Abstract
Dosage compensation in Drosophila is mediated by a histone-modifying complex that upregulates transcription of genes on the single male X chromosome. The male-specific lethal (MSL) complex contains at least five proteins and two noncoding roX (RNA on X) RNAs. The mechanism by which the MSL complex targets the X chromosome is not understood. Here we use a sensitized system to examine the function of roX genes on the X chromosome. In mutants that lack the NURF nucleosome remodeling complex, the male polytene X chromosome is severely distorted, appearing decondensed. This aberrant morphology is dependent on the MSL complex. Strikingly, roX mutations suppress the Nurf mutant phenotype regionally on the male X chromosome. Furthermore, a roX transgene induces disruption of local flanking autosomal chromatin in Nurf mutants. Taken together, these results demonstrate the potent capability of roX genes to organize large chromatin domains in cis, on the X chromosome. In addition to interacting functions at the level of chromosome morphology, we also find that NURF complex and MSL proteins have opposing effects on roX RNA transcription. Together, these results demonstrate the importance of a local balance between modifying activities that promote and antagonize chromatin compaction within defined chromatin domains in higher organisms.
THE compaction of eukaryotic DNA into nucleosomes and higher-order chromatin structure has profound effects on gene transcription. By modifying nucleosome structure or altering nucleosome positioning, histone-modifying enzymes and ATP-dependent chromatin-remodeling factors modulate chromatin architecture and thereby regulate gene expression. A major question is how the disparate activities of these chromatin-organizing enzymes are deployed and integrated.
In Drosophila, X-chromosome dosage compensation is mediated by a histone-modifying complex called the male-specific-lethal (MSL) complex (Lucchesi 1998; Meller and Kuroda 2002; Gilfillan et al. 2004). The MSL complex binds specifically to genes along the male X chromosome, resulting in an approximately twofold increase in the transcription of X-linked genes in males (X) relative to females (XX), thereby compensating for the difference in X-chromosome number between the sexes. The MSL complex contains at least five proteins (MSL1, MSL2, MSL3, MLE, and MOF) and two noncoding RNAs (roX1 and roX2). MOF is a histone H4 lysine 16 (H4K16) acetyltransferase that catalyzes enriched H4K16 acetylation on the male X chromosome. This histone modification facilitates transcription, perhaps by specifically disrupting charge-based internucleosomal interactions between histone H4 and histone H2A, thus partially decondensing chromatin structure (Shogren-Knaak et al. 2006).
The biochemical roles of the two noncoding RNAs in the MSL complex, roX1 and roX2, are largely unknown. Without the roX RNAs, MSL targeting to the X chromosome is very poor (Meller and Rattner 2002). The RNAs are functionally redundant although they differ in both sequence and size. The genes that encode the two roX RNAs are positioned at distinct locations, near the tip (roX1) and the middle (roX2) of the euchromatic portion of the X chromosome (Figure 1A). Our previous studies suggest that nascent roX RNA transcripts serve as local nucleation sites for assembly of the complex (Park et al. 2002; Bai et al. 2004). This can be seen when roX transgenes are inserted at ectopic positions on the autosomes, particularly when the endogenous X chromosome lacks both roX genes. Therefore, the location of roX genes on the X chromosome could facilitate initial binding to that chromosome. Whether roX genes contribute regionally to MSL targeting on the X chromosome is not evident, however, from the final colocalization of roX RNAs and MSL proteins along the length of the chromosome in wild type or roX1 or roX2 single mutant males.
Figure 1.—
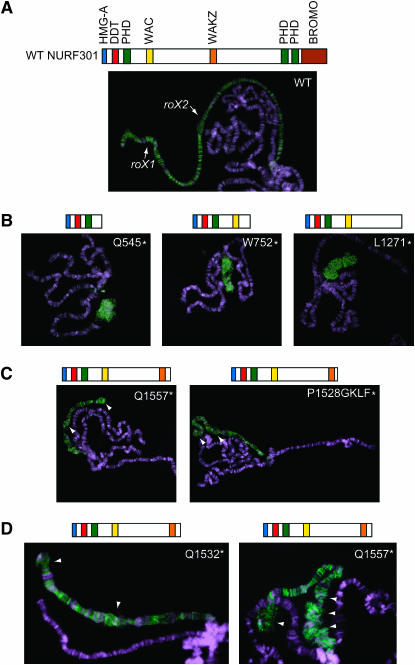
An allelic series of Nurf301 mutants display regional puffing on the male X chromosome. (A) Top: domain structure of NURF301 protein. Bottom: wild-type male X chromosome stained by anti-MSL2 antibody (green). DNA is counterstained by DAPI (purple). The approximate cytological positions of roX1 and roX2 are indicated by arrows. (B) Severely truncated alleles of Nurf301 (top) dramatically disrupt the morphology of the male X chromosome (bottom). (C) Overall X morphology is retained in the two hypomorphic Nurf301 mutant males, although regional puffing is observed (arrowheads). (D) Higher magnification images of hypomorphic Nurf301alleles highlighting regional puffing.
The MSL complex must bind its targets on the X chromosome in the context of general regulators of chromatin organization in Drosophila. A major regulator is the nucleosome remodeling factor (NURF). NURF is the founding member of the imitation switch (ISWI) family of ATP-dependent chromatin-remodeling factors. These are multisubunit protein complexes that use the energy of ATP hydrolysis to slide nucleosomes. NURF contains four protein subunits (Xiao et al. 2001). In addition to the catalytic ISWI ATPase subunit, NURF comprises NURF301, a large protein that contains multiple domains involved in chromatin regulation such as PHD motifs and a bromodomain; NURF55, a WD-40 repeat protein found in many histone-metabolizing enzymes; and NURF38, an inorganic pyrophosphatase. NURF301 is the only subunit exclusively found in NURF and is obligatory for the assembly of NURF (Xiao et al. 2001; Barak et al. 2003). All other NURF subunits are present in other chromatin-modifying complexes. For example, the ISWI ATPase is also the catalytic subunit of chromatin accessibility complex (CHRAC) (Varga-Weisz et al. 1997) and ATP-utilizing chromatin assembly and remodeling factor (ACF) (Ito et al. 1997), two other ATP-dependent chromatin regulators. Reconstitution experiments reveal that only NURF301 and ISWI are essential for the chromatin-remodeling activities of NURF, suggesting that they are the major subunits (Xiao et al. 2001).
Genetic ablation of NURF301 and ISWI confirms that NURF is essential for viability (Deuring et al. 2000; Badenhorst et al. 2002). Moreover, NURF-mediated nucleosome sliding appears to be involved in both transcriptional activation and repression. In Nurf301 and Iswi mutants the activation of heat-shock genes, the homeotic genes Ubx and engrailed, and ecdysone responsive genes is blocked (Deuring et al. 2000; Badenhorst et al. 2002, 2005). Conversely, loss of NURF leads to activation of the JAK/STAT pathway, resulting in melanotic tumors caused by overproliferation of larval blood cells (Badenhorst et al. 2002). Consistent with NURF functioning both as an activator or repressor of transcription, whole genome expression profiling indicates that large numbers of genes are both up- and downregulated in Nurf301 mutants although the direct target loci remain unknown (Badenhorst et al. 2005).
The activities of the MSL and NURF complexes intersect on the male X chromosome, revealing a potentially complementary or antagonistic relationship. The male polytene X chromosome in either Nurf301 or Iswi mutants is severely distorted (Deuring et al. 2000; Badenhorst et al. 2002). In the absence of NURF, the male X chromosome becomes disorganized and highly decondensed, appearing very wide at the expense of length. MSL staining of this aberrant polytene X chromosome reveals strong MSL localization without the typical banded pattern seen on wild-type X chromosomes. Interestingly, the aberrant male-specific X morphology can be suppressed by disrupting dosage compensation (in mle mutants) and, conversely, can be induced in females by forced expression of the MSL complex (Corona et al. 2002). One explanation for this phenotype is that NURF is essential for chromatin organization on all chromosomes, but that as the maternal supply of NURF is dissipated in homozygous Nurf301 or Iswi mutants, the male X chromosome becomes the first to lose its defined morphology due to antagonism between the MSL and NURF complexes. Consistent with this idea, biochemical studies demonstrate that acetylation of histone H4K16 reduces interaction of ISWI with nucleosomes, suggesting that MSL-mediated histone modification may directly disrupt the chromosomal organization function of NURF (Clapier et al. 2002; Corona et al. 2002). Conversely, ISWI has been proposed to block the positive effects of H4K16ac on the X chromosome in an alternative model for dosage compensation (Bhadra et al. 2005).
In this study, we utilize the Nurf301 mutant phenotype to assay the importance of roX gene location on the X chromosome. We find that the aberrant morphology of the male X chromosome in Nurf301 mutants can be regionally suppressed by deletion of either roX1 or roX2, providing strong evidence that roX genes function over very long distances along the X chromosome. In addition to interacting functions at the level of chromosome morphology, we also find that NURF and MSL proteins have opposing effects on roX gene transcription, with NURF repressing roX transcription in females. Together, these results demonstrate the importance of a local balance between factors that promote and antagonize chromatin compaction.
MATERIALS AND METHODS
Fly stocks and genetic crosses:
The following Nurf301 mutant strains were used; the molecular nature of the lesions are indicated in parentheses: Nurf3012 (Q545 to stop codon substitution), Nurf3013 (W752 to stop codon substitution), Nurf3014 [splice donor mutant that introduces four additional amino acids (GKLF) and causes premature truncation of the protein after P1528], Nurf3015 (L1271 to stop codon substitution), Nurf30110 (Q1557 to stop codon substitution), Nurf30112 (Q1532 to stop codon substitution), and Df(3L)3643 (deletion that removes Nurf301). To check reporter expression in Nurf 301 mutants, yw; [reporter]; Nurf3013/Tb was crossed to yw; Nurf3013/Tb. Non-Tb larvae were compared to Tb larvae in the next generation. The roX1+roX2− X chromosome was generated by recombination between y w and y+ roX1ex6 Df (1) roX252 [w+ 4Δ4.3] X chromosomes. yw roX1+roX2−; Nurf3013 male larvae were generated by a brother-sister cross of yw roX1+roX2−; Nurf3013/Tb flies. roX1−roX2+; Nurf3013 male larvae were generated using yw roX1ex6; Nurf3013/Tb flies.
Polytene chromosome immunostaining:
Polytene chromosomes were prepared and immunostained using anti-MSL1 or anti-MSL2 antibodies as previously described (Kelley et al. 1999). The Nurf301 mutant male X-chromosome allelic series was generated using larvae hemizygous for, respectively, Nurf3012, Nurf3013, Nurf3014, Nurf3015, Nurf30110, and Nurf30112. Hemizygotes were generated by crossing the respective Nurf301 allele to Df(3L)3643. All strains were balanced using TM6B, Tb, allowing mutant larvae to be selected as non-Tb.
Quantitative real-time RT–PCR:
DNaseI-treated total RNA was subjected to first-strand cDNA synthesis using SuperScriptII reverse transcriptase (Invitrogen) and primed by oligo dT. Real-time PCR was performed using the ABI PRISM 7000 sequence detection system (Applied Biosystems). The Pka gene was used as the internal reference for normalizing variance in the quality of RNA and the amount of input cDNA. PCR amplification was performed as previously described (Bai et al. 2004). Relative quantification of tested transcriptions was determined by the comparative CT method based on the manufacturer's instruction (Applied Biosystems). A standard curve for each set of primers was constructed using a serial dilution of cDNA to verify equal amplification efficiency. The primers used for real-time PCR are Pka: forward 5′-TTCTCGGAGCCGCACTCGCGCTTCTAC3′ and reverse 5′-CAATCA GCAGATTCTCCGGCT-3′; Reporter: forward 5′-AGCACGACTTCTTCAAGTCCG-3′ and reverse 5′-GTGTCGCCCTCGA ACTTCAC-3′; roX1: forward 5′-ATGCGAGCGAGACAATGATACT-3′ and reverse 5′-GACTTGCAGTCCGCCCTATG-3′; roX2: forward 5′-AGCTCGGATGGCCATCGAAA-3′ and reverse 5′-CGTTACTCTTGCTTGATTTTGCTTCG-3′; glutamine synthetase 2 (Gs2): forward 5′-TGCAGGAGAACATCGTTCAG-3′ and reverse 5′-TCCATCGTAGTTCCAAACGG-3′; phosphogluconate dehydrogenase (Pgd): forward 5′GAACACACGAAACATGAGCG-3′ and reverse 5′-CTCGTCCATGTTGAGTATCAGG-3′; and Zwischenferment (Zw): forward 5′-TGACCGTCGATAGCATCAAG-3′ and reverse 5′-CCAGAACTCCTCGTACTTCTTC-3′.
Chromatin immunoprecipitation:
Chromatin immunoprecipitation (ChIP) was performed as described in Badenhorst et al. (2005). As ChIP-grade antibodies against the NURF-specific subunit NURF301 are unavailable, antibodies against the catalytic subunit ISWI were used. ISWI ChIP signals that are due to NURF were resolved by comparing ChIP profiles of wild-type and Nurf3012 mutant tissues (that lack NURF). Third instar larval salivary glands were used in ChIP. For most ChIP experiments, mixed samples of both male and female larvae were used. For sex-specific ChIP experiments, larvae were sexed by observing the gonad under a stereo microscope and sorted into female-only and male-only populations. All subsequent procedures were identical. Salivary glands were dissected on ice, pooled, and fixed in batches of 25 pairs of glands. After fixation, formaldehyde was removed by extensive washing with ice-cold PBS and glands stored as pellets at −70° until further use. For each ChIP experiment 75 pairs of wild type and 150 pairs of Nurf3012 mutant salivary glands were used. Immune complexes were precipitated using protein A-conjugated magnetic beads (Dynal). Target DNA abundance in ChIP eluates was assayed by quantitative PCR with addition of 0.2 μCi [α-32P]-deoxyadenosine 5′-triphosphate (Perkin-Elmer, specific activity 6000 Ci/mmol) as a tracer before the amplification step. Quantification of PCR products was conducted using a PhosphorImager screen. The ratio of ChIP sample to input sample was determined after titration of samples, and variation of PCR cycle number was conducted to ensure that all PCR products quantified are in the linear range. Typically, 0.5% of the final input sample and 3% of the final immunoprecipitation (IP) sample are used for quantitation. The final input sample is 5% of the total material added to the IP. Data presented are the mean of three separate determinations. Variation between ChIP analyses was <10%. Primer pairs for PCR were roX1 set 1: 5′-ACATGGGCGTAGTTTCATATACG-3′ and 5′-TACATCTTGCCAGAGATTTCG-3′; roX1 set 2: 5′-ACGCGTACGCATACCTCTATC-3′ and 5′-GAGCGGAGCGGTATTCGTGAG-3′; roX2 set 1: 5′ATATGAGCCACGTCATGGGT-3′ and 5′-GGGAACTGCATGAATGCGAA-3′; roX2 set 2: 5′-AGCTCGGATGGCCATCGAAA-3′ and 5′-CGTTACTCTTGCTTGATTTTGCTTCG-3′; and roX2 set 3: 5′-ACTAGTGAAATGTTATACGAAAC-3′ and 5′-TGTAATTTAAGTGTCAGTTC-3′.
RESULTS
Regional puffing on the male X chromosome in hypomorphic Nurf301 mutants:
NURF301 is a large 300-kDa protein containing multiple conserved domains implicated in regulation of chromatin organization (Figure 1A). As demonstrated previously, loss of NURF function causes the male X chromosome to become disorganized and highly decondensed, appearing very wide at the expense of length (Figure 1B). MSL staining of this aberrant polytene X chromosome reveals strong MSL localization, but without the typical banded pattern always seen on wild-type X chromosomes (Figure 1A). Among the four subunits of NURF, NURF301 is the only NURF-specific subunit and is obligatory for the assembly of NURF. A series of EMS-induced mutations were previously generated in Nurf301, all of which produce truncated NURF301 protein (Badenhorst et al. 2002). The hypomorphic alleles of Nurf301 that produce progressively longer truncated proteins can survive beyond the pupal stage, and some heteroallelic combinations survive to the adult stage with developmental abnormalities and are sterile (Badenhorst et al. 2005). We tested the male X morphology in these hypomorphic mutants by staining the polytene chromosomes with anti-MSL2 antibody. The overall structure of the male X chromosome is retained, as shown in Figure 1C. However, regional puffing can still be seen in most of the mutants (Figure 1, C and D), suggesting that there may be some sites on the X chromosome that are more sensitive to the loss of NURF function. Interestingly, we found that these locally puffed regions are often close to the tip, near roX1 at cytological position 3F, or in the middle of the X chromosome near roX2 at cytological position 10C (Figure 1D).
roX mutations suppress the Nurf301 mutant phenotype in cis on the male X chromosome:
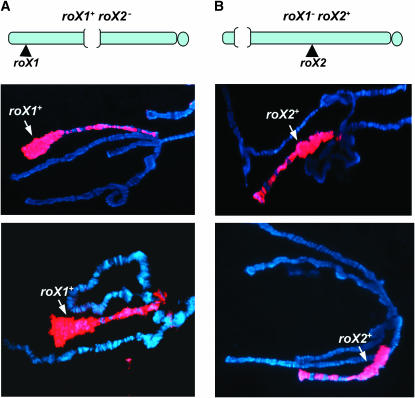
The local puffing on the X chromosome in Nurf301 mutants prompted us to examine whether roX genes could modulate the Nurf301 phenotype. Previous studies showed that disrupting MSL function by a mutation in mle could suppress the puffed X morphology caused by loss of NURF function (Corona et al. 2002). Although deletion of a single roX gene normally has no effect on the distribution or function of the MSL complex, we found that mutation of either roX1 or roX2 could regionally inhibit the disrupted X morphology in Nurf301 mutants (Figure 2, A and B). There was a striking inverse correlation between the regional puffing retained on the X chromosome and the site of roX mutation. As shown in Figure 2A, when roX2 was missing on the male X chromosome, MSL complexes were found to accumulate around the roX1+ locus and the surrounding region displayed a very puffed morphology with a dramatic loss of the normal banding pattern, while the rest of the X chromosome displayed more normal levels of MSL staining. In contrast, if roX1 was missing, the bulk of MSL complexes were restricted on the proximal half of the X chromosome surrounding the roX2+ locus, resulting in chromosome puffing only in that region (Figure 2B). These phenotypes were consistent from nucleus to nucleus and were present in multiple individual larvae. These data strongly support the idea that the MSL complex opposes compaction mediated by NURF to regulate male X-chromosome morphology. More importantly, these results suggest that roX genes, which are proposed to be MSL assembly and spreading initiation sites (Park et al. 2002; Oh et al. 2003), can regulate the X chromosome in cis.
Figure 2.—
roX mutations result in a regional reversal of the Nurf mutant phenotype on the male X chromosome. Male polytene chromosomes were stained with anti-MSL1 antibody (red) and counterstained with DAPI (blue). (A) In Nurf301 mutant males, deletion of roX2 strongly reduces chromosome puffing of a large portion of the proximal X, including around the roX2 locus. (B) Similarly, puffing of the distal X including the roX1 locus is lost when roX1 is deleted. The genotypes are (A) yw Df (1) roX252 [w+4Δ4.3]; nurf3013 and (B) yw roX1ex6; nurf3013.
A roX transgene enhances aberrant morphology in Nurf301 mutants at ectopic locations on autosomes:
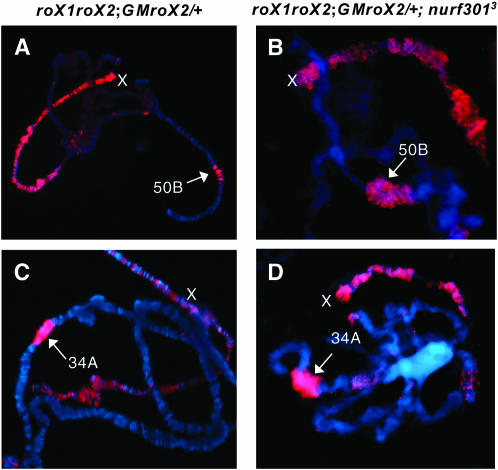
Our previous studies have shown that a genomic roX transgene (GMroX) can recruit the MSL complex to an autosome. In the absence of endogenous roX genes, the GMroX transgene often displays spreading of the MSL complex into flanking autosomal regions (Park et al. 2002) (Figure 3A). We therefore asked if such an ectopic, autosomal roX insertion could induce chromosomal disruption in flanking chromatin in Nurf301 mutants. We tested this by crossing roX1roX2 double mutant flies that carry a GMroX2 transgene into the Nurf301 mutant background. Chromosomal decondensation surrounding the autosomal GMroX2 transgene was indeed detected (Figure 3, B and D). Conversely, the morphology of the X chromosome was largely improved compared to a wild-type, roX+ X chromosome (Figure 3, B and D). These results indicate that the ectopic MSL complex recruited by a GMroX transgene can induce autosomal puffing in Nurf mutants, consistent with the MSL complex counteracting NURF function. Furthermore, these data suggest that the site of roX transcription and MSL nucleation are spatially linked to this antagonism. The improvement of X morphology may be explained by a lower concentration of MSL complexes on the roX mutant X, as some complexes are recruited to the autosome by the transgene. Relatively weaker MSL staining on the roX-deleted X chromosome in transgenic males suggests that this is the case (Figure 3, A and C).
Figure 3.—
Chromosomal decondensation induced by a GMroX2 transgene in nurf301 mutant males. In the absence of X-linked roX genes, local spreading is seen at the GMroX2 autosomal insertion sites at cytological positions 50B and 34A (A and C, respectively, arrows). Loss of NURF function causes local chromosomal puffing at these same transgene insertion sites (B and D, arrows), and X-chromosome puffing is less severe. Male polytene chromosomes were stained with anti-MSL1 antibody (red) and counterstained with DAPI (blue). The genotype of the double mutant X chromosome is yw roX1ex6 Df (1) roX252 [w+4Δ4.3].
NURF represses roX RNA transcription:
Previously, we reported that in the absence of functional MSL complexes (for example, in females or in msl mutant males) roX1 transcription is repressed by an unknown factor (Bai et al. 2004). Since MSL and NURF complexes appear to be antagonistic at the level of chromatin compaction, and MSL proteins positively regulate roX transcription, we examined whether NURF might be a repressor of roX genes. Derepression of roX transcription, leading to a higher local concentration of MSL complexes, might explain the local puffing observed in the vicinity of roX genes in Nurf301 hypomorphs.
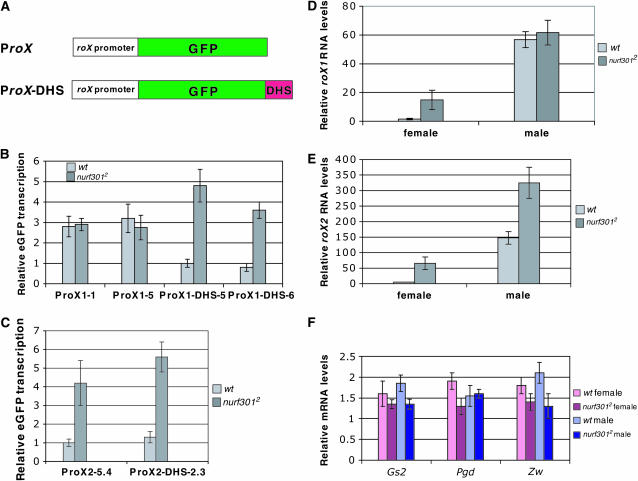
To investigate whether NURF represses roX transcription we first analyzed expression of heterologous roX reporter constructs in Nurf mutants. In these experiments, expression of a GFP reporter is driven by the endogenous roX promoter, to which its cognate MSL binding site is either added or omitted, 3′ to the GFP reporter (Figure 4A). These regulatory sites, previously shown to function as MSL-dependent enhancers of roX transcription, are male-specific DNase hypersensitive sites (DHS). The constructs analyzed were ProX1 vs. ProX1-DHS and ProX2 vs. ProX2-DHS (Bai et al. 2004). Previous studies have shown that the roX1 reporter is repressed in females through the same region that binds the MSL complex, forms the DHS, and activates roX1 in males. In contrast, repression of the roX2 reporter is independent of its MSL binding site (DHS). Both the roX1 and roX2 reporter systems were tested in Nurf301 mutant larvae by real-time RT–PCR. As shown in Figure 4B, in Nurf301 females the ProX1-DHS reporter was derepressed, reaching an expression level comparable to that of the ProX1 reporter. In addition, the expression of both ProX2 and ProX2-DHS were increased in Nurf mutant females (Figure 4C), demonstrating that both reporters are repressed by NURF in wild-type females. Similar effects were observed in Iswi mutant females (data not shown). The expression of roX reporters in male Nurf301 and Iswi mutant larvae was unchanged (data not shown). Thus, NURF clearly functions to repress expression from roX gene reporters in females. While these data do not explain the local effects of roX genes on puffing of the X chromosome in males, they provide a second example of the opposing functions of NURF and MSL complexes.
Figure 4.—
NURF represses transcription of roX genes in females. (A) Schematic of roX gene reporter constructs with and without the DHS. Real-time RT–PCR was used to measure the transcription from either the roX reporters (B and C) or the endogenous roX genes (D and E) in Nurf301 mutants. RNA levels of three other X-linked genes were not affected in Nurf mutants (F). For each sample, data represents the mean ± SD from three independent experiments.
We also examined whether NURF represses endogenous, steady-state roX RNA levels. In both Nurf301 and Iswi mutants, roX1 and roX2 RNA levels were significantly increased in females (Figure 4, D and E). Although increased, the level of roX RNAs in mutant females was still lower than in wild-type males, probably due to RNA instability in females lacking the MSL complex. In Nurf mutant males, the roX1 RNA level was not altered (Figure 4D), but the roX2 level was modestly increased (approximately twofold) (Figure 4E). We did not detect any significant changes in levels of several other X-linked transcripts in Nurf mutants, suggesting that NURF exerts a specific effect on roX genes (Figure 4F). We also tested 11 other chromatin modifiers that are generally involved in gene repression, including polycomb group proteins HP1 and SU(VAR)3-9 (Table 1). None affected roX transcription. These results demonstrate that wild-type NURF negatively regulates roX1 and roX2 RNA transcription in females, with an approximately twofold effect on endogenous roX2 levels in males. Our results also suggest some differences between roX1 and roX2 regulation. Both roX1 activation by MSL complex and repression by NURF act through the DHS, while repression of roX2 by NURF is mediated by 5′ sequences rather than by the DHS.
TABLE 1.
Summary of genes tested for their regulation of ProX-DHS reporter gene transcription
| Chromatin regulators tested | Dominant effect | Homozygous mutants |
|---|---|---|
| Pc | − | ND |
| Psc | − | ND |
| Pcl | − | ND |
| Su(z)2 | − | ND |
| Scm | − | ND |
| Asx | − | ND |
| HP1 | − | ND |
| Su(var)3-9 | ND | − |
| Pr-set-7 | ND | − |
| PIWI | ND | − |
| GAGA factor | ND | − |
At least two lines of ProX1-DHS or ProX2-DHS were crossed into each genetic background. The transcription level of the GFP reporter gene was measured by real-time RT–PCR and compared with that of a wild-type genetic background. A change less than twofold was scored as negative (−). Transcription was assayed in a heterozygous background for HP1 mutant and PcG mutants and in a homozygous background for all the other mutants. ND, not determined.
In vivo binding of ISWI, the catalytic component of NURF, at roX genes:
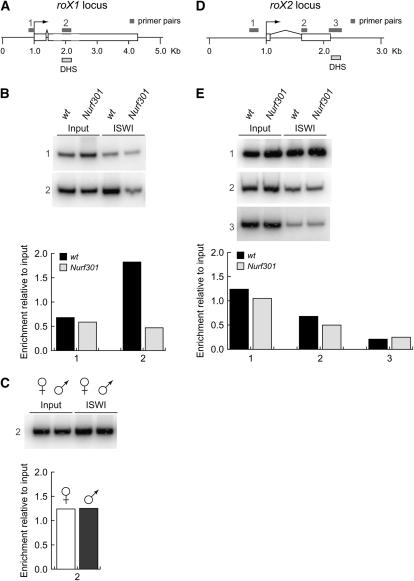
NURF may regulate roX transcription by directly binding to roX genes or act indirectly through other factors. To address this question, we performed ChIP using antibodies against ISWI, the catalytic subunit of NURF. ISWI is essential for NURF function and our transcription analyses have indicated an important role for ISWI in roX transcription (see above). As shown in Figure 5B, we detected specific binding of ISWI at the roX1-DHS. ISWI binding is reduced in Nurf301 mutants, indicating that binding is due to the recruitment of NURF. The 5′ region of roX1 did not show high levels of ISWI binding, consistent with our reporter assays that indicated that repression of roX1 by NURF is mediated through the roX1-DHS. ISWI binding to the roX1-DHS was detected in both males and females, suggesting that NURF-mediated regulation could occur in both sexes (Figure 5C), although repression was evident only in females by our roX reporter assays.
Figure 5.—
ChIP using antibodies against the ISWI subunit of NURF indicates that NURF binds to the DHS of roX1. (A) Organization of the roX1 locus showing primer pairs used in ChIP and location of the MSL-binding DHS. (B) Comparison of ISWI ChIP signals in wild type and Nurf3012 mutant tissue reveals that NURF binds to the DHS (primer pair 2), but is not localized to the promoter (primer pair 1) of roX1. (C) ISWI ChIP was performed in sorted samples of male and female salivary glands and demonstrates that NURF is recruited to the roX1 DHS in both males and females. (D) Organization of the roX2 locus showing primer pairs used in ChIP and location of the MSL-binding DHS. (E) NURF is not detected at the roX2 DHS or on the body of the gene. B, C, and E show representative ChIP PCR data. Graphs show the fold enrichment of ChIP signals relative to the input and are derived from the ratio of ChIP to input band signal intensities averaged over three separate determinations. Input and IP samples were titrated and PCR cycle number was optimized to ensure that PCR products quantified using a PhosphorImager were in the linear range.
Unlike roX1, ISWI binding was not enriched at the roX2 DHS region (Figure 5E). This suggests that NURF acts through another element to regulate the roX2 gene, consistent with the results of roX2-reporter studies that indicated that repression of roX2 is independent of the DHS. Further analysis using primer sets that span the roX2 locus revealed ISWI binding over the promoter region (Figure 5E). However, binding was not abolished in Nurf301 mutants. Therefore, it may be that other ISWI-containing complexes such as CHRAC or ACF bind this region, potentially masking an additional, NURF-dependent recruitment.
DISCUSSION
The roX noncoding RNAs are critical components that regulate targeting of MSL complexes to the male X chromosome. roX RNAs are not stably expressed in wild-type females, and here we show that NURF, an ATP-dependent chromatin-remodeling enzyme, is a repressor of roX transcription in females. Furthermore, we found that regional decondensation of the male X chromosome in Nurf mutants was dependent on the presence of a linked roX gene, roX1 for the distal X and roX2 for the proximal X. These results support a model in which the MSL complex assembles at roX genes and can act long distances along the X chromosome.
NURF controls roX transcription:
Previous analyses indicated that MSL proteins are required for transcription of roX genes (Bai et al. 2004). In the case of roX1, the MSL complex binds to the DHS and counteracts the activity of a constitutive repressor, establishing the male-specific pattern of roX1 transcription. Our analysis of endogenous roX transcript levels and heterologous roX reporter constructs indicate that NURF mediates this repression and that, for roX1, NURF acts through the DHS. This is confirmed by ChIP analysis that shows NURF is recruited to the roX1 DHS, demonstrating that regulation is direct.
The requirement for NURF in roX repression was detected in females that do not ordinarily express roX genes. In the absence of NURF, endogenous levels of both roX1 and roX2 are increased in females. In males, steady-state transcript levels are either unaffected (roX1) or increased approximately twofold (roX2). It is clear from the extreme decondensation of the NURF mutant male X chromosome and its suppression by roX and MSL complex mutants that the male X chromosome is very sensitive to the loss of the NURF complex. One model that may reconcile the apparent contradiction between lack of roX gene derepression in Nurf mutant males and the extreme male X chromosome phenotype is as follows: NURF may affect key initial levels of roX RNAs at embryonic stages when MSL complex binding is first initiated. The effect of improperly regulated complex assembly during initial stages could be progressively amplified during development resulting in a chromosome morphology defect at a time when roX gene expression is no longer regulated by NURF in males. Alternatively, antagonism at the level of roX transcription and at the level of chromatin morphology may be functionally independent events.
Mechanism of NURF and MSL antagonism:
A principal activity of NURF is ATP-dependent nucleosome sliding in cis on DNA without displacement (Hamiche et al. 1999). It is not difficult to envisage how nucleosome sliding can be used to expose or block binding sites for transcription factors and thereby control transcription. Indeed there is much evidence from studies of the orthologous yeast Isw2 remodeling complex that nucleosome sliding can be used to repress transcription. Isw2 is needed to repress early meiotic genes and targets of the Tup1-Ssn6 complex (Goldmark et al. 2000; Kent et al. 2001; Ruiz et al. 2003; Zhang and Reese 2004). However, in these cases, repression is mediated through 5′ regulatory elements at the level of transcription initiation. As we have seen above, NURF represses roX1 through a binding site present in the coding region, ∼1 kb 3′ of the transcription initiation site.
The location of NURF binding within the roX1 gene becomes more pertinent when the known distribution of the MSL complex is considered. Recent whole genome profiling of MSL complex distribution on X-chromosome targets indicates that the complex shows a strong preference for the 3′ ends of gene targets (Alekseyenko et al. 2006; Gilfillan et al. 2006). This correlates with a previous, more restricted, analysis showing that acetylation of H4K16, the epigenetic mark established by the MSL complex, follows a similar distribution (Smith et al. 2001). One implication of this distribution is that the MSL complex may regulate transcription of targets (including roX genes) not at the level of transcription initiation, but by improving elongation. It is tempting to speculate that NURF may also control elongation, as suggested by studies of the yeast ISW1 complex (Morillon et al. 2003).
Model for regional action of roX genes on the X chromosome:
Here we find that the aberrant morphology of the male X chromosome in Nurf301 mutants is regionally suppressed by deletion of either roX1 or roX2, providing strong visual evidence that roX genes can function in cis over long distances (>1 Mb). We believe that our results are consistent with a model in which nascent roX RNAs normally assemble and nucleate “spreading” of MSL complexes along the X chromosome. The term “spreading” has been controversial as it is subject to a myriad of interpretations. What we mean by spreading is that following assembly, the MSL complex is much more likely to act regionally, in cis, than to be unconstrained. We previously proposed that in addition to roX genes, specific MSL interaction occurs at “high affinity sites” (also termed “chromatin entry sites”) whose identifying characteristics are yet to be defined (Lyman et al. 1997; Kelley et al. 1999; Demakova et al. 2003; Oh et al. 2004; Dahlsveen et al. 2006). “Spreading” from roX genes and high affinity sites to the full MSL binding pattern could occur by scanning along the chromosome in a linear mode, but it could also occur solely by local release and recapture of the complex by regions in close physical proximity. Movement from one DNA molecule to another clearly can occur when roX genes or various segments of X are moved to autosomes (Meller et al. 1997; Kelley et al. 1999; Fagegaltier and Baker 2004; Oh et al. 2004; Dahlsveen et al. 2006).
High affinity sites and roX genes might normally function together to constrain the MSL complex largely to the X chromosome. An “affinities” model developed in a series of reports (Demakova et al. 2003; Fagegaltier and Baker 2004; Dahlsveen et al. 2006) posits that there is a continuum of affinity sites for MSL complexes, ranging from high to low, and that only when high sites are locally concentrated can low affinity sites be recognized. In our opinion, this clearly falls under the general premise of the spreading model. In both cases, MSL targeting is a multistep process in which many binding sites are not recognized independently, in the absence of influence of neighboring sites in cis. While the image of X-chromosome morphology regionally controlled by the presence or absence of a roX gene is, to us, strong evidence for function of roX genes over long distances in cis, a more mechanistic view of MSL targeting clearly awaits additional data on the molecular nature of MSL–chromatin interactions.
Acknowledgments
We thank R. Kelley, M. Gelbart, A. Gortchakov, and A. Alekseyenko for critical reading of the manuscript. This work was supported by a grant from the National Institutes of Health (GM45744). E.L. is a Leukemia and Lymphoma Society postdoctoral fellow. S.Y.K. and P.B. are supported by the BBSRC. M.I.K. is a Howard Hughes Medical Institute investigator.
References
- Alekseyenko, A. A., E. Larschan, W. R. Lai, P. J. Park and M. I. Kuroda, 2006. High-resolution ChIP-chip analysis reveals that the Drosophila MSL complex selectively identifies active genes on the male X chromosome. Genes Dev. 20: 848–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badenhorst, P., M. Voas, I. Rebay and C. Wu, 2002. Biological functions of the ISWI chromatin remodeling complex NURF. Genes Dev. 16: 3186–3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badenhorst, P., H. Xiao, L. Cherbas, S. Y. Kwon, M. Voas et al., 2005. The Drosophila nucleosome remodeling factor NURF is required for Ecdysteroid signaling and metamorphosis. Genes Dev. 19: 2540–2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai, X., A. A. Alekseyenko and M. I. Kuroda, 2004. Sequence-specific targeting of MSL complex regulates transcription of the roX RNA genes. EMBO J. 23: 2853–2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barak, O., M. A. Lazzaro, W. S. Lane, D. W. Speicher, D. J. Picketts et al., 2003. Isolation of human NURF: a regulator of Engrailed gene expression. EMBO J. 22: 6089–6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhadra, M. P., U. Bhadra, J. Kundu and J. A. Birchler, 2005. Gene expression analysis of the function of the male-specific lethal complex in Drosophila. Genetics 169: 2061–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapier, C. R., K. P. Nightingale and P. B. Becker, 2002. A critical epitope for substrate recognition by the nucleosome remodeling ATPase ISWI. Nucleic Acids Res. 30: 649–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corona, D. F., C. R. Clapier, P. B. Becker and J. W. Tamkun, 2002. Modulation of ISWI function by site-specific histone acetylation. EMBO Rep. 3: 242–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlsveen, I. K., G. D. Gilfillan, V. I. Shelest, R. Lamm and P. B. Becker, 2006. Targeting determinants of dosage compensation in Drosophila. PLoS Genet. 2: e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demakova, O. V., I. V. Kotlikova, P. R. Gordadze, A. A. Alekseyenko, M. I. Kuroda et al., 2003. The MSL complex levels are critical for its correct targeting to the chromosomes in Drosophila melanogaster. Chromosoma 112: 103–115. [DOI] [PubMed] [Google Scholar]
- Deuring, R., L. Fanti, J. A. Armstrong, M. Sarte, O. Papoulas et al., 2000. The ISWI chromatin-remodeling protein is required for gene expression and the maintenance of higher order chromatin structure in vivo. Mol. Cell 5: 355–365. [DOI] [PubMed] [Google Scholar]
- Fagegaltier, D., and B. S. Baker, 2004. X chromosome sites autonomously recruit the dosage compensation complex in Drosophila males. PLoS Biol. 2: e341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilfillan, G. D., I. K. Dahlsveen and P. B. Becker, 2004. Lifting a chromosome: dosage compensation in Drosophila melanogaster. FEBS Lett. 567: 8–14. [DOI] [PubMed] [Google Scholar]
- Gilfillan, G. D., T. Straub, E. de Wit, F. Greil, R. Lamm et al., 2006. Chromosome-wide gene-specific targeting of the Drosophila dosage compensation complex. Genes Dev. 20: 858–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldmark, J. P., T. G. Fazzio, P. W. Estep, G. M. Church and T. Tsukiyama, 2000. The Isw2 chromatin remodeling complex represses early meiotic genes upon recruitment by Ume6p. Cell 103: 423–433. [DOI] [PubMed] [Google Scholar]
- Hamiche, A., R. Sandaltzopoulos, D. A. Gdula and C. Wu, 1999. ATP-dependent histone octamer sliding mediated by the chromatin remodeling complex NURF. Cell 97: 833–842. [DOI] [PubMed] [Google Scholar]
- Ito, T., M. Bulger, M. J. Pazin, R. Kobayashi and J. T. Kadonaga, 1997. ACF, an ISWI-containing and ATP-utilizing chromatin assembly and remodeling factor. Cell 90: 145–155. [DOI] [PubMed] [Google Scholar]
- Kelley, R. L., V. H. Meller, P. R. Gordadze, G. Roman, R. L. Davis et al., 1999. Epigenetic spreading of the Drosophila dosage compensation complex from roX RNA genes into flanking chromatin. Cell 98: 513–522. [DOI] [PubMed] [Google Scholar]
- Kent, N. A., N. Karabetsou, P. K. Politis and J. Mellor, 2001. In vivo chromatin remodeling by yeast ISWI homologs Isw1p and Isw2p. Genes Dev. 15: 619–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucchesi, J. C., 1998. Dosage compensation in flies and worms: the ups and downs of X-chromosome regulation. Curr. Opin. Genet. Dev. 8: 179–184. [DOI] [PubMed] [Google Scholar]
- Lyman, L. M., K. Copps, L. Rastelli, R. L. Kelley and M. I. Kuroda, 1997. Drosophila male-specific lethal-2 protein: structure/function analysis and dependence on MSL-1 for chromosome association. Genetics 147: 1743–1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meller, V. H., and M. I. Kuroda, 2002. Sex and the single chromosome. Adv. Genet. 46: 1–24. [DOI] [PubMed] [Google Scholar]
- Meller, V. H., and B. P. Rattner, 2002. The roX genes encode redundant male-specific lethal transcripts required for targeting of the MSL complex. EMBO J. 21: 1084–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meller, V. H., K. H. Wu, G. Roman, M. I. Kuroda and R. L. Davis, 1997. roX1 RNA paints the X chromosome of male Drosophila and is regulated by the dosage compensation system. Cell 88: 445–457. [DOI] [PubMed] [Google Scholar]
- Morillon, A., N. Karabetsou, J. O'Sullivan, N. Kent, N. Proudfoot et al., 2003. Isw1 chromatin remodeling ATPase coordinates transcription elongation and termination by RNA polymerase II. Cell 115: 425–435. [DOI] [PubMed] [Google Scholar]
- Oh, H., Y. Park and M. I. Kuroda, 2003. Local spreading of MSL complexes from roX genes on the Drosophila X chromosome. Genes Dev. 17: 1334–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh, H., J. R. Bone and M. I. Kuroda, 2004. Multiple classes of MSL binding sites target dosage compensation to the X chromosome of Drosophila. Curr. Biol. 14: 481–487. [DOI] [PubMed] [Google Scholar]
- Park, Y., R. L. Kelley, H. Oh, M. I. Kuroda and V. H. Meller, 2002. Extent of chromatin spreading determined by roX RNA recruitment of MSL proteins. Science 298: 1620–1623. [DOI] [PubMed] [Google Scholar]
- Ruiz, C., V. Escribano, E. Morgado, M. Molina and M. J. Mazon, 2003. Cell-type-dependent repression of yeast a-specific genes requires Itc1p, a subunit of the Isw2p-Itc1p chromatin remodelling complex. Microbiology 149: 341–351. [DOI] [PubMed] [Google Scholar]
- Shogren-Knaak, M., H. Ishii, J. M. Sun, M. J. Pazin, J. R. Davie et al., 2006. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science 311: 844–847. [DOI] [PubMed] [Google Scholar]
- Smith, E. R., C. D. Allis and J. C. Lucchesi, 2001. Linking global histone acetylation to the transcription enhancement of X-chromosomal genes in Drosophila males. J. Biol. Chem. 276: 31483–31486. [DOI] [PubMed] [Google Scholar]
- Varga-Weisz, P. D., M. Wilm, E. Bonte, K. Dumas, M. Mann et al., 1997. Chromatin-remodelling factor CHRAC contains the ATPases ISWI and topoisomerase II. Nature 388: 598–602. [DOI] [PubMed] [Google Scholar]
- Xiao, H., R. Sandaltzopoulos, H. M. Wang, A. Hamiche, R. Ranallo et al., 2001. Dual functions of largest NURF subunit NURF301 in nucleosome sliding and transcription factor interactions. Mol. Cell 8: 531–543. [DOI] [PubMed] [Google Scholar]
- Zhang, Z., and J. C. Reese, 2004. Ssn6-Tup1 requires the ISW2 complex to position nucleosomes in Saccharomyces cerevisiae. EMBO J. 23: 2246–2257. [DOI] [PMC free article] [PubMed] [Google Scholar]