Abstract
Migration of border cells during Drosophila melanogaster oogenesis is a good model system for investigating the genetic requirements for cell migration in vivo. We present a sensitized loss-of-function screen used to identify new genes required in border cells for their migration. Chromosomes bearing FRTs on all four major autosomal arms were mutagenized by insertions of the transposable element PiggyBac, allowing multiple parallel clonal screens and easy identification of the mutated gene. For border cells, we analyzed homozygous mutant clones positively marked with lacZ and sensitized by expression of dominant-negative PVR, the guidance receptor. We identified new alleles of genes already known to be required for border cell migration, including aop/yan, DIAP1, and taiman as well as a conserved Slbo-regulated enhancer downstream of shg/DE–cadherin. Mutations in genes not previously described to be required in border cells were also uncovered: hrp48, vir, rme-8, kismet, and puckered. puckered was unique in that the migration defects were observed only when PVR signaling was reduced. We present evidence that an excess of JNK signaling is deleterious for migration in the absence of PVR activity at least in part through Fos transcriptional activity and possibly through antagonistic effects on DIAP1.
CELL migrations are essential events that occur during embryonic development and throughout the life of animals. Many detailed studies in tissue culture cells have identified and characterized molecular components involved in the migration of individual cultured cells. Less is known about the cellular and molecular mechanisms acting in vivo. A number of characterized cell migrations have attracted attention and have been developed as models to study cell migration in vivo. Well-known examples are neural crest cells in vertebrates, lateral line primordium in zebrafish, germ cells in both vertebrates and Drosophila, as well as tracheal cells and border cells in Drosophila.
Due to the sophisticated genetics tools in Drosophila, including the ability to easily generate clones of mutant cells, border cells have become a powerful system for studying cell migration in vivo. Border cells are a group of ∼8 cells that arise from the follicular epithelium of the egg chamber. The follicular epithelium is a monolayer of somatic cells that covers the germline-derived cells, the 15 nurse cells, and the oocyte. Border cells delaminate from the anterior part of this epithelium and migrate first posteriorly between the nurse cells toward the oocyte, and then dorsally along the border between the nurse cells and the oocyte (Montell et al. 1992). Migrating upon other cells, border cells use the cell–cell adhesion molecule DE–cadherin, encoded by the shotgun (shg) locus, for substrate adhesion (Niewiadomska et al. 1999). Thus, expression of shg is required both in border cells and in the germline cells for migration to occur. Localization of DE–cadherin protein within border cells as well as protein turnover appears be regulated at multiple levels and by different factors: the nonconventional myosinVI, the polarity proteins Par3 and Par6, as well as the transcriptional regulators Taiman and aop/yan (Bai et al. 2000; Geisbrecht and Montell 2002; Pinheiro and Montell 2004; Schober et al. 2005). Also, sequences within the C-terminal tail of DE–cadherin are specifically required for border cell migration (Pacquelet and Rorth 2005).
Loss-of-function screens have previously been performed and have identified genes whose function is required for border cells to migrate properly (Montell et al. 1992; Liu and Montell 1999). Among them, the transcription regulators Slbo, Taiman, Jing, and Stat92E are all expressed in border cells and required for migration (Montell et al. 1992; Bai et al. 2000; Liu and Montell 2001; Silver and Montell 2001; Beccari et al. 2002). Slbo was shown to upregulate DE–cadherin mRNA levels in border cells, as well as expression of jing (Niewiadomska et al. 1999; Liu and Montell 2001). The transcription profiles of wild-type and slbo mutant border cells have recently been described (Borghese et al. 2006; Wang et al. 2006). These studies allowed the discovery of additional genes regulated by Slbo and important for border cell migration, such as stathmin and kuzbanian. However, the targets of the other transcriptional regulators are still unknown and many gaps remain in our understanding of the transcriptional changes important for border cell migration.
Through gain-of-function screens, Gurken and PVF1 were identified as guidance cues for border cells. They signal through the tyrosine kinase receptors (RTKs) EGFR and PVR (PDGF/VEGF receptor), respectively. These RTKs have redundant activity in guiding the posterior migration toward the oocyte, whereas only the EGFR pathway is essential for dorsal migration as only EGFR ligands are unequally distributed between dorsal and ventral sides (Duchek and Rorth 2001; Duchek et al. 2001). However, it remains unclear which pathways act downstream of these RTKs in guidance. The unconventional GTP exchange factor Myoblast city and the small GTPase Rac are important for border cell migration and act downstream of PVR in regulating actin polymerization (Duchek et al. 2001). The inhibitor of apoptosis DIAP1 is also involved in border cell migration and regulates F-actin levels downstream of, or in parallel to, Rac (Geisbrecht and Montell 2004).
To identify new players required for border cell migration, we developed and conducted a loss-of-function screen for mutants affecting border cell migration. Mutagenesis in this screen was based on mobilization of a PiggyBac transposon onto double FRT-bearing chromosomes. The FRT allowed us to perform a clonal screen on lethal loci, as we could analyze the phenotype of homozygous mutant cells in an otherwise heterozygous animal. We developed a modified version of the MARCM system (Lee and Luo 2001), referred to here as MdnP (for MARCM with dominant-negative PVR). In this system, lacZ was used to allow direct visualization of mutant border cells, even if they were rare. In addition, the MdnP system allowed us to sensitize the mutant border cells by perturbing their guidance with a dominant-negative form of PVR (dnPVR). Here, we report the results obtained from this screen and analysis of some of the uncovered mutants.
MATERIALS AND METHODS
Drosophila melanogaster stocks:
P{neoFRT}FRT40A, P{neoFRT}FRT42D (Vegh and Basler 2003) was obtained from K. Basler. P{neoFRT}FRT80, P{neoFRT}FRT82B was generated by recombination and tested for neor and functionality of both FRTs. These double-FRT chromosomes were isogenized and tested before the generation of the screen stocks.
P{w+,ovoD1},P{neoFRT}FRT80 and P{neoFRT}FRT42D, P{w+,ovoD1} chromosomes were generated as follows: w1118; P{neoFRT}FRT80/P{w+,ovoD1},P{FRT(whs)}2A and w1118; P{neoFRT}FRT42D/P{FRT(whs)}G13,P{w+,ovoD1} second instar larvae were X-ray irradiated at a dose of 1000 rad (4 mA, 100 kV, 3′18′′, Philips MG102) to induce mitotic recombination, including in the germline. Adult male progeny were crossed to eyFLP,w1118; P{neoFRT}FRT42D or eyFLP,w1118; P{neoFRT}FRT80 virgins. Appearance of mosaic eye color in progeny allowed identification of the likely correct recombinant chromosomes, subsequently tested for female sterility and generation of germline clones.
The PiggyBac (PBac) transposase chromosome (Horn and Wimmer 2000) was obtained from E. Wimmer and was recombined with the If dominant marker. hsFLP122 and UASp-lacZ were recombined on a w1118 X chromosome, and slbo-Gal4 (Rorth et al. 1998) and UAS-dnPVR (Duchek et al. 2001) were recombined on the second and the third chromosome. The Pvf11624 mutant and EPg11235 (EP-Pvf1) are described in Duchek et al. (2001), and kek-LacZ15A6 (Musacchio and Perrimon 1996) was obtained from N. Perrimon. UAS-DIAP1 (Wang et al. 1999) and pucE69 (Ring and Martinez Arias 1993) were obtained from S. Cohen. UAS-hepACT (Weber et al. 2000) was obtained from M. Mlodzik. UAS-fosRNAi and UAS-junRNAi were kind gifts from D. Bohmann. pucA251 was obtained from L. Raftery. bsk2, jraIA109, kay1 as well as recombinant chromosomes with tub-Gal80 and FRT40A, FRT42D, FRT80, or FRT82B were obtained from the Bloomington Stock Center.
The genotypes of the MdnP stocks are w1118,hsFLP,UAS-lacZ; tub-Gal80,FRT40; slbo-Gal4,UAS-dnPVR/TM3,Ser (MdnP40), w1118,hsFLP,UAS-lacZ; FRT42,tub-Gal80; slbo-Gal4,UAS-dnPVR/TM3,Ser (MdnP42), w1118,hsFLP,UAS-lacZ; slbo-Gal4,UAS-dnPVR/CyO; tub-Gal80,FRT80 (MdnP80), and w1118,hsFLP,UAS-lacZ; slbo-Gal4,UAS-dnPVR/CyO; FRT82,tub-Gal80 (MdnP80).
We initially attempted to recover new PBac insertions based on GFP expression in late embryos/early larvae using a fluorescently activated embryo sorter, but had a very low frequency of positives. We assumed that GFP expression was inefficient as also reflected in the recovered lines (see Table 1) and instead used white expression to identify new insertions.
TABLE 1.
Summary of the selected lines
| PBac line | Insertion site | Gene | Molecular function | PBac orientation relative to gene | GFP expression | References (border cells) |
|---|---|---|---|---|---|---|
| PBac 79 | 2R:18871317 | virilizer | RNA binding | Sense, in intron | − | |
| PBac456 | 2L:6919366 | hrp48 | RNA binding | Sense, in 5′-UTR | + | |
| PBac1527 | 2L:6922912 | hrp48 | RNA binding | Antisense, in intron | − | |
| PBac104 | 2L:2178018 | anterior open/yan | Transcription factor | Sense, in intron | − | Schober et al. (2005) |
| PBac378 | 2L:2176647 | anterior open/yan | Transcription factor | Antisense, in promoter | − | Schober et al. (2005) |
| PBac911 | 2L:247422 | kismet | Elongation factor | Sense, in intron | − | |
| PBac5201 | 2L:242721 | kismet | Elongation factor | Antisense, in intron | − | |
| PBac1916 | 2R:4682442 | rme-8 | Chaperone | Sense, in intron | − | |
| PBac2148 | 3L:16013380 | DIAP1 | Ubiquitin–protein ligase | Sense, in intron | + | Geisbrecht and Montell (2004) |
| PBac3840 | 2L:9202838 | taiman | Transcription co-activator | Sense, in intron | − | Bai et al. (2000) |
| PBac4936 | 2L:9204904 | taiman | Transcription co-activator | Antisense, in intron | − | Bai et al. (2000) |
| PBac4624 | 2L:9206379 | taiman | Transcription co-activator | Antisense, in intron | − | Bai et al. (2000) |
| PBac4354 | 2R:16549897 | shg | Cell–cell adhesion | 8 kb downstream of shg | − | Niewiadomska et al. (1999) |
| PBac3929 | 3R:3933469 | puckered | Phosphatase | Antisense, in 5′-UTR | − | |
| PBac4251a | 3R:16954630 | slimb | Ubiquitin–protein ligase | Sense, in intron | − | Muzzopappaet al. (2005) |
The insertion site (second column) gives the chromosome arm location, and the first base from the left foot of the PiggyBac. The PiggyBac orientation relative to the gene (fifth column) is relative to that shown in Figure 1A.
Clones recovered only with adult heat shock.
All fly cultures were performed at 25°. Heat shocks were performed for 1 hr at 37°, either on larvae (L2–L3) or on adult females.
Molecular mapping of novel PBac insertions:
DNA was prepared using the DNAeasy kit (QIAGEN, Chatsworth, CA), and inverse PCR and DNA sequencing were performed as described in Hacker et al. (2003).
Generation of constructs:
PiggyBac vector:
A NheI site was cloned in place of the PstI site in pPBac{3xP3-EGFPaf} (Horn and Wimmer 2000). A linker containing the splice acceptor site, three stop codons, and the start site was cloned into NheI/NcoI (5′-GCTAGCCTTTCTCTTACAGGTCGAATTGATGTGATGCTCGAGCCCATGG-3′). A linker containing the splice donor site and the white 3′-end was cloned into AvrII/NgoMI (5′-CCTAGGCTTCGGGCCCGACGCAAGGAGTAGAAGGTAAGTAGCCGG-3′). Mini-white from pCasper3 (XbaI/ApaI) was then cloned into ApaI/AvrII.
1.3 LacZ:
Genomic DNA was amplified with the oligonucleotides 5′-CAAAGCATGCTTGCTAGCAAGAAAC-3′ and 5′-AGGCAAACTTCTAACTGAAAAGGTG-3′. The PCR product was cloned into pCR-TOPO (Invitrogen, San Diego) and subcloned at the EcoRI site into P{CaSpeR-AUG-betagal}.
X-gal staining:
To monitor lacZ expression, ovaries were dissected in PBS, fixed in 0.5% glutaraldehyde in PBS for 10 min, rinsed in PBS-0.1%Triton (PT), and stained in 10 mm NaH2PO4/Na2HPO4 (pH 7.2), 150 mm NaCl, 1 mm MgCl2, 3.1 mm K4(FeII(CN)6), 3.1 mm K3(FeIII(CN)6), 0.3% Triton X-100, and 0.2% X-Gal at room temperature.
Immunocytochemistry:
Ovaries were dissected in Grace's medium or PBS, fixed in 4% paraformaldehylde for 20 min at room temperature, rinsed in PBS+ 0.1% Triton X-100 (PT), blocked and incubated overnight with the primary antibody in PT + 5% normal goat serum, rinsed in PT, incubated 2 hr with secondary antibody in PT, rinsed in PT and PBS, and mounted for microscopy. Confocal images were collected sequentially on a Leica TCS-NT microscope.
Primary antibodies were rat anti-DE–cadherin (Oda et al. 1994), rabbit anti-β-galactosidase (Cappel), and rat anti-Slbo (P. Rorth, unpublished results). Fluorescent secondary antibodies were from Jackson Immunoresearch; rhodamin–phalloidin and DAPI were from Molecular Probes (Eugene, OR).
RESULTS
Mutagenesis and screening strategy:
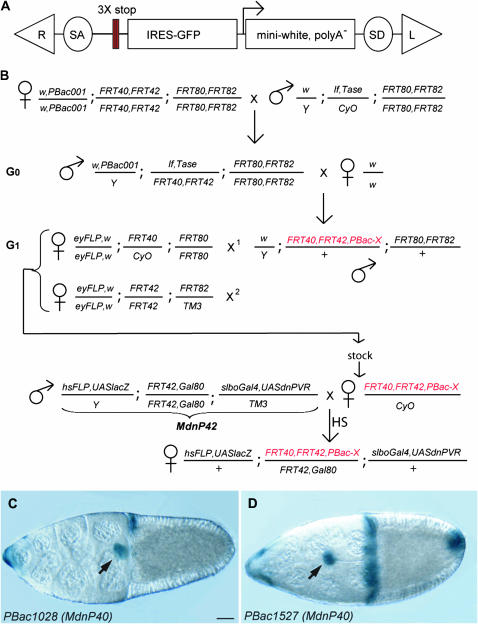
Flies were mutagenized by transposition of a PBac transposon modified to include a gene-trap cassette (Figure 1A). A splice acceptor and three stop signals were introduced to increase the chance of severely disrupting a gene upon insertion. They were followed by a modified version of an existing gene-trap P element (Lukacsovich et al. 2001), including internal ribosome entry site (IRES)-GFP and a mini-white gene with its own promoter but lacking a 3′-UTR and natural stop signal. Upon insertion of the transposon in a transcribed region, a GFP protein might be translated from the IRES, and the mRNA encoding the white gene product may be stabilized if there is an endogenous poly-adenylation signal downstream. Thus, the insertions giving GFP and/or white expression should have a high probability of being located in transcribed regions and of severely mutating the genes in which they are inserted. The starter line had a PBac insertion on the X chromosome (PBac001) and second and third chromosomes carrying FRTs on both sides of the centromere (Figure 1B). PBac001 was recovered due to its GFP expression, but was phenotypically w−. By mobilizing this PBac in w− mutant flies, we could therefore easily identify new insertions on the basis of the appearance of colored eyes (Figure 1B, chromosome bearing new PBac-X insertion in red). New PBac insertions were mapped to a chromosome arm by testcrosses with flies expressing FLPase in the eye primordium (eyFLP) and carrying specific FRT chromosomes, looking for mosaic eyes (Figure 1B). Temporary stocks were generated and used in several screens: a growth screen using the eyFLP system (Thompson et al. 2005, screen not described in article), a maternal effect screen using the ovoD system, and the border cell migration screen using the MdnP system described in detail below. Overall, this type of screen allows mutagenesis of all four major autosomal arms at once, direct clonal screens with the new insertions, and, if a phenotype is seen, rapid identification of the affected gene.
Figure 1.—
The PiggyBac screen. (A) The PiggyBac vector used as a mutagen. R and L are the right and left feet of the PBac transposon. SA and SD are the splice acceptor and splice donor. The three stop codons, one in each frame, are represented as red bars. (B) The crossing scheme. PBac001 indicates the jump starter. Tase indicates the piggyBac transposase. G0, generation of new insertion. G1, mapping. The new mutated chromosome, carrying a w+ insertion, is represented in red. FRT40,FRT42 is indicated as an example but it could also be FRT80,FRT82, as the crosses are identical. X1 and X2 are the two successive crosses done with single males, progeny of G0, to map the insertion to a chromosome arm. In the MdnP cross, virgins from the PBac stocks are crossed with MdnP males. The genotype of the MdnP42 male is shown as example for a PBac insertion distal to FRT42. Heat shock (HS) is applied to the progeny, either at larval stage or at adult stage. Females are dissected and ovaries are stained with X-Gal. An example of insertion with a border cell migration delay phenotype is shown in D; compare to an insertion with no phenotype (C). (C and D) X-gal-stained stage 10 egg chambers from females of the genotype hsFLP,UAS-lacZ/+;PBac1028,FRT40,FRT42/tub-Gal80,FRT40; slbo-Gal4,UASdnPVR/+ (C) and hsFLP,UAS-lacZ/+ ; PBac1527,FRT40,FRT42/tub-Gal80,FRT40;slbo-Gal4,UASdnPVR/+ (D). Arrow indicates the border cell cluster. Bar, 20 μm. Here and in all figure panels, anterior is to the left.
Overview of the PBac screen:
From a small pilot screen, we estimated that ∼20% (7 of 32) of new PBac insertions were lethal. This frequency was similar to that obtained in the generation of other PBac collections (Thibault et al. 2004; S. Thibault, personal communication). The lethality was primarily due to the PBac insertion since precise excision reverted the lethality in most cases (6 of 7 lines tested). The jump efficiency (defined as the percentage of crosses with the white positive insertion recovered using one male carrying the PBac starter and the PBac transposase and two virgins per cross) of our starter PBac was 21%. In the screen, we generated 5908 new jumps and recovered 3298 lines (56%) where the PBac could be clearly mapped to a chromosome arm. Partial recovery was expected due to a combination of death of the PBac carrier male before completion of mapping crosses and many “unmappable” insertions: insertions proximal to the FRTs, insertions on the fourth chromosome, and multiple insertions. With an expected lethal hit frequency of 20%, testing ∼3300 lines corresponded to ∼660 lethal insertions, not a saturating screen.
The border cell screen:
To identify genes important for border cell migration, all mapped PBac insertions were analyzed in homozygous mutant border cell clones using the MdnP system. With the MdnP system (Figure 1B, bottom), homozygous mutant border cells were marked with expression of lacZ, which allowed easy identification of mutant clones (Figure 1, C and D), even if they were rare. In addition, homozygous mutant cells specifically expressed a dominant-negative form of the guidance receptor PVR (dnPVR), which induces a weak phenotype on its own but was expected to sensitize border cell migration to further perturbations. In the initial screen where we recovered mutant clones after larval heat shock, 3182 of the lines tested had no detectable migration phenotype, 14 exhibited a migration phenotype (Table 1), and we could not recover any clones for 102 lines (3%). The genes in this last category could be important for cell viability or for functionality of the follicular stem cells and thus not allow recovery of mutant clones 7–10 days after clone induction as would be required after larval heat shock. We therefore generated clones with these PBac insertions by heat-shocking adult females and analyzing them 3–4 days afterward. This treatment revealed one more insertion (PBac4251) with a border cell migration phenotype and an additional 75 PBac insertions with no apparent effect in homozygous mutant border cells; for 26 PBac insertions, homozygous mutant clones could still not be obtained.
A summary of the 15 lines showing a border cell phenotype, the insertion site of the PBac, and the gene that they affect is given in Table 1. To complement the MdnP approach, all 15 lines were subsequently tested for border cell migration defects in standard clonal analysis without dnPVR expression, and 13 of 15 (all but PBac378 and PBac3929) displayed comparable phenotypes in such clones. PBac378 is an insertion in yan/aop and had a very weak phenotype in our screen. PBac104 also mutates yan/aop and, similarly to previously described alleles, it has a border cell migration phenotype on its own (Schober et al. 2005). Of the 15 lines, 5 additional lines had the PBac inserted in a gene previously found to affect border cell migration (PBac3840, PBac4624, and PBac4936 in taiman, PBac2148 in DIAP1, and PBac4251 in slimb), and two genes were represented by two independent insertions each (PBac456 and PBac1527 in hrp48 and PBac911 and PBac5201 in kismet). Further analysis of the two lines PBac4354 (a shg regulatory mutant) and PBac3929 (puckered) is described below. The two remaining lines were not investigated in more detail (PBac79 in virilizer, PBac1916 in rme-8).
A border cell enhancer for shg:
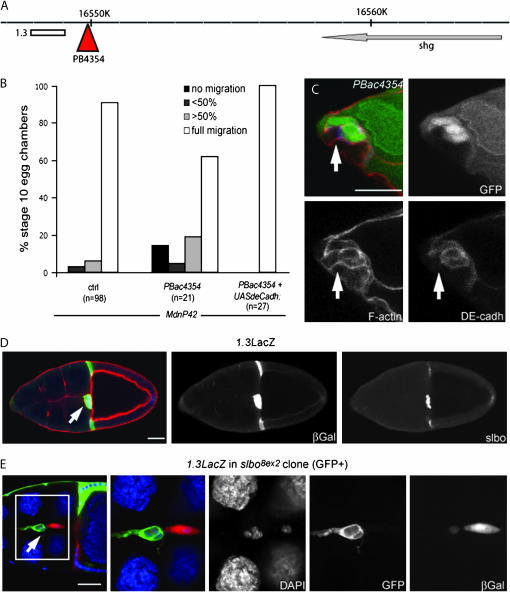
PBac4354 is a homozygous viable mutant in which the PBac is not inserted within a gene, but 8 kb downstream of the shg locus (Figure 2A). As shg, which encodes DE–cadherin, is required in border cells for their migration (Niewiadomska et al. 1999), we considered that PBac4354 might disrupt some regulatory sequences required for shg expression in border cells. In agreement with this idea, a clear reduction in DE–cadherin levels was observed in PBac4354 homozygous mutant border cells (Figure 2C). In addition, the migration phenotype could be rescued by expressing shg cDNA in PBac4354 mutant border cells (Figure 2B). To characterize this apparent regulatory mutant, we analyzed the genomic region surrounding the PBac4354 insertion site (Figure 2A). Within a region of ∼1.3 kb, sequence analysis revealed the presence of multiple consensus binding sites for the transcription factor Slbo, known to regulate shg mRNA level in border cells (Niewiadomska et al. 1999). This region also showed stretches of high sequence similarity to a D. pseudoobscura genomic region downstream of its shg ortholog, suggesting that it is a conserved enhancer (supplemental Figure 1 at http://www.genetics.org/supplemental/). We generated transgenic flies expressing lacZ under the control of this potential enhancer (1.3-lacZ). In stage 10 egg chambers, β-galactosidase expression was detected in the border cells and centripetal cells (Figure 2D, middle), similar to the expression pattern of Slbo (Figure 2D, right). In slbo mutant border cells, expression of the 1.3-lacZ transgene was severely reduced (Figure 2E, mutant cells marked by the presence of GFP). Thus we have identified a conserved, Slbo-responsive enhancer for the shg gene. The insertion of PBac4354 seems to disrupt the regulatory properties of the enhancer, perhaps by interfering with its ability to interact with the promoter of the shg gene. DE–cadherin is expressed in all cells of the follicular epithelium as well as in the germline, raising the question of why a border-cell-specific enhancer for shg would be functionally important. Specific transcriptional upregulation mediated by this enhancer may be advantageous for ensuring that the cells can retain strong DE–cadherin-dependent adhesion while having faster turnover of DE–cadherin protein during the dynamic process of migration.
Figure 2.—
PBac4354 identifies a shotgun/DEcadherin enhancer. (A) Scheme of the genomic region around PBac4354. The PBac (red triangle) is inserted 8 kb downstream from the 3′-end of the shg gene. The white bar corresponds to the fragment used to generate the 1.3LacZ construct. (B) Quantification of migration in control border cell clones, PBac4354 clones, and PBac4354 border cell clones expressing a shg cDNA generated with the MdnP system. The genotypes of the dissected females were hsFLP,UAS-LacZ/+ ; FRT40,FRT42/FRT42,tubGal80; slbo-Gal4,UAS-dnPVR/+ (control, left) hsFLP,UAS-LacZ/+ ; FRT40,FRT42, PBac4354/ FRT42,tubGal80; slbo-Gal4,UAS-dnPVR/+ (PBac4354, middle) and hsFLP,UAS-LacZ/UAS-shg; FRT40,FRT42, PBac4354/FRT42,tubGal80; slbo-Gal4,UAS-dnPVR/+ (PBac4354+ UAS-shg, right). (C) Stage 10 egg chamber from a female of the genotype hsFLP; FRT40,FRT42,PBac4354/FRT42,UbiGFP subjected to adult heat shock. Mutant cells are marked by the absence of GFP (top right and green in overlay). Phalloidin labels F-actin (bottom left and red in overlay) and an antibody labels DE–cadherin (bottom right and blue in overlay). The membrane staining (arrow) of DE–cadherin between two mutant border cells is strongly reduced. (D) Stage 10 egg chambers from female carrying the 1.3LacZ reporter, stained with phalloidin (red in overlay), anti-β-galactosidase (β-gal, middle, green in overlay), and anti-Slbo (right, blue in overlay). Left is the overlay. Arrow indicates the border cell cluster in D and E. (E) Stage 10 egg chambers from female with the 1.3LacZ reporter and slbo8ex2 mutant clones (GFP-positive cells, green in overlay, are homozygous mutant), stained with DAPI to show nuclei (blue in overlay) and with anti-β-galactosidase (right, red in overlay) to reveal the expression of 1.3LacZ. The region in the white box is enlarged in the right panels. The genotype is hsFLP,UAS-GFP/+; FRT42,slbo8ex2/FRT42,tubGal80; tubGal4/1.3lacZ. (C–E) Bar, 20 μm.
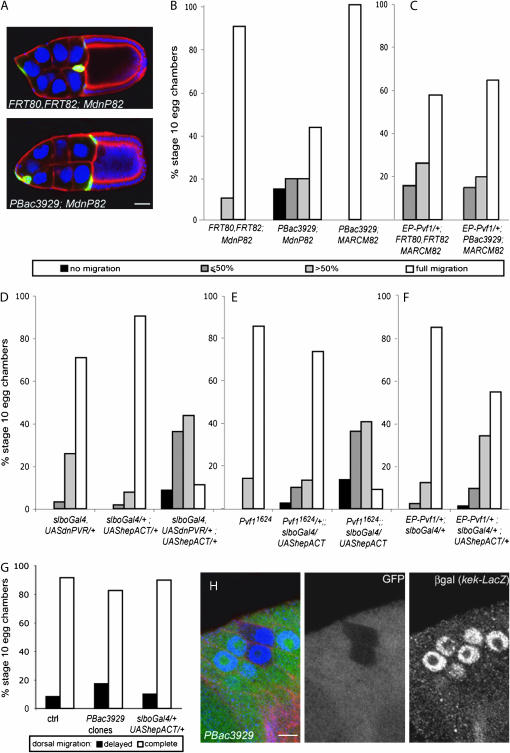
puckered is required in border cells with low PVR signaling:
PBac3929 was the only insertion that exhibited a prominent phenotype in the sensitized background of the screen but no detectable phenotype in standard clonal analysis without dominant-negative PVR (Figure 3, A and B). PBac3929 mutates puckered (puc), as confirmed by complementation test. puckered encodes a dual-specificity phosphatase that acts on the Drosophila Jun kinase (JNK) basket (bsk) and thereby negatively regulates the activity of the JNK signaling pathway (Martin-Blanco et al. 1998). Gain of function of JNK signaling can also be achieved by expressing a constitutively active form of hemipterous (hepACT), an activating kinase for bsk (Glise et al. 1995; Sluss et al. 1996; Weber et al. 2000). hepACT had no significant effect when expressed on its own in border cells (Figure 3D). However, like pucPBac3929 loss of function, hepACT had a strong effect on border cell migration when coexpressed with dnPVR (Figure 3D). The effect of hepACT was stronger than that of pucPBac3929 loss of function, possibly reflecting that pucPBac3929 is not a complete loss-of-function allele. The PVR receptor is activated in border cells by its ligand PVF1 (Duchek et al. 2001). As with expression of dnPVR in border cells, loss of function of Pvf1 had only mild effect on migration by itself, but seriously impaired border cell migration when combined with activated JNK signaling (Figure 3E). Thus elevated JNK signaling is deleterious for migration of border cells with reduced or absent PVR signaling.
Figure 3.—
puc is required in border cells with low PVR signaling. (A) Stage 10 egg chambers, stained with DAPI (blue) to show nuclei, anti-β-galactosidase antibody (green) to reveal mutant cells, and phalloidin (red) from females of the following genotypes: hsFLP,UAS-lacZ/+; slbo-Gal4,UAS-dnPVR/+; FRT80,FRT82/FRT82,tub-Gal80 (top) and hsFLP,UAS-lacZ/+; slbo-Gal4,UAS-dnPVR/+; FRT80,FRT82, pucPBac3929/FRT82,tubGal80 (bottom). Bar, 20 μm. (B–F) Quantification of border cell migration toward the oocyte (posterior migration) in stage 10 egg chambers. (B) pucPBac3929 clones with or without dnPVR and control MdnP clones from females of the genotypes hsFLP,UAS-lacZ/+; slbo-Gal4,UAS-dnPVR/+; FRT80,FRT82/FRT82,tub-Gal80 (left, n = 10); hsFLP,UAS-lacZ/+; slbo-Gal4,UAS-dnPVR/+; FRT80,FRT82, pucPBac3929/FRT82,tub-Gal80 (middle, n = 21); and hsFLP,UAS-GFP/+; tub-Gal4/+; FRT80,FRT82, pucPBac3929/FRT82,tub-Gal80 (right, n = 10). (C) Control border cells overexpressing Pvf1 (left; genotype hsFLP,UAS-GFP/EP-Pvf1; tubGal4/+; FRT80,FRT82/FRT82,tubGal80, n = 19) and pucPBac3929 clones overexpressing Pvf1 (right; genotype hsFLP,UAS-GFP/EP-Pvf1; tubGal4/+; FRT80,FRT82,pucPBac3929/FRT82,tubGal80; n = 20). (D) Border cell clusters expressing dnPVR, hepACT, or both (n = 93, 272, 80). (E) Clusters expressing hepACT in control, Pvf1/+, or Pvf1 mutant background (n = 42, 149, 22). (F) Clusters expressing uniform Pvf1, alone or with hepACT (n = 203, 73). Genotypes are indicated and include the controls in each experiment. (G) Quantification of dorsal migration in pucPBac3929 border cell clones (from hsFLP/+; FRT80,FRT82, pucPBac3929/FRT82,UbiGFP females, n = 23), border cells expressing hepACT (slbo-Gal4/+; UAS- hepACT/+, n = 20), and the control (slbo-gal4/+, n = 12). (H) Follicular epithelium of stage 10 egg chamber from female of the genotype hsFLP/+; kek-lacZ/+; FRT80,FRT82, pucPBac3929/FRT82,UbiGFP, stained with anti-β-galactosidase antibody (β-gal, blue in overlay) to reveal kek-lacZ reporter expression and phalloidin (red in overlay). pucPBac3929mutant cells are marked by absence of GFP (green in the overlay). Dorsal is to the top. Bar, 5 μm.
For posterior migration of border cells, guidance is mediated by the redundant activity of PVR and EGFR. Loss of activity of a single receptor does not severely affect border cell migration, while perturbing the activity of both at the same time disrupts border cell migration (Duchek et al. 2001). The observation that activated JNK signaling specifically affected border cell migration in the background of reduced PVR signaling suggested that it might disrupt the guidance activity of EGFR. EGFR signaling in the border cells is essential for their final, dorsal migration (Duchek and Rorth 2001). To test whether elevated JNK signaling affects EGFR guidance activity, we therefore analyzed dorsal migration of pucPBac3929 border cell clones and of hepACT-expressing border cells (Figure 3G). In both cases, border cell clusters did migrate dorsally. In addition, the classical MAPK/ERK signaling downstream of EGFR is not affected in pucPBac3929 clones in the dorsal follicular epithelium, as indicated by the unchanged level of expression of the kek-lacZ reporter (Figure 3H; Musacchio and Perrimon 1996; Ghiglione et al. 1999). These results suggested that pucPBac3929 mutant cells were not deficient in EGFR signaling.
Another feature of PVR and EGFR as guidance receptors is that uniformly activating them, for example, by mis-expressing a ligand throughout the tissue that the cells migrate through, also impairs migration. To further investigate whether the gain of JNK signaling interferes with the guidance of border cells, we analyzed the phenotype of pucPBac3929 mutant border cells (Figure 3C) and of border cells expressing hepACT (Figure 3F) in which PVR signaling is uniform due to overexpression of Pvf1. Excess of JNK signaling did slightly enhance the migration phenotype due to unlocalized PVR signaling but did not show the strong synergy seen with PVR loss of function. This contrasts with the effect of perturbing EGFR signaling, which strongly synergizes with misexpression of Pvf1 (Duchek et al. 2001). Overall, this indicated that excess JNK signaling was deleterious for border cell migration if PVR activity was low or absent, but excess JNK signaling still allowed EGFR to function as a guidance receptor.
JNK signaling activity in the border cells:
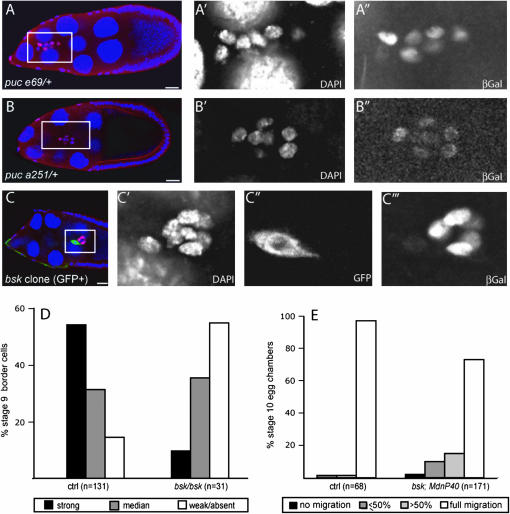
Puc is a negative regulator of JNK signaling, but is also a transcriptional target of the JNK pathway. For example, the expression of the pucE69 reporter, a lacZ insertion in the puc locus that reflects puc transcription in the dorsal epidermis of the embryo (Martin-Blanco et al. 1998), is lost in hep and bsk mutants (Glise et al. 1995; Riesgo-Escovar et al. 1996). This negative feedback loop is also present during thorax closure in the Drosophila pupae (Zeitlinger and Bohmann 1999) and in the follicular epithelium of the ovary at late stages (Suzanne et al. 2001). We asked whether such a feedback loop was present in border cells using pucE69 reporter as transcriptional readout.
We first examined the expression of pucE69 in a control egg chamber (puc E69/+) by antibody staining. We could detect some β-galactosidase staining in migrating border cells, but the expression level was not uniform among the cells (Figure 4A). The central cells, the anterior polar cells, generally had weak staining. For the migratory outer border cells, staining could be strong, intermediate, or weak and we did not detect any obvious correlation between the staining intensity and the position of the cell in the cluster. To confirm this observation, we analyzed the expression level of β-galactosidase in another puc reporter, pucA251 (Martin-Blanco et al. 1998). Although the overall expression level of the reporter was weaker than that of pucE69, we also saw some unequal distribution of the β-galactosidase staining in pucA251/+ border cells (Figure 4B). Analysis of pucE69 expression in many border cell clusters revealed that in a control situation most of the outer border cells have a strong or intermediate level of staining (Figure 4D). To determine if the expression of puc depends on JNK signaling in border cells, we analyzed the expression of pucE69 in bsk mutant border cells. Mutant cells showed a significant reduction in staining (Figure 4, C and D), indicating that puc expression is positively regulated by the JNK pathway in border cells as in other tissues.
Figure 4.—
JNK signaling in border cells. (A–C) Stage 9 egg chambers, stained with DAPI (blue nuclei in overlay) and anti-β-galactosidase antibody (β-gal, red in overlay) to reveal puc-lacZ reporters (pucE69 in A and C, pucA251 in B) in control flies (A and B) and bsk2 mutant border cell clone (C; mutant cells are GFP positive and green in overlay). Females were of the following genotypes: (A) pucE69/+, (B) pucA251/+, and (C) hsFLP,UAS-GFP; bsk2,FRT40/tubGal80; tubGal4/pucE69. Left panels show overlays, outlined box indicates the region enlarged in the panels to the right showing the indicated single channel images. Bar, 20 μm. (D) Quantification of β-galactosidase staining in control cells (ctrl) and bsk2 homozygous mutant border cells from female with genotype as in C. (E) Migration at stage 10 of bsk2 mutant and control border cells with the MdnP system from the following genotypes: hsFLP,UAS-lacZ/+; FRT40,FRT42/tubGal80,FRT40; slbo-Gal4,UAS-dnPVR/+ (ctrl) and hsFLP,UAS-lacZ/+; bsk2,FRT40/tubGal80,FRT40; slbo-Gal4,UAS-dnPVR/+.
The expression of the puc reporter per se, as well the altered expression in bsk clones, indicated that the JNK pathway was somewhat active in border cells. Although hep mutant border cells were described to migrate properly (Suzanne et al. 2001), we wondered whether JNK signaling would be required for border cell migration in the absence of PVR signaling. To address this question, we quantified the migration phenotype of bsk mutant clones generated with the MdnP system (Figure 4E). In this situation, some clusters were delayed, although the phenotype was less severe than that of pucPBac3929 with dnPVR. Thus the JNK pathway has some activity in border cells, and loss of JNK signaling affects migration if PVR signaling is also reduced. Overall, increased JNK signaling coupled with decreased/absent PVR signaling was quite deleterious for border cell migration (Figure 3, B, D, and E), whereas loss of function for both pathways (Figure 4E) or gain of function for both pathways (Figure 3, C and F) gave milder phenotypes without clear indication of synergy.
Fos and DIAP1 may mediate the puc phenotype in border cells:
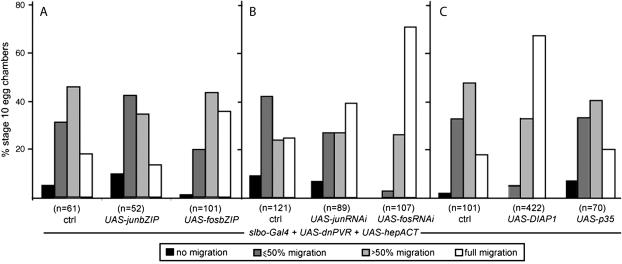
JNK is well known to stimulate the activity of the transcription factors Fos and Jun (in Drosophila encoded by kayak/fos and jra/jun, respectively; Peverali et al. 1996; Zeitlinger et al. 1997). But JNK has also been shown to bind and phosphorylate cytoskeletal regulators such as Spire (Otto et al. 2000). To test whether the effects observed upon overactivation of the JNK pathway were due to the transcriptional effects, we reduced the level of active Jun or Fos in border cells with elevated JNK signaling and reduced PVR. This was achieved by coexpressing hepACT and dnPVR with either dominant-negative Jun or Fos (Figure 5A) or with RNA interference (RNAi) directed against jun or fos (Figure 5B). Reducing Fos activity, by either approach, suppressed the border cell migration phenotype induced by elevated JNK signaling. Reducing Jun activity had a mild or no suppressive effect. This suggested that the Fos transcriptional activity stimulated by the JNK pathway contributes to the border cell phenotype.
Figure 5.—
Suppression of a high-JNK, low-PVR phenotype by decreasing Fos/Jun or increasing DIAP1 expression. (A–C) Quantification of migration at stage 10 of border cells expressing hepACT and dnPVR (ctrl in all experiments), as well as (A) dominant-negative versions of Jun (slbo-Gal4,UAS-dnPVR/+; UAS-hepACT/UAS-junbZIP) or Fos (slbo-Gal4,UAS-dnPVR/+; UAS-hepACT/UAS-fosbZIP), (B) expressing RNAi against jun (UAS-junRNAi/+; slbo-Gal4,UAS-dnPVR/UAS-hepACT) or fos (UAS-fosRNAi/+; slbo-Gal4,UAS-dnPVR/UAS-hepACT) in border cells, and (C) overexpressing DIAP1 (slbo-Gal4,UAS-dnPVR/+; UAS-hepACT/UAS-DIAP1) or p35 (slbo-Gal4,UAS-dnPVR/+; UAS-hepACT/UAS-p35) in border cells. The control flies in A and C had the genotype slbo-Gal4,UAS-dnPVR/+ UAS-hepACT/+ and, in B, slbo-Gal4,UAS-dnPVR/UAS-hepact. Controls were done in parallel with each experiment.
Although some studies have investigated the transcriptional changes resulting from a modulation of JNK signaling in embryos, larvae, or adult heads (Jasper et al. 2001; Etter et al. 2005), no information is available in border cells. Given that we observed a puc phenotype only in the dnPVR context, we reasoned that the targets of JNK responsible for the border cell phenotype might be regulated by RTK signaling in the opposite direction. One general role of JNK signaling is to promote apoptosis, while RTK signaling generally promotes cell survival. Indeed, the pro-apoptotic factor Hid was shown to be positively regulated by the JNK pathway (Moreno et al. 2002) and negatively regulated by MAPK signaling (Bergmann et al. 1998, 2002; Kurada and White 1998). Hid is a regulator of DIAP1, a ubiquitin ligase that inhibits caspase activity and thereby inhibits apoptosis. DIAP1 is also essential for border cell migration (Geisbrecht and Montell 2004) and an additional allele was recovered in this PBac screen. If the phenotype observed in border cells expressing hepACT and dnPVR was due to a misregulation of the hid/DIAP1 pathway, one might expect that it could be suppressed by increasing DIAP1 levels. We tested this hypothesis by overexpressing DIAP1 together with hepACT and dnPVR in border cells and found that this indeed improved their migration (Figure 5C). We next determined whether this effect of DIAP1 was due to a simple suppression of apoptosis by expressing the caspase inhibitor p35 instead. Expression of p35 together with hepACT and dnPVR did not show any suppressive effect (Figure 5C). Also, border cells expressing hepACT and dnPVR were not visibly apoptotic. These results indicate that the suppression observed with DIAP1 is specific and likely unrelated to its anti-apoptotic function.
DISCUSSION
Technical evaluation of the screen:
In this study, we presented a piggyBac insertional mutagenesis coupled with a modified version of the MARCM system, called MdnP, to perform a clonal screen for border cell migration phenotypes. Due to the expression of β-galactosidase in mutant clones, we could easily detect and score mutant clones under the dissecting microscope after X-Gal staining. This allowed us to detect clones even when they were rare, which was the case for PBac104 (aop), PBac3929 (puc), and PBac4251 (slmb). The choice of piggyBac as a mutagen in this study was dictated by the need to directly mutagenize FRT-bearing chromosomes for immediate generation of clones. Although it was reported that piggyBac does not share hotspots of integration with the P element (Hacker et al. 2003; Thibault et al. 2004), all the mutants that we identified and studied affected genes that had already been targeted by one or more P-element insertions. This reflects the high degree of saturation with P elements but also indicates that on a genome level the mutagenic insertion sites of P elements and PBac elements are perhaps not so different. The presence of PBac “hotspots” was indicated by the finding that of the 10 genes identified as mutating to a border cell migration phenotype, three genes were hit twice and one gene hit three times. This is significant, considering that it was very far from a saturating screen with 3300 lines tested and a projected 660 (20%) lethal insertions. In total, 15 of 3300 tested lines, or ∼0.5%, had border cell migration defects and 26, or 1%, were apparent cell lethals. In a clonal screen for border cell migration defects using EMS as mutagen (Liu and Montell 1999), 16 complementation groups were found on 2R, compared to three genes in our screen. From these considerations, it is likely that we have identified <20% of the genes that can mutate to give viable clones but border cell migration defects in this screen.
Among the mutants that we recovered in the border cell screen, most were located on the second chromosome. We think that this is an artificial bias introduced by the fact that the Slbo-Gal4, UAS-dnPVR combination located on the third chromosome had a weaker effect on border cell migration than its counterpart on the second chromosome. It was therefore easier to score migration defects above background when using the MdnP40 and MdnP42 stocks. For example, mutants affecting migration only slightly (such as PBac911, PBac5201/kis, and PBac79/vir) might have looked like the background effects with the stronger slbo-Gal4,UAS-dnPVR. In addition, expression of Gal80 from chromosome arm 3L was not strong enough to fully block the activity of Gal4. This led to leaky β-galactosidase expression in some nonmutant cells and thus imperfect identification of mutant cells such that weak phenotypes might have been missed.
Designing the screen such that mutant border cells would also express dnPVR was intended to sensitize the mutant border cells to further mild perturbations. However, expression of dnPVR did not appear to generally increase the number of mutants recovered, as most of the mutants recovered had a comparable phenotype when tested in standard clones without dnPVR. The approach did allow us to uncover an allele of puc, which would not have been identified otherwise. Finally, we noted that, where analyzed, most of the PBac mutants were apparent hypomorphs with weaker phenotypes than those published for strong alleles. This includes PBac2148 (DIAP1), PBac3929 (puc), PBac378, and PBac104 (yan/aop) as well as PBac4354, the shg regulatory mutant.
The RNA-binding proteins vir and hrp48:
Among the 10 genes identified in this screen as affecting border cell migration, 2 of them encode the RNA-binding proteins virilizer and hrp48. These two genes have been shown to be involved in the regulation of alternative splicing of Ubx mRNA (Burnette et al. 1999). We do not have any evidence that Ubx loss of function affects border cell migration, but vir and hrp48 could also cooperatively regulate the splicing of other targets with important functions in border cells. In addition, hrp48 has recently been shown to regulate mRNA localization/translation in the Drosophila oocyte (Huynh et al. 2004; Yano et al. 2004). It could play a similar role in border cell migration, regulating mRNA localization or translation. Localization of β-actin mRNA has been described in chicken embryo fibroblasts, where it appears to be important for cell motility (Kislauskis et al. 1997). RNAs encoding components of the Arp2/3 complex, a nucleator of actin assembly, have also been reported to be localized in these cells although through a different mechanism than the one responsible for β-actin mRNA localization (Mingle et al. 2005). The components of the Arp2/3 complex are required for normal border cell motility (K. Somogyi and P. Rorth, unpublished results). Thus it is possible that the mRNAs of actin and/or some of the Arp2/3 components need to be localized and regulated by Hrp48 in border cells for proper migration.
A convergence of JNK and PVR signaling in border cells:
pucPBac3929 exhibited a phenotype only in combination with dnPVR. Given the redundant activity of EGFR and PVR during border cell guidance, a simple explanation for the observed phenotype would be that loss of puc activity, and hence gain of JNK signaling, specifically blocked or perturbed EGFR signaling. Finding that pucPBac3929 mutant border cells migrated dorsally and that excess JNK signaling did not synergize with delocalized PVR signaling, we rejected this hypothesis. As we observed the most pronounced border cell migration defects in the context of reduced PVR signaling and excessive JNK signaling, it seemed likely that these two pathways had convergent but antagonistic effects on a protein or a process important for border cell motility.
In vertebrate cells and in Drosophila, convergence between the ERK and JNK MAPK pathways has been described at the level of the transcription factor AP-1, composed of a homo- or a hetero-dimer of Fos and Jun (Peverali et al. 1996; Kockel et al. 1997; Leppa et al. 1998; Ciapponi et al. 2001). In Drosophila, Fos was found to be phosphorylated on different sites by rl/ERK compared to bsk/JNK, and it was proposed to be in different activated states, depending on whether it was phosphorylated by ERK or JNK (Ciapponi et al. 2001). One might imagine an antagonism of the two pathways on the basis of competing activities acting on a fixed pool of Fos protein. In such a scenario, one role of PVR/ERK signaling would be to block potentially excessive JNK/Fos transcriptional activity. The suppression of the dnPVR and hepACT border cell phenotype by fos RNAi or dominant negative (fosbZIP) is in agreement with such a scenario. However, this is unlikely to be the explanation since strong enhancement of ERK signaling in border cells (overexpression of activated Raf) did not suppress the phenotype of dnPVR and hepACT (data not shown). Thus, the convergent but antagonistic effect of the pathways is more likely to happen downstream of Fos and Jun.
The suppression experiment suggests that DIAP1 may act as an important convergence point of PVR and JNK signaling in border cells. JNK signaling can downregulate the DIAP1 level whereas PVR signaling, through MAPK or other pathways, may increase the DIAP1 level or activity. Specifically, DIAP1 overexpression has previously been shown to suppress JNK-induced apoptosis (Igaki et al. 2002; Moreno et al. 2002). DIAP1 overexpression also suppressed the weak border cell migration phenotype induced by dnPVR (Geisbrecht and Montell 2004). In these two situations, DIAP1 acts as a negative regulator of the caspase DRONC. The function of DIAP1 in border cells is not related to apoptosis but rather to the control of actin assembly (Geisbrecht and Montell 2004). Such nonapoptotic function of caspases is not unique to border cells but was also observed in sperm individualization (Arama et al. 2003, 2006; Huh et al. 2004; Muro et al. 2006). In addition, DIAP1 levels and activity in relation to control of the actin cytoskeleton can be regulated by different kinases (Kuranaga et al. 2006; Oshima et al. 2006). Our genetic results suggest that the antagonism of JNK and RTK signaling on DIAP1 function is not only relevant in the context of apoptosis, but also in the regulation of the actin cytoskeleton.
Acknowledgments
We are grateful to Ernst Wimmer, Dirk Bohmann, Norbert Perrimon, Konrad Basler, Marek Mlodzik, Laurel Raftery, Steve Cohen, and the Bloomington Stock Center for flies, to Pascal Heitzler for help with the X-ray irradiation, to Ann-Marie Voie for embryo injections, and to Adam Cliffe for comments on the manuscript. J.M. was supported by the Human Frontier Science Program LT00443/2003.
References
- Arama, E., J. Agapite and H. Steller, 2003. Caspase activity and a specific cytochrome C are required for sperm differentiation in Drosophila. Dev. Cell 4: 687–697. [DOI] [PubMed] [Google Scholar]
- Arama, E., M. Bader, M. Srivastava, A. Bergmann and H. Steller, 2006. The two Drosophila cytochrome C proteins can function in both respiration and caspase activation. EMBO J. 25: 232–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai, J., Y. Uehara and D. J. Montell, 2000. Regulation of invasive cell behavior by taiman, a Drosophila protein related to AIB1, a steroid receptor coactivator amplified in breast cancer. Cell 103: 1047–1058. [DOI] [PubMed] [Google Scholar]
- Beccari, S., L. Teixeira and P. Rorth, 2002. The JAK/STAT pathway is required for border cell migration during Drosophila oogenesis. Mech. Dev. 111: 115–123. [DOI] [PubMed] [Google Scholar]
- Bergmann, A., J. Agapite, K. McCall and H. Steller, 1998. The Drosophila gene hid is a direct molecular target of Ras-dependent survival signaling. Cell 95: 331–341. [DOI] [PubMed] [Google Scholar]
- Bergmann, A., M. Tugentman, B. Z. Shilo and H. Steller, 2002. Regulation of cell number by MAPK-dependent control of apoptosis: a mechanism for trophic survival signaling. Dev. Cell 2: 159–170. [DOI] [PubMed] [Google Scholar]
- Borghese, L., G. Fletcher, J. Mathieu, A. Atzberger, W. C. Eades et al., 2006. Systematic analysis of the transcriptional switch inducing migration of border cells. Dev. Cell 10: 497–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnette, J. M., A. R. Hatton and A. J. Lopez, 1999. Trans-acting factors required for inclusion of regulated exons in the Ultrabithorax mRNAs of Drosophila melanogaster. Genetics 151: 1517–1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciapponi, L., D. B. Jackson, M. Mlodzik and D. Bohmann, 2001. Drosophila Fos mediates ERK and JNK signals via distinct phosphorylation sites. Genes Dev. 15: 1540–1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchek, P., and P. Rorth, 2001. Guidance of cell migration by EGF receptor signaling during Drosophila oogenesis. Science 291: 131–133. [DOI] [PubMed] [Google Scholar]
- Duchek, P., K. Somogyi, G. Jekely, S. Beccari and P. Rorth, 2001. Guidance of cell migration by the Drosophila PDGF/VEGF receptor. Cell 107: 17–26. [DOI] [PubMed] [Google Scholar]
- Etter, P. D., R. Narayanan, Z. Navratilova, C. Patel, D. Bohmann et al., 2005. Synaptic and genomic responses to JNK and AP-1 signaling in Drosophila neurons. BMC Neurosci. 6: 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisbrecht, E. R., and D. J. Montell, 2002. Myosin VI is required for E-cadherin-mediated border cell migration. Nat. Cell Biol. 4: 616–620. [DOI] [PubMed] [Google Scholar]
- Geisbrecht, E. R., and D. J. Montell, 2004. A role for Drosophila IAP1-mediated caspase inhibition in Rac-dependent cell migration. Cell 118: 111–125. [DOI] [PubMed] [Google Scholar]
- Ghiglione, C., K. L. Carraway, III, L. T. Amundadottir, R. E. Boswell, N. Perrimon et al., 1999. The transmembrane molecule kekkon 1 acts in a feedback loop to negatively regulate the activity of the Drosophila EGF receptor during oogenesis. Cell 96: 847–856. [DOI] [PubMed] [Google Scholar]
- Glise, B., H. Bourbon and S. Noselli, 1995. hemipterous encodes a novel Drosophila MAP kinase kinase, required for epithelial cell sheet movement. Cell 83: 451–461. [DOI] [PubMed] [Google Scholar]
- Hacker, U., S. Nystedt, M. P. Barmchi, C. Horn and E. A. Wimmer, 2003. piggyBac-based insertional mutagenesis in the presence of stably integrated P elements in Drosophila. Proc. Natl. Acad. Sci. USA 100: 7720–7725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn, C., and E. A. Wimmer, 2000. A versatile vector set for animal transgenesis. Dev. Genes Evol. 210: 630–637. [DOI] [PubMed] [Google Scholar]
- Huh, J. R., S. Y. Vernooy, H. Yu, N. Yan, Y. Shi et al., 2004. Multiple apoptotic caspase cascades are required in nonapoptotic roles for Drosophila spermatid individualization. PLoS Biol. 2: E15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh, J. R., T. P. Munro, K. Smith-Litiere, J. A. Lepesant and D. St Johnston, 2004. The Drosophila hnRNPA/B homolog, Hrp48, is specifically required for a distinct step in osk mRNA localization. Dev. Cell 6: 625–635. [DOI] [PubMed] [Google Scholar]
- Igaki, T., H. Kanda, Y. Yamamoto-Goto, H. Kanuka, E. Kuranaga et al., 2002. Eiger, a TNF superfamily ligand that triggers the Drosophila JNK pathway. EMBO J. 21: 3009–3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasper, H., V. Benes, C. Schwager, S. Sauer, S. Clauder-Munster et al., 2001. The genomic response of the Drosophila embryo to JNK signaling. Dev. Cell 1: 579–586. [DOI] [PubMed] [Google Scholar]
- Kislauskis, E. H., X. Zhu and R. H. Singer, 1997. beta-Actin messenger RNA localization and protein synthesis augment cell motility. J. Cell Biol. 136: 1263–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kockel, L., J. Zeitlinger, L. M. Staszewski, M. Mlodzik and D. Bohmann, 1997. Jun in Drosophila development: redundant and nonredundant functions and regulation by two MAPK signal transduction pathways. Genes Dev. 11: 1748–1758. [DOI] [PubMed] [Google Scholar]
- Kurada, P., and K. White, 1998. Ras promotes cell survival in Drosophila by downregulating hid expression. Cell 95: 319–329. [DOI] [PubMed] [Google Scholar]
- Kuranaga, E., H. Kanuka, A. Tonoki, K. Takemoto, T. Tomioka et al., 2006. Drosophila IKK-related kinase regulates nonapoptotic function of caspases via degradation of IAPs. Cell 126: 583–596. [DOI] [PubMed] [Google Scholar]
- Lee, T., and L. Luo, 2001. Mosaic analysis with a repressible cell marker (MARCM) for Drosophila neural development. Trends Neurosci. 24: 251–254. [DOI] [PubMed] [Google Scholar]
- Leppa, S., R. Saffrich, W. Ansorge and D. Bohmann, 1998. Differential regulation of c-Jun by ERK and JNK during PC12 cell differentiation. EMBO J. 17: 4404–4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y., and D. J. Montell, 1999. Identification of mutations that cause cell migration defects in mosaic clones. Development 126: 1869–1878. [DOI] [PubMed] [Google Scholar]
- Liu, Y., and D. J. Montell, 2001. Jing: a downstream target of slbo required for developmental control of border cell migration. Development 128: 321–330. [DOI] [PubMed] [Google Scholar]
- Lukacsovich, T., Z. Asztalos, W. Awano, K. Baba, S. Kondo et al., 2001. Dual-tagging gene trap of novel genes in Drosophila melanogaster. Genetics 157: 727–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Blanco, E., A. Gampel, J. Ring, K. Virdee, N. Kirov et al., 1998. puckered encodes a phosphatase that mediates a feedback loop regulating JNK activity during dorsal closure in Drosophila. Genes Dev. 12: 557–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mingle, L. A., N. N. Okuhama, J. Shi, R. H. Singer, J. Condeelis et al., 2005. Localization of all seven messenger RNAs for the actin-polymerization nucleator Arp2/3 complex in the protrusions of fibroblasts. J. Cell Sci. 118: 2425–2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montell, D. J., P. Rorth and A. C. Spradling, 1992. slow border cells, a locus required for a developmentally regulated cell migration during oogenesis, encodes Drosophila C/EBP. Cell 71: 51–62. [DOI] [PubMed] [Google Scholar]
- Moreno, E., M. Yan and K. Basler, 2002. Evolution of TNF signaling mechanisms: JNK-dependent apoptosis triggered by Eiger, the Drosophila homolog of the TNF superfamily. Curr. Biol. 12: 1263–1268. [DOI] [PubMed] [Google Scholar]
- Muro, I., D. L. Berry, J. R. Huh, C. H. Chen, H. Huang et al., 2006. The Drosophila caspase Ice is important for many apoptotic cell deaths and for spermatid individualization, a nonapoptotic process. Development 133: 3305–3315. [DOI] [PubMed] [Google Scholar]
- Musacchio, M., and N. Perrimon, 1996. The Drosophila kekkon genes: novel members of both the leucine-rich repeat and immunoglobulin superfamilies expressed in the CNS. Dev. Biol. 178: 63–76. [DOI] [PubMed] [Google Scholar]
- Niewiadomska, P., D. Godt and U. Tepass, 1999. DE-cadherin is required for intercellular motility during Drosophila oogenesis. J. Cell Biol. 144: 533–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda, H., T. Uemura, Y. Harada, Y. Iwai and M. Takeichi, 1994. A Drosophila homolog of cadherin associated with armadillo and essential for embryonic cell-cell adhesion. Dev. Biol. 165: 716–726. [DOI] [PubMed] [Google Scholar]
- Oshima, K., M. Takeda, E. Kuranaga, R. Ueda, T. Aigaki et al., 2006. IKK epsilon regulates F actin assembly and interacts with Drosophila IAP1 in cellular morphogenesis. Curr. Biol. 16: 1531–1537. [DOI] [PubMed] [Google Scholar]
- Otto, I. M., T. Raabe, U. E. Rennefahrt, P. Bork, U. R. Rapp et al., 2000. The p150-Spir protein provides a link between c-Jun N-terminal kinase function and actin reorganization. Curr. Biol. 10: 345–348. [DOI] [PubMed] [Google Scholar]
- Pacquelet, A., and P. Rorth, 2005. Regulatory mechanisms required for DE-cadherin function in cell migration and other types of adhesion. J. Cell Biol. 170: 803–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peverali, F. A., A. Isaksson, A. G. Papavassiliou, P. Plastina, L. M. Staszewski et al., 1996. Phosphorylation of Drosophila Jun by the MAP kinase rolled regulates photoreceptor differentiation. EMBO J. 15: 3943–3950. [PMC free article] [PubMed] [Google Scholar]
- Pinheiro, E. M., and D. J. Montell, 2004. Requirement for Par-6 and Bazooka in Drosophila border cell migration. Development 131: 5243–5251. [DOI] [PubMed] [Google Scholar]
- Riesgo-Escovar, J. R., M. Jenni, A. Fritz and E. Hafen, 1996. The Drosophila Jun-N-terminal kinase is required for cell morphogenesis but not for DJun-dependent cell fate specification in the eye. Genes Dev. 10: 2759–2768. [DOI] [PubMed] [Google Scholar]
- Ring, J. M., and A. Martinez Arias, 1993. puckered, a gene involved in position-specific cell differentiation in the dorsal epidermis of the Drosophila larva. Dev. Suppl.: 251–259. [PubMed]
- Rorth, P., K. Szabo, A. Bailey, T. Laverty, J. Rehm et al., 1998. Systematic gain-of-function genetics in Drosophila. Development 125: 1049–1057. [DOI] [PubMed] [Google Scholar]
- Schober, M., I. Rebay and N. Perrimon, 2005. Function of the ETS transcription factor Yan in border cell migration. Development 132: 3493–3504. [DOI] [PubMed] [Google Scholar]
- Silver, D. L., and D. J. Montell, 2001. Paracrine signaling through the JAK/STAT pathway activates invasive behavior of ovarian epithelial cells in Drosophila. Cell 107: 831–841. [DOI] [PubMed] [Google Scholar]
- Sluss, H. K., Z. Han, T. Barrett, R. J. Davis and Y. T. Ip, 1996. A JNK signal transduction pathway that mediates morphogenesis and an immune response in Drosophila. Genes Dev. 10: 2745–2758. [DOI] [PubMed] [Google Scholar]
- Suzanne, M., N. Perrimon and S. Noselli, 2001. The Drosophila JNK pathway controls the morphogenesis of the egg dorsal appendages and micropyle. Dev. Biol. 237: 282–294. [DOI] [PubMed] [Google Scholar]
- Thibault, S. T., M. A. Singer, W. Y. Miyazaki, B. Milash, N. A. Dompe et al., 2004. A complementary transposon tool kit for Drosophila melanogaster using P and piggyBac. Nat. Genet. 36: 283–287. [DOI] [PubMed] [Google Scholar]
- Thompson, B. J., J. Mathieu, H. H. Sung, E. Loeser, P. Rorth et al., 2005. Tumor suppressor properties of the ESCRT-II complex component Vps25 in Drosophila. Dev. Cell 9: 711–720. [DOI] [PubMed] [Google Scholar]
- Vegh, M., and K. Basler, 2003. A genetic screen for hedgehog targets involved in the maintenance of the Drosophila anteroposterior compartment boundary. Genetics 163: 1427–1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, S. L., C. J. Hawkins, S. J. Yoo, H. A. Muller and B. A. Hay, 1999. The Drosophila caspase inhibitor DIAP1 is essential for cell survival and is negatively regulated by HID. Cell 98: 453–463. [DOI] [PubMed] [Google Scholar]
- Wang, X., J. Bo, T. Bridges, K. D. Dugan, T. C. Pan et al., 2006. Analysis of cell migration using whole-genome expression profiling of migratory cells in the Drosophila ovary. Dev. Cell 10: 483–495. [DOI] [PubMed] [Google Scholar]
- Weber, U., N. Paricio and M. Mlodzik, 2000. Jun mediates Frizzled-induced R3/R4 cell fate distinction and planar polarity determination in the Drosophila eye. Development 127: 3619–3629. [DOI] [PubMed] [Google Scholar]
- Yano, T., S. Lopez de Quinto, Y. Matsui, A. Shevchenko and A. Ephrussi, 2004. Hrp48, a Drosophila hnRNPA/B homolog, binds and regulates translation of oskar mRNA. Dev. Cell 6: 637–648. [DOI] [PubMed] [Google Scholar]
- Zeitlinger, J., and D. Bohmann, 1999. Thorax closure in Drosophila: involvement of Fos and the JNK pathway. Development 126: 3947–3956. [DOI] [PubMed] [Google Scholar]
- Zeitlinger, J., L. Kockel, F. A. Peverali, D. B. Jackson, M. Mlodzik et al., 1997. Defective dorsal closure and loss of epidermal decapentaplegic expression in Drosophila fos mutants. EMBO J. 16: 7393–7401. [DOI] [PMC free article] [PubMed] [Google Scholar]