at |
| Cldn5 | Claudin 56 | 1.46 | Integral to membrane | U82758 | 12741 | 104516_at | |
| Btg2 | B-cell translocation gene 2, antiproliferative1,3,6 | 1.52 | Antiproliferative protein | M64292 | 12277 | 101583_at | |
| No Assignment to Functional Group at GO Level 2 at p < 0.05 | Nell2 | Nel-like 2 homolog (chicken) | 1.55 | Calcium ion binding | U59230 | 54003 | 92358_at |
| Plp | Proteolipid protein (myelin) | −1.59 | Myelination; synaptic transmission | M16472 | 18823 | 92802_s_at | |
| Islr | Immuoglobulin superfamily containing leucine-rich repeat | 1.45 | Extracellular space | AB024538 | 26968 | 99010_at | |
| C920001 C06Rik | RIKEN cDNA C920001C06 gene | 1.33 | AB023622 | 92462_at |
Functional groups as determined by DAVID Bioinformatics Functional Group Analysis (http://niaid.abcc.ncifcrf.gov/home.jsp):
Cellular Physiological Processes;
Tube Development;
Response to Stress;
Intracellular/Intracellular Organelle;
Membrane-Bound Organelle;
Protein Binding;
Nucleotide Binding.
Eleven genes were changed in both genotypes (Table 3) and 4 ESTs were changed in both mutant and wild-type mice (data not shown). Of these genes, 6 were up-regulated in both genotypes, 2 were down-regulated in both genotypes, and 3 genes demonstrated interactions between genotype and diet with up-regulation in one genotype and down-regulation in the other. A complete list of genes are available upon request.
TABLE 3.
Gene expression changes in both PKCγ mutant and wild-type mice due to chronic ethanol diet. Cellular process with fold change ≥ ±1.3 in at least one group; both changes p < 0.05. Fold changes with positive values indicate up-regulation of expression; negative values indicate down-regulation of expression.
| Second Level GO Term | Gene Name | Gene Description | MUT Fold Change | WT Fold Change | Sixth Level GO Term | Genbank Accession | Locuslink Accession | Affymetrix Accession |
|---|---|---|---|---|---|---|---|---|
| Cell Communication/Differentiation | Ptprn | Protein tyrosine phosphatase, receptor type, N | 1.36 | 1.26 | Integral to membrane; transmembrane receptor protein tyrosine phosphatase signaling pathway | U11812 | 19275 | 104422_at |
| Cellular Physiological Process/Cell Death | Myo10 | Myosin X | 1.32 | 1.62 | Actin binding; cytoskeleton organization and biogenesis | AJ249706 | 17909 | 100932_at |
| Junb | Jun-B oncogene | 4.16 | 2.22 | Transcription factor activity; regulation of cell cycle; DNA binding | U20735 | 16477 | 102362_i_at | |
| Nr4a1 | Nuclear receptor subfamily 4, group A, member 1 (Nur77) | 2.23 | −2.52 | Transcription factor activity; inhibition of caspase activation; steroid hormone receptor activity | X16995 | 15370 | 102371_at | |
| Prkdc | Protein kinase, DNA activated, catalytic polypeptide | −1.54 | −1.3 | DNA repair; apoptosis; protein serine/threonine kinase activity | AB011543 | 19090 | 93476_at | |
| Igfbp2 | Insulin-like growth factor binding protein 2 | 1.24 | 1.46 | Growth factor binding; regulation of cell growth | X81580 | 16008 | 98627_at | |
| Metabolism | Hk2 | Hexokinase 2 | −1.58 | −1.76 | ATP binding; glycolysis; transferase activity | Y11666 | 15277 | 94375_at |
| Ptp4a2 | Protein tyrosine phosphatase 4a2 | 1.38 | 1.24 | Protein amino acid dephosphorylation | AF035644 | 19244 | 100595_at | |
| Morphogenesis | Efnb2 | Ephrin B2 | −1.14 | 1.37 | Development; integral to membrane; neurogenesis | U30244 | 13642 | 160857-at |
| Unclassified | Nip-snap1 | 4-nitrophenylphosphatase domain and non-neuronal SNAP25-like protein homolog 1 (C.elegans) | 1.4 | −1.26 | Biological process unknown | AJ001260 | 18082 | 93251_at |
| Tex261 | Testis expressed gene 261 | 1.29 | 1.37 | Integral to membrane; extracellular space | X81580 | 21766 | 98627_at |
The microarray data generated a putative list of genes whose expression may be related to 1) tolerance development, 2) resistance to tolerance, and 3) the effects of chronic ethanol treatment. Confirmation was done in the second stage of analysis using an independent method of gene expression quantification, qRT-PCR. From the microarray results, three genes that changed in mutant mice only (Hsp70.2, Bdnf, Th), three genes that changed in wild-type mice only (Twik-1, Plp, and Adk2), one gene whose expression was altered in the same direction in both genotypes (JunB), and one gene that changed in opposite directions after chronic ethanol ( Nur77) were chosen for further analysis. These genes were selected based on their possible significance in the neurobiology of addiction (Janak et al., 2006; Nakahara et al., 2002; Nestler, 2001,2005). Some of the genes have been reported in gene expression studies of human alcoholics (Flatscher-Bader et al., 2005; Lewohl et al., 2000; Liu et al., 2004; Mayfield et al., 2002) and in gene expression studies of chronic ethanol in mice (Daniels and Buck, 2002).
3.2 qRT-PCR Verifications
Results from the microarray experiments indicated that gene expression levels for Hsp70.2 and Th were significantly increased only in mutant cerebellum after chronic ethanol diet (p < 0.003; p < 0.009, respectively)( Figure 1a,c); whereas, Bdnf gene expression decreased in mutant cerebellum after ethanol diet (p < 0.02). However, qRT-PCR confirmations using 9–10 independent samples from wild-type and mutant cerebellum after control and ethanol diet revealed that for Hsp70.2, expression levels were increased in both mutant and wild-type mice after ethanol diet (t18 = 2.68, p < 0.008; t16 = 3.17, p < 0.003; one-tailed, respectively), indicating that changes in this gene were more likely the result of exposure to chronic ethanol diet in both genotypes (F1,37 = 17.56, p < 0.0001, diet effect) (Figure 1b). The increase in gene expression for Th in mutant cerebellum after ethanol diet was confirmed by qRT-PCR (t18 = 2.37, p< 0.02, one-tailed) (Figure 1d). The results of a two-way ANOVA (genotype x diet) also indicated significant genotype (F1,37 = 16.92, p < 0.0001) and diet (F1,37 = 7.55, p < 0.02) effects. The overall diet effect was due to a small, but significant increase in gene expression in wild-type ethanol-treated mice (t16 = 2.49, p < 0.02, one-tailed). Although the impact of the ethanol diet was greater in mutant mice, an overall effect of chronic ethanol cannot be ruled out. For Bdnf, the decrease in gene expression seen in the arrays for ethanol-treated mutant mice was not confirmed by qRT-PCR.
FIGURE 1.
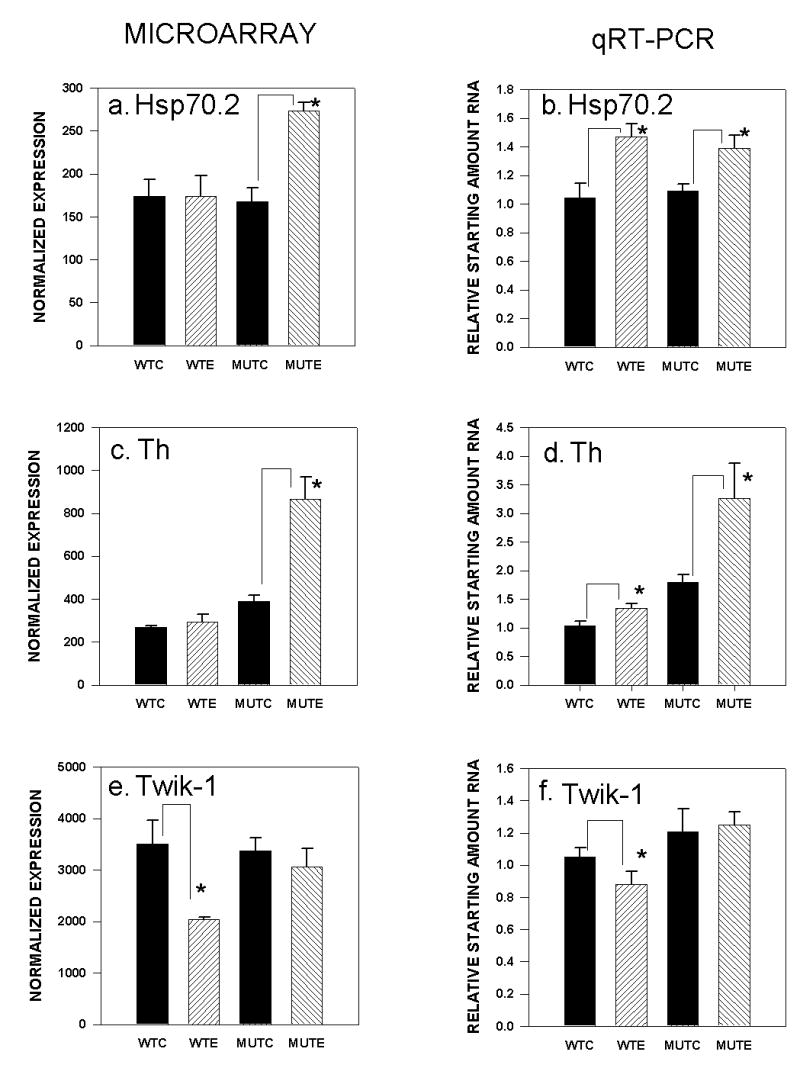
Gene expression changes in PKCγ mutant and wild-type mice after chronic ethanol diet as measured by microarray chip analyses (panels a., c., e.) with confirmation by qRT-PCR analyses of independent samples (panels b., d., f.). WTC: wild type control diet; WTE: wild type ethanol diet; MTC: mutant control diet; MTE: mutant ethanol diet. a.) Heat shock protein 70.2 microarray; * p < 0.05 increase after ethanol diet); b.) Heat shock protein 70.2 qRT-PCR; * p < 0.001 increase after ethanol diet); c.) Tyrosine hydroxylase microarray, *p < 0.05 increase after ethanol diet; d.) Tyrosine hydroxylase qRT-PCR, * p < 0.02 increase after ethanol diet; e.) Twik-1 microarray, * p < 0.05 decrease after ethanol diet; f.) Twik-1 qRT-PCR, * p < 0.03 decrease after ethanol diet.
Of the three genes chosen from the micorarrays for confirmation by qRT-PCR that were changed in wild-type mice after chronic ethanol diet, only one, the gene for the potassium channel, Twik-1 (Kcnk1), was verified as a gene regulated by chronic diet in ethanol-treated wild-type mice, but not mutants (Figure 1e). The microarray results indicated that Twik-1 gene expression was decreased in ethanol-treated wild-type mice (p < 0.03). This was confirmed using 14 independent samples for the qRT-PCR analyses (t26 = 1.71, p < 0.05, one-tailed) (Figure 1f). Data from the microarrays indicated that expression of the gene encoding the proteolipid protein, Plp, was significantly decreased in wild-type mice (p < 0.03), but not mutants (p = 0.43), after chronic ethanol diet; however, qRT-PCR results from 13–14 independent RNA samples demonstrated that Plp expression was decreased in both genotypes as a function of ethanol diet ( F1,54 = 16.59, p < 0.0001, diet effect) (Figure 2a,b). The increase in expression found in the microarrays in wild-type ethanol-treated mice for Adk2 (p < 0.04) was not confirmed by qRT-PCR (n = 9–10 independent samples; data not shown).
FIGURE 2.
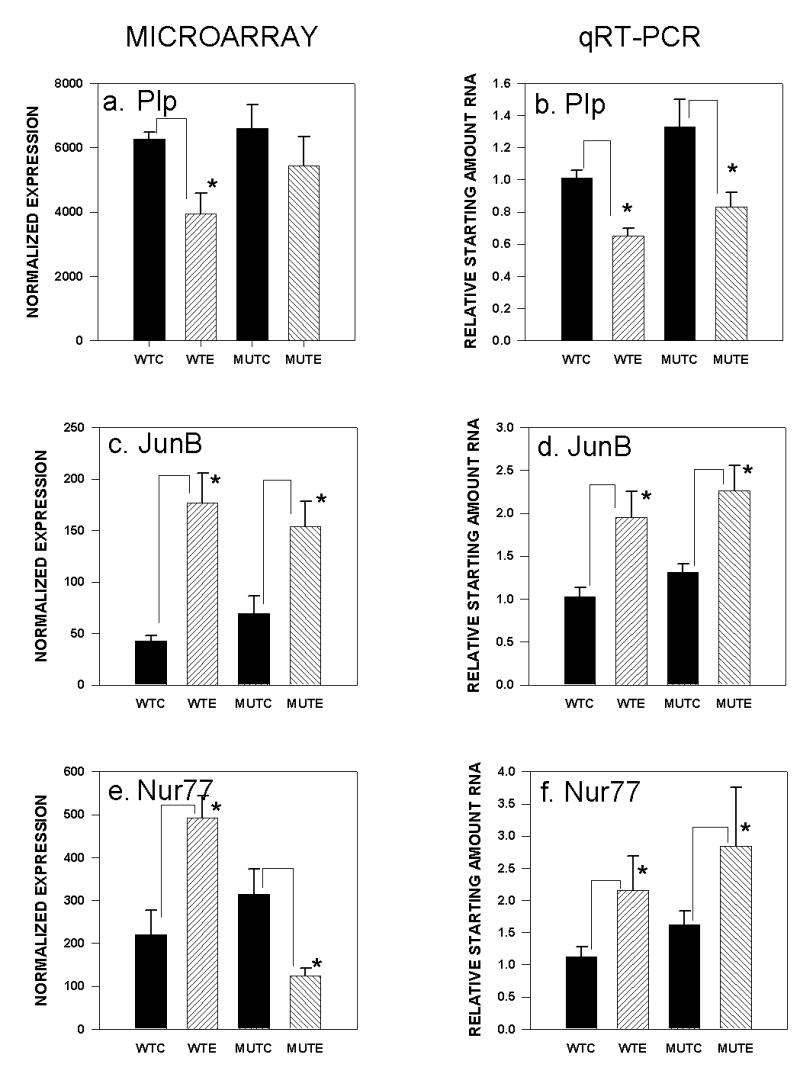
Gene expression changes in PKCγ mutant and wild-type mice after chronic ethanol diet as measured by microarray chip analyses (panels a., c., e.) with confirmation by qRT-PCR analyses of independent samples (panels b., d., f.). WTC: wild type control diet; WTE: wild type ethanol diet; MTC: mutant control diet; MTE: mutant ethanol diet. a.) Phospholipid protein microarray, * p < 0.05 decrease after ethanol diet; b.) Phospholipid protein qRT-PCR, * p < 0.001 decrease after ethanol diet; c.) JunB microarray, *p < 0.05 increase after ethanol diet; d.) JunB qRT-PCR, * p < 0.001 increase after ethanol diet; e.) Nur77 microarray, * p < 0.05 increase after ethanol diet in wild-type mice and decrease in mutant mice after ethanol diet; f.) Nur77 qRT-PCR, * p < 0.05 increase in both genotypes after ethanol diet.
One gene from the microarrays that changed in the same direction in both genotypes after chronic ethanol diet was chosen for confirmation as a gene associated with a response to chronic ethanol exposure. The transcription factor, JunB, was up-regulated in both genotypes (p < 0.05; p < 0.01; mutant and wild-type, respectively). qRT-PCR confirmation with 10 independent RNA samples verified the up-regulation due to chronic ethanol diet in both mutants and wild-type mice (F1,39 = 14.06, p < 0.001, diet effect) (Figure 2c,d]).
One additional gene encoding the transcription factor Nur77 (Nr4a1), was chosen for confirmation because the results from the microarrays indicated that an interaction between genotype and diet had occurred. Data from the microarrays indicated that the gene encoding the nuclear receptor subfamily 4, group A, member 1(Nr4a1), referred to as Nur77, was significantly up-regulated 2.23 fold (p = 0.02) in wild-type mice, and was significantly down-regulated 2.52 fold in mutant mice (p = 0.04) after chronic ethanol treatment. Verification experiments using 10–15 independent RNA samples from PKCγ mutant and wild-type mice confirmed the up-regulation (1.92 fold) seen in the wild-type mice (t27 = 1.87, p < 0.04, one-tailed), however, the down-regulation in mutant mice was not confirmed (Figure 2e,f). Analysis by ANOVA indicated that there was a significant diet effect (F1,46 = 4.82, p < 0.04), but no genotype x diet interaction. This was due to a non-significant up-regulation in mutant mice in the qRT-PCR samples. Therefore Nur77 may also be involved in the effects of chronic ethanol treatment, unrelated to PKCγ.
DISCUSSION
In the microarray experiments, the expression of several genes was altered due to chronic ethanol treatment that may be potentially associated with 1) the development of tolerance; i.e., those genes that were changed in wild-type mice only, 2) the resistance to tolerance; i.e. those genes that were changed in mutant mice only, or 3) the effects of chronic ethanol treatment; i.e., those genes that changed in both genotypes. Liberal criteria were used to identify genes that might be associated with these phenotypes. However, until the changes in expression were verified by an independent method, the genes identified in the microarrays were considered putative candidates only. In the present study, the expression of eight genes were selected for verification based on their relevance to ethanol responses or to addiction, in general. Of these eight genes, six were confirmed by qRT-PCR (Twik-1, JunB, Nur77, Hsp70.2, Th and Plp). Three of these (Hsp70.2, Plp, and Th) that had appeared to be associated with either tolerance development or resistance to the development of tolerance based on microarray results, were found later to be more generally associated with the effects of exposure to chronic ethanol based on qRT-PCR studies. Two genes that were altered on the microarrays, Bdnf, and Adk2, were not confirmed.
Alterations in gene expression produced by chronic treatment were examined at one time point taken immediately after removal of ethanol or control diet and prior to the development of withdrawal symptoms. It is likely that many changes related to the development of ethanol tolerance may occur within a very specific time window during ethanol treatment. Thus, transient changes or early changes in expression would be missed without detailed time course studies. The genes whose changes in expression were detected in the present study probably reflect rather stable or late changes in gene expression. In addition, because tissue was taken immediately after removal of the ethanol diet, the presence of ethanol cannot be ruled out as a factor in gene expression changes. As will be discussed, there is some overlap with previous studies of the short-term and long-term effects of ethanol treatment on gene expression in mice, cell cultures, and brain tissue from human alcoholics.
4.1 Expression Changes Associated with Tolerance Development
Twik-1
Potassium channels have been shown to be affected by ethanol exposure. Recently it has been reported that G-protein-coupled inwardly rectifying potassium channel function is increased in cerebellar granule cells after acute ethanol exposure (Lewohl et al., 1999). In a microarray analysis of gene expression in the frontal cortex of human alcoholics, Lewohl et al. (2000) found a down-regulation of an ATP-regulated inwardly rectifying potassium channel, suggesting that chronic alcohol may decrease potassium channel expression and may be a factor in tolerance development. In the present study, microarray analysis demonstrated that the gene encoding a weak inwardly rectifying potassium channel (Twik-1 or Kcnk1) was found to be down-regulated in wild-type mice only. This effect was confirmed by qRT-PCR indicating that this type of potassium channel may also play a role in tolerance development. In mouse, Twik-1 protein is found in abundance in the cerebellar granule cell layer and is activated by PKC (Lesage et al., 1997). Results from the present study indicate that PKCγ may play a significant role in Twik-1 activation. Twik-1 is structurally different than other inwardly rectifying potassium channels in that it belongs to a family of 2 pore-domain potassium channels and functions to set the resting membrane potential in many cell types, including cerebellar granule cells (Lesage and Lazdunski, 2000). Expression of Twik-1 appears to be sensitive to other brain perturbations. The results of two microarray analyses in rat have shown that Twik-1 is also down-regulated in response to brain injury (Rao et al., 2003) and after chronic nicotine exposure (Konu et al., 2001).
4.2 Genes Up-Regulated by Chronic Ethanol Treatment
JunB, Nur77, Hsp70.2, Th
It is not surprising that changes in the mRNA concentrations of some transcriptional factors occur in the cerebellum as a function of chronic ethanol treatment. Both JunB and Nur77 expression were increased as a function of chronic treatment, but in contrast to the usual transient increases seen in immediate early gene expression, these two factors had increases that must have persisted, or had been induced late in treatment, in order to be detected here.
Both JunB and Nur77 have been previously implicated in the actions of alcohol or neuroadaptations to alcohol treatment (Beckman et al., 1997; Ogilvie et al., 1998). Members of the immediate early gene Fos (c-Fos, FosB, Fra-1 and Fra-2) and Jun (c-Jun, JunB and JunD) families code for proteins that dimerize to form the AP-1 transcription factor which recognizes the binding motif TGACTCA. Supershift assays of AP-1 DNA binding activity in rat cortex, hippocampus, and cerebellum assayed at various times after chronic ethanol vapor treatment demonstrated increased AP-1 binding in all of these regions. Immediately after cessation of ethanol treatment, all members of the Jun family including JunB as well as cFos and Fos-B proteins were elevated in all brain regions studied (Beckman et al., 1997). Elevated JunB expression in both wild-type and mutant PKCγ mice after chronic ethanol treatment reproduces these effects of ethanol treatment observed in rat cerebellum observed immediately after removal from ethanol vapor. Human epidermal keratinocytes also respond to ethanol exposure with increased expression of JunB, and this effect is associated with increased PKC activation (Kharbanda et al., 1993). In addition, carbachol-stimulated expression of JunB in SH-SY5Y cells is potentiated in response to ethanol. This effect is inhibited by blocking activation of PKC, suggesting a role for PKC in ethanol potentiation of JunB expression (Ding et al., 1998). However, results from the present study would indicte that the isotype of PKC involved in this regulation of JunB expression is not PKCγ. Nur77, also known as NR4A2 and NGFI-B, is a member of the nuclear receptor family of steroid/thyroid-like receptors (Paulsen et al., 1995). As a transcriptional factor, it regulates the expression of genes by binding as monomers or dimers to hormone response elements (HREs) with two canonical sequences, 5′-AGGTCA and 5′-AGAACA, or by binding at the NGRI-B response element (NBRE) at 5′-(A/T)AAAGGTCA as a monomer (Meinke and Sigler, 1999). Nur77 is highly expressed in many brain regions including the Purkinje cell layer and the lateral deep nucleus of the cerebellum, in hippocampus, frontal cortex, striatal complex, and hypothalamus (Xiao et al., 1996). Recently, Daniels and Buck (2002) demonstrated increased Nur77 expression in the hippocampus during withdrawal from chronic ethanol treatment suggesting that Nur77 has some role in the hippocampus related to the hyperexcitability that causes handling induced seizures.
Quantitative RT-PCR experiments in the present study identified two additional genes that were up-regulated in response to chronic ethanol diet, Hsp70.2 and Th. Therefore, it was of interest to see if the up-regulation of the transcription factors, JunB and Nur77 could be associated with the up-regulation of Hsp70.2 and Th. A search of 5’ untranslated regions (UTR) of these genes for JunB and Nur77 binding sites identified two potential sites for Nur77 in 506 bp of the 5’ UTR of the Th gene. This suggests a potential relationship between the up-regulation of these two genes. Further studies would need to be done to confirm whether Th is regulated by Nur77. No binding sites for either transcription factor were found in 630 bp of the 5’ UTR of the Hsp70.2 gene. This does not preclude unidentified binding sites further upstream of Hsp70.2, however.
Heat shock proteins (HSP) are considered molecular chaperones that regulate a variety of cellular functions and act as neuroprotective factors against environmental stressors such as heat shock, some chemical compounds, and brain injury (Allen and Chase, 2001; Ohtsuka et al., 2005). Alterations in expression of several members of the diverse heat shock gene family were observed in rat brain after chronic treatment with morphine (Ammon et al., 2003), after acute ethanol treatment of C. elegans (Kwon et al., 2004), and mouse hippocampus during withdrawal from chronic ethanol treatment (Daniels and Buck, 2002). Data from the microarray experiments presented here indicated that gene expression of one member of the 70 k Da HSP family, Hsp70.2, was up-regulated in mutant mice only after ethanol diet, suggesting that it may play a role in the resistance to tolerance. However, confirmation with independent samples showed that Hsp70.2 was up-regulated in both genotypes after ethanol exposure. This implies that Hsp70.2 responded to the deleterious effects of the chronic ethanol diet, unrelated to the development of tolerance or to regulation by PKCγ.
From previous studies, it might be expected that the catecholamine biosynthesis pathway would be impacted by exposure to ethanol. Tyrosine hydroxylase is the rate-limiting enzyme in the synthesis of dopamine (Kumer and Vrana, 1996). An up-regulation of Th was observed in prefrontal cortex in DBA/2J mice after acute ethanol treatment (Kerns et al., 2005). However, it was somewhat unexpected to find an increase in Th gene expression in the cerebellum, a brain region not usually associated with a dopamine response to reward. Studies have shown that dopamine innervation, tyrosine hydroxylase immunoreactivity, and mRNA are found in the cerebellum of humans, primates and rodents (Fujii et al., 1994; Hurley et al., 2003; Melchitzky and Lewis, 2000). In humans, it has been suggested that changes in dopaminergic markers in the cerebellum of Parkinson’s patients may explain the poor motor coordination found in this disease (Hurley et al., 2003). Further evidence that tyrosine hydroxylase is involved in abnormal motor activity is the observation that abnormal expression of Th immunoreactivity is found ectopically in the cerebellum of several ataxic mouse mutants, including weaver mutants, rolling mouse Nagoya, dilute-lethal mice, and pogo mutant mice (Abbott and Sotelo, 2000; Jeong et al., 2001; Sawada et al., 1999). This may be relevant to the PKCγ mutant mice, since they demonstrate ataxia, due to abnormal development of the innervation of multiple climbing fibers onto Purkinje cells in the cerebellum (Chen et al., 1995). In the qRT-PCR experiments, increased expression of Th was found in mutant control-diet mice compared to wild-type control-diet mice (see Figure 1c,d) suggesting that endogenous Th expression is increased in PKCγ mutant cerebellum. Gayer et al. (1991) have shown that Th gene expression is increased in neuroblastoma cells after ethanol exposure. Although expression was increased in both genotypes, suggesting a general effect of chronic ethanol exposure, the effect was greater in mutant mice compared to wild-types. This may reflect a greater sensitivity in mutant mice to the interaction of ethanol and the stress of the chronic ethanol paradigm. Tyrosine hydroxylase mRNA has been shown to increase in response to stress, drug administration, and food restriction (Kumer and Vrana, 1996; Lindblom et al., 2006). Because the mutant mice do not become tolerant to the effects of ethanol (Bowers et al., 1999), they are physiologically stressed throughout the exposure and they consume less ethanol diet than wild-type mice; therefore, experiencing self-imposed food restriction.
4.3 One Gene Down-Regulated by Chronic Ethanol Treatment
Plp
Acute ethanol treatment induced an increase in expression of a cluster of myelin and myelin-related genes including proteolipid protein (Plp) in mouse prefrontal cortex (Kerns et al., 2005). Long-term exposure to ethanol produces the opposite effect in humans. As reported here, chronic treatment in the mouse also down-regulates Plp expression. Microarray analyses of brain tissue from human alcoholics have consistently revealed down-regulation of myelin-related genes (Flatscher-Bader et al., 2005; Lewohl et al., 2000; Liu et al., 2004; Mayfield et al., 2002; however, see Iwamoto et al., 2004). This implies that chronic alcohol abuse decreases the transcription of these genes resulting in a decrease in myelin-related proteins. Neuroimaging studies of alcoholic men and women have shown reduced white and gray matter volumes in several brain regions compared to non-drinking controls that may be the result of the down-regulation of these genes (Fein et al., 2002; Hommer et al., 2001; Pfefferbaum et al., 1992). Specifically, cerebellar atrophy is one of the hallmarks of alcohol abuse (Estrin, 1987; Karhune et al., 1994; Torvik and Torp, 1986). Proteolipid protein (Plp) is one of the most abundant proteins in central nervous system myelin and accounts for nearly 50% of myelin-related proteins. Regulation of expression of the Plp gene is very important for normal brain structure and plays a role in the interaction between oligodendrocytes and axons (Wight and Dobretsova, 2004). Results from the microarray experiments in the present study initially indicated that Plp expression was significantly down-regulated in wild-type cerebellum after ethanol exposure; however, a nonsignificant decrease was also observed in PKCγ mutant mice suggesting that Plp expression is more likely related to chronic ethanol diet exposure than to tolerance development. Confirmation using independent RNA samples and qRT-PCR supported this observation since significant down-regulation was seen in both genotypes after ethanol diet. Levels of expression in mutant and wild-type control mice were similar indicating that the regulation of transcription of Plp is not dependent on PKCγ.
In conclusion, the data presented here are the results of a two-stage process for identifying genes that may be associated with the development of tolerance to the sedative-hypnotic effects of ethanol in mice. The premise of these experiments was to find genes differentially expressed between PKCγ wild-type mice that do develop tolerance and PKCγ mutant mice that do not. The preliminary screen of gene expression using microarray technology identified several candidate genes. Using relatively liberal criteria, a subset of genes was selected for confirmation by qRT-PCR in the second stage. This two stage process resulted in the following findings: 1) a relationship between PKCγ and tolerance development was confirmed for one gene, Twik-1; whereas, 2) the expression of five of the remaining genes were found to be associated with the effects of exposure to chronic ethanol independent of any regulation by PKCγ or tolerance development. Alterations in the transcriptional regulation of several of these genes including: Twik-1, JunB, Nur77, and Plp have been observed in a number of studies and are likely candidates for further study regarding acute and chronic effects of ethanol.
Acknowledgments
This work was supported by AA13901 to BJB; AA13177 to RAR; a NIH traineeship, MH53668 to AMS; and AA03527 and AA13018 to JMW.
References
- Abbott LC, Sotelo C. Ultrastructural analysis of catecholaminergic innervation in Weaver and normal mouse cerebellar cortices. J Comp Neurol. 2000;426:316–329. [PubMed] [Google Scholar]
- Abeliovich A, Paylor R, Chen C, Kim JJ, Wehner JM, Tonegawa S. PKCγ mutant mice exhibit mild deficits in spatial and contextual learning. Cell. 1993;75:1263–1271. doi: 10.1016/0092-8674(93)90614-v. [DOI] [PubMed] [Google Scholar]
- Allen GV, Chase T. Induction of heat shock proteins and motor function deficits after focal cerebellar injury. Neurosci. 2001;102:603–614. doi: 10.1016/s0306-4522(00)00519-4. [DOI] [PubMed] [Google Scholar]
- Ammon S, Mayer P, Riechert U, Tischmeyer H, Holt V. Microarray analysis of genes expressed in the frontal cortex of rats chronically treated with morphine and after naloxone precipitated withdrawal. Mol Brain Res. 2003;112:113–125. doi: 10.1016/s0169-328x(03)00057-3. [DOI] [PubMed] [Google Scholar]
- Andersen BB. Reduction of Purkinje cell volume in cerebellum of alcoholics. Brain Res. 2004;1007:10–18. doi: 10.1016/j.brainres.2004.01.058. [DOI] [PubMed] [Google Scholar]
- Beaudry G, Langlois MC, Weppe I, Rouillard C, Levesque D. Contrasting patterns and cellular specificity of transcriptional regulation of the nuclear receptor nerve growth factor-inducible B by haliperidol and clozapine in the rat forebrain. J Neurochem. 2000;75:1694–1702. doi: 10.1046/j.1471-4159.2000.0751694.x. [DOI] [PubMed] [Google Scholar]
- Beckman AM, Matsumoto I, Wilce PA. AP-1 and Egr DNA-binding activities are increased in rat brain during ethanol withdrawal. J Neurochem. 1997;69:306–314. doi: 10.1046/j.1471-4159.1997.69010306.x. [DOI] [PubMed] [Google Scholar]
- Bowers BJ, Owen EH, Collins AC, Abeliovich A, Tonegawa S, Wehner JM. Decreased ethanol sensitivity and tolerance development in γ-Protein Kinase C null mutant mice is dependent on genetic background. Alc Clin Exp Res. 1999;23:387–397. [PubMed] [Google Scholar]
- Calabrese V, Renis M, Calderone A, Russo A, Reale S, Barcellona ML, Rizza V. Stress proteins and SH-groups in oxidant-induced cellular injury after chronic ethanol administration in rat. Free Radical Biol Med. 1998;24:1159–1167. doi: 10.1016/s0891-5849(97)00441-3. [DOI] [PubMed] [Google Scholar]
- Chen C, Kano M, Abeliovich A, Chen L, Bao S, Kim JJ, Hashimoto K, Thompson RF, Tonegawa S. Impaired motor coordination correlates wit persistent multiple climbing fiber innervation in PKCγ mutant mice. Cell. 1995;83:1233–1242. doi: 10.1016/0092-8674(95)90148-5. [DOI] [PubMed] [Google Scholar]
- Daniels GM, Buck KJ. Expression profiling identifies strain-specific changes associated with ethanol withdrawal in mice. Genes, Brain, Behav. 2002;1:36–45. doi: 10.1046/j.1601-1848.2001.00008.x. [DOI] [PubMed] [Google Scholar]
- Ding W-Q, Fried U, Larsson C, Alling C. Ethanol exposure potentiates fosB and junB expression induced by muscarinic receptor stimulation in neuroblastoma SH-SY5Y cells. Alc Clin Exp Res. 1998:225–235. [PubMed] [Google Scholar]
- Edenberg HJ, Strother WN, McClintock JN, Tian H, Stephens M, Jerome RE, Lumeng L, Li T-K, McBride WJ. Gene expression in the hippocampus of inbred alcohol-preferring and–nonpreferring rats. Genes, Brain, Behav. 2005;4:20–30. doi: 10.1111/j.1601-183X.2004.00091.x. [DOI] [PubMed] [Google Scholar]
- Estrin WJ. Alcoholic cerebellar degeneration is not a dose-dependent phenomenon. Alc Clin Exp Res. 1987;11:372–375. doi: 10.1111/j.1530-0277.1987.tb01327.x. [DOI] [PubMed] [Google Scholar]
- Fein G, Di Sclafani V, Cardenas VA, Goldmann H, Tolou-Shams M, Meyerhoff DJ. Cortical gray matter loss in treatment-naïve alcohol dependent individuals. Alc Clin Exp Res. 2002;26:558–564. [PMC free article] [PubMed] [Google Scholar]
- Flatscher-Bader T, van der Brug M, Hwang JW, Gochee PA, Matsumoto I, Niwa S, Wilce PA. Alcohol-responsive genes in the frontal cortex and nucleus accumbens of human alcoholics. J Neurochem. 2005;93:359–370. doi: 10.1111/j.1471-4159.2004.03021.x. [DOI] [PubMed] [Google Scholar]
- Flatscher-Bader T, van der Brug MP, Landis N, Hwang JW, Harrison E, Wilce PA. Comparative gene expression in brain regions of human alcoholics. Genes, Brain, Behav. 2006;5:78–84. doi: 10.1111/j.1601-183X.2006.00197.x. [DOI] [PubMed] [Google Scholar]
- Freeman WM, Brebner K, Lynch WJ, Patel KM, Robertson DJ, Roberts DCS, Vrana KE. Changes in rat frontal cortex gene expression following chronic cocaine. Mol Brain Res. 2002;104:11–20. doi: 10.1016/s0169-328x(02)00197-3. [DOI] [PubMed] [Google Scholar]
- Fujii T, Sakai M, Nagatsu I. Immunohistochemical demonstration of expression of tyrosine hydroxylase in cerebellar Purkinje cells of the human and mouse. Neurosci Lett. 1994;165:161–163. doi: 10.1016/0304-3940(94)90734-x. [DOI] [PubMed] [Google Scholar]
- Gayer GG, Gordon A, Miles MF. Ethanol increases tyrosine hydroxylase gene expression in N1E-115 neroblastoma cells. J Biol Chem. 1991;266:22279–22284. [PubMed] [Google Scholar]
- Gervais J, Soghomonian JJ, Richard D, Rouillard C. Dopamine and serotonin interactions in the modulation of the expression of the immediate-early transcription factor, nerve growth factor-inducible B, in the striatum. Neurosci. 1999;91:1045–1054. doi: 10.1016/s0306-4522(98)00688-5. [DOI] [PubMed] [Google Scholar]
- Harris RA, McQuilkin SJ, Paylor R, Abeliovich A, Tonegawa S, Wehner JM. Mutant mice lacking the γ isoform of protein kinase C show decreased behavioral actions of ethanol and altered function of γ-aminobutyrate type A receptors. Proc Nat’l Acad Sci. 1995;92:3658–3662. doi: 10.1073/pnas.92.9.3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hommer DW, Momenan R, Kaiser E, Rawlings RR. Evidence for gender-related effect of alcoholism on brain volumes. Am J Psychiat. 2001;158:198–204. doi: 10.1176/appi.ajp.158.2.198. [DOI] [PubMed] [Google Scholar]
- Hurley MJ, Mash DC, Jenner P. Markers for dopaminergic neurotransmission in the cerebellum in normal individuals and patients with Parkinson’s disease examined by RT-PCR. Eur J Neurosci. 2003;18:2668–2672. doi: 10.1046/j.1460-9568.2003.02963.x. [DOI] [PubMed] [Google Scholar]
- Iwamoto K, Bundo M, Yamamoto M, Qzawa H, Saito T, Kato T. Decreased expression of NEFH and PCP4/PEP19 in the prefrontal cortex of alcoholics. Neurosci Res. 2004:379–385. doi: 10.1016/j.neures.2004.04.002. [DOI] [PubMed] [Google Scholar]
- Janak PH, Wolf FW, Heberlein U, Pandey SC, Logrip ML, Ron D. BIG news in alcohol addiction: new findings on growth factor pathways BDNF, insulin, and GDNF. Alc Clin Exp Res. 2006;30:214–221. doi: 10.1111/j.1530-0277.2006.00026.x. [DOI] [PubMed] [Google Scholar]
- Jeong YG, Kim MK, Hawkes R. Ectopic expression of tyrosine hydroxylase in Zebrin II immunoreactive Purkinje cells in the cerebellum of the ataxic mutant mouse, pogo. Develop Brain Res. 2001;129:201–209. doi: 10.1016/s0165-3806(01)00212-7. [DOI] [PubMed] [Google Scholar]
- Kalant H. Current state of knowledge about the mechanisms of alcohol tolerance. Addict Biol. 1996;1:133–141. doi: 10.1080/1355621961000124756. [DOI] [PubMed] [Google Scholar]
- Kalivas PW. How do we determine which drug-induced neuroplastic changes are important? Nature Neurosci. 2005;8:1440–1441. doi: 10.1038/nn1105-1440. [DOI] [PubMed] [Google Scholar]
- Karhune PJ, Erkinjuntti T, Laippala P. Moderate alcohol consumption and loss of cerebellar Purkinje cells. BMJ. 1994;308:1663–1667. doi: 10.1136/bmj.308.6945.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley AE, Schiltz CA, Landry CF. Neural systems recruited by drug- and food-related cues: studies of gene activation in corticolimbic regions. Physiol Behav. 2005;86:11–14. doi: 10.1016/j.physbeh.2005.06.018. [DOI] [PubMed] [Google Scholar]
- Kerns RT, Ravindranathan A, Hassan S, Cage MP, York T, Sikela JM, Williams RW, Miles MJ. Ethanol-responsive brain region expression networks: implications for behavioral responses to acute ethanol in DBA/2J versus C57BL/6J. mice. J Neurosci. 2005;25:2255–2266. doi: 10.1523/JNEUROSCI.4372-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharbanda S, Nakamura T, Kufe D. Induction of the c-jun proto-oncogene by a protein kinase C-dependent mechanism during exposure of human epidermal keratinocytes to ethanol. Biochem Pharmacol. 1993;45:675–681. doi: 10.1016/0006-2952(93)90142-j. [DOI] [PubMed] [Google Scholar]
- Konu O, Kane JK, Barrett T, Vawter MP, Chang R, Ma JZ, Donovan DM, Sharp B, Becker KG, Li MD. Region-specific transcriptional response to chronic nicotine in rat brain. Brain Res. 2001;909:194–203. doi: 10.1016/s0006-8993(01)02685-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumer SC, Vrana KE. Intricate regulation of tyrosine hydroxylase activity and gene expression. J Neurochem. 1996;67:443–462. doi: 10.1046/j.1471-4159.1996.67020443.x. [DOI] [PubMed] [Google Scholar]
- Kwon JY, Hong M, Choi MS, Kang S, Duke K, Kim S, Lee S, Lee J. Ethanol-responsive genes and their regulation analyzed by a microarray and comparative genomic approach in the nematode Caenorhabditis elegans. 2004. [DOI] [PubMed] [Google Scholar]
- Lesage F, Lauritzen I, Duprat F, Reyes R, Fink M, Heurteaux C, Lazdunski M. The structure, function and distribution of the mouse TWIK-1 K+ channel. FEBS Lett. 1997;402:28–32. doi: 10.1016/s0014-5793(96)01491-3. [DOI] [PubMed] [Google Scholar]
- Lesage F, Lazdunski M. Molecular and functional properties of two-pore domain potassium channels. Am J Physiol Renal Physiol. 2000;279:F793–F801. doi: 10.1152/ajprenal.2000.279.5.F793. [DOI] [PubMed] [Google Scholar]
- Lewohl JM, Wang L, Miles MF, Zhang L, Dodd PR, Harris RA. Gene expression in human alcoholism: micorarray analysis of frontal cortex. Alc Clin Exp Res. 2000;24:1873–1882. [PubMed] [Google Scholar]
- Lewohl JM, Wilson WR, Mayfield RD, Brozowski SJ, Morrisett RA, Harris RA. G-protein-coupled inwardly rectifying potassium channels are targets of alcohol action. Nature Neurosci. 1999;2:1084–1090. doi: 10.1038/16012. [DOI] [PubMed] [Google Scholar]
- Lindblom J, Johansson A, Holmgren A, Grandin E, Nederg rd C, Fredriksson R, Schioth HB. Increased mRNA levels of tyrosine hydroxylase and dopamine transporter in the VTA of male rats after chronic food restriction. Eur J Neurosci. 2006;23:180–186. doi: 10.1111/j.1460-9568.2005.04531.x. [DOI] [PubMed] [Google Scholar]
- Liu JL, Lewohl JM, Dodd PR, Randall PK, Harris RA, Mayfield RD. Gene expression profiling of individual cases reveals consistent transcriptional changes in alcoholic human brain. J Neurochem. 2004;90:1050–1058. doi: 10.1111/j.1471-4159.2004.02570.x. [DOI] [PubMed] [Google Scholar]
- Mayfield RD, Lewohl JM, Doss PR, Herlihy A, Liu J, Harris RA. Patterns of gene expression are altered in the frontal and motor cortices of human alcoholics. J Neurochem. 2002;81:802–813. doi: 10.1046/j.1471-4159.2002.00860.x. [DOI] [PubMed] [Google Scholar]
- McBride WJ, Kerns RT, Rodd ZA, Strother WN, Edenberg HJ, Hashimoto JG, Wiren KM, Miles MF. Alcohol effects on central nervous system gene expression in genetic animal models. Alc Clin Exp Res. 2005;29:167–175. doi: 10.1097/01.alc.0000153539.40955.42. [DOI] [PubMed] [Google Scholar]
- Meinke G, Sigler PB. DNA-binding mechanism of the monomeric orphan nuclear receptor NGFI-B. Nature Struct Biol. 1999;6:471–477. doi: 10.1038/8276. [DOI] [PubMed] [Google Scholar]
- Melchitzky DS, Lewis DA. Tyrosine hydroxylase-and dopamine transporter-immunoreactive axons in the primate cerebellum. Neuropsychopharm. 2000;22:466–472. doi: 10.1016/S0893-133X(99)00139-6. [DOI] [PubMed] [Google Scholar]
- Mulligan MK, Ponomarev I, Hitzeman RJ, Belknap JK, Tabakoff B, Harris RA, Crabbe JC, Blednov YA, Grahame NJ, Phillips TJ, Finn DA, Hoffman PL, Iyer VR, Koob GF, Bergeson SE. Toward understanding the genetics of alcohol drinking through transcriptsome meta-analysis. Proc Nat’l Acad Sci. 2006;103:6368–6373. doi: 10.1073/pnas.0510188103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy BC, Chiu T, Harrison M, Uddin RK, Singh SM. Examination of ethanol responsive liver and brain specific gene expression, in the mouse strains with variable ethanol preferences, using cDNA expression arrays. Biochem Gen. 2002;40:395–410. doi: 10.1023/a:1020777528602. [DOI] [PubMed] [Google Scholar]
- Naik MU, Benedikz E, Hernandez I, Libien J, Hrabe J, Valsamis M, Dow-Edwards D, Osman M, Sacktor TC. Distribution of protein kinase Mζ and the complete protein kinase C isoform family in rat brain. J Comp Neurol. 2000;426:243–258. doi: 10.1002/1096-9861(20001016)426:2<243::aid-cne6>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- Nakahara T, Hirano M, Uchimura H, Shirali S, Martin CR, Bonner AB, Preedy VR. Chronic alcohol feeding and its influence on cFos and heat shock protein-70 gene expression in different brain regions of male and female rats. Metabolism. 2002;51:1562–1568. doi: 10.1053/meta.2002.35595. [DOI] [PubMed] [Google Scholar]
- Nestler EJ, Aghajanian GK. Molecular and cellular basis of addiction. Science. 1997;278:58–63. doi: 10.1126/science.278.5335.58. [DOI] [PubMed] [Google Scholar]
- Nestler EJ. Molecular basis of long-term plasticity underlying addiction. Nature Rev Neurosci. 2001;2:119–128. doi: 10.1038/35053570. [DOI] [PubMed] [Google Scholar]
- Nestler EJ. Is there a common molecular pathway for addiction? Nature Neurosci. 2005;8:1445–1449. doi: 10.1038/nn1578. [DOI] [PubMed] [Google Scholar]
- Oberdoerster J, Rabin RA. Enhanced caspase activity during ethanol-induced apoptosis in rat cerebellar granule cells. Eur J Pharm. 1999;385:273–282. doi: 10.1016/s0014-2999(99)00714-1. [DOI] [PubMed] [Google Scholar]
- Ogilvie K, Lee S, Rivier C. Effect of three different modes of alcohol administration on the activity of the rat hypothalamic-pituitary-adrenal axis. Alc Clin Exp Res. 1997;21:467–476. doi: 10.1111/j.1530-0277.1997.tb03792.x. [DOI] [PubMed] [Google Scholar]
- Ogilvie KM, Lee S, Rivier C. Divergence in the expresson of molecular markers of neuronal activation in the parvocellular paraventricular nucleus of the hypothalamus evoked by alcohol administration via different routes. J Neurosci. 1998;18:4344–4352. doi: 10.1523/JNEUROSCI.18-11-04344.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsuka K, Kawashima D, Gu Y, Saito K. Inducers and co-inducers of molecular chaperones. Int J Hypotherm. 2005;21:703–711. doi: 10.1080/02656730500384248. [DOI] [PubMed] [Google Scholar]
- Paulsen RE, Granas K, Johnsen H, Rolseth V, Sterri S. Three related brain nuclear receptors, NGF1-B, Nurr1, and NOR-1, as transcriptional activators. 1995. [DOI] [PubMed] [Google Scholar]
- Pfefferbaum A, Lim KO, Zipursky RB, Mathalon DH, Rosenbloom MJ, Lane B, Ha CN, Sullivan EV. Brain gray and white matter volume loss accelerates with aging in chronic alcoholics: a quantitative MRI study. Alc Clin Exp Res. 1992;16:1078–1089. doi: 10.1111/j.1530-0277.1992.tb00702.x. [DOI] [PubMed] [Google Scholar]
- Phillips SC, Harper CG, Kril J. A quantitative histological study of the cerebellar vermis in alcoholic patients. Brain. 1987;110:301–314. doi: 10.1093/brain/110.2.301. [DOI] [PubMed] [Google Scholar]
- Proctor WR, Poelchen W, Bowers BJ, Wehner JM, Messing RO, Dunwiddie TV. Ethanol differentially enhances hippocampal GABAA receptor-mediated responses in protein kinase Cγ (PKCγ) and PKCε null mice. J Pharm Exp Ther. 2003;305:264–270. doi: 10.1124/jpet.102.045450. [DOI] [PubMed] [Google Scholar]
- Rahman S, Miles MJ. Identification of novel ethanol-sensitive genes by expression profiling. Pharm Therap. 2001;92:123–134. doi: 10.1016/s0163-7258(01)00163-2. [DOI] [PubMed] [Google Scholar]
- Rao VLR, Dhodda VK, Song G, Bowen KK, Dempsey RJ. Traumatic brain injury-induced acute gene expression changes in rat cerebral cortex identified by genechip analysis. J Neurosci Res. 2003;71:208–219. doi: 10.1002/jnr.10486. [DOI] [PubMed] [Google Scholar]
- Saito M, Smiley J, Toth R, Vadasz C. Microarray analysis of gene expression in rat hippocampus after chronic ethanol treatment. Neurochem Res. 2002;27:1221–1229. doi: 10.1023/a:1020937728506. [DOI] [PubMed] [Google Scholar]
- Sawada K, Komatsu S, Haga H, Sun XZ, Hisano S, Fukui Y. Abnormal expression of tyrosine hydroxylase immunoreactivity in cerebellar cortex of ataxic mutant mice. Brain Res. 1999;829:107–112. doi: 10.1016/s0006-8993(99)01347-5. [DOI] [PubMed] [Google Scholar]
- Schochet TL, Kelley AE, Landry CF. Differential expression of arc mRNA and other plasticity-related genes induced by nicotine in adolescent rat forebrain. Neurosci. 2005;135:285–297. doi: 10.1016/j.neuroscience.2005.05.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AM, Bowers BJ, Radcliffe RA, Wehner JM. Microarray analysis of the effects of a γ-protein kinase C null mutation on gene expression in striatum: a role for transthyretin in mutant phenotypes. Behav Gen. 2006 doi: 10.1007/s10519-006-9083-6. Online First: DOI:10.1007/s10519-006-9083-6. [DOI] [PubMed] [Google Scholar]
- Smolen A, Marks MJ, Smolen TN, Collins AC. Dose and route of administration alter relative elimination of ethanol by long-sleep and short-sleep mice. Alc Clin Exp Res. 1986;10:198–204. doi: 10.1111/j.1530-0277.1986.tb05071.x. [DOI] [PubMed] [Google Scholar]
- Sorensen S, Palmer M, Dunwiddie T, Hoffer B. Electrophysiological correlates of ethanol-induced sedation in differentially sensitive lines of mice. Science. 1980;210:1143–1144. doi: 10.1126/science.7444444. [DOI] [PubMed] [Google Scholar]
- Spuhler K, Hoffer B, Weiner N, Palmer M. Evidence for genetic correlation of hypnotic effects and cerebellar Purkinje neuron depression in response to ethanol in mice. Pharm Biochem Behav. 1982;17:569–578. doi: 10.1016/0091-3057(82)90320-3. [DOI] [PubMed] [Google Scholar]
- Torvik A, Torp S. The prevalence of alcoholic cerebellar atrophy. A morphometric and histological study of an autopsy material. J Neurol Sci. 1986;75:43–51. doi: 10.1016/0022-510x(86)90049-3. [DOI] [PubMed] [Google Scholar]
- Wight PA, Dobretsova A. Where, when and how much: regulation of myelin proteolipid protein gene expression. Cell Mol Life Sci. 2004;61:810–821. doi: 10.1007/s00018-003-3309-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Q, Castillo SO, Nikodem VM. Distribution of messenger RNAs for the orphan nuclear receptors NURR1 and NUR77 (NGFI-B) in adult rat brain using in situ hybridization. Neurosci. 1996;75:221–230. doi: 10.1016/0306-4522(96)00159-5. [DOI] [PubMed] [Google Scholar]


