Abstract
In mammals, male fertility is a quantitative feature determined by numerous genes. Until now, several wide chromosomal regions involved in fertility have been defined by genetic mapping approaches; unfortunately, the underlying genes are very difficult to identify. Here, 53 interspecific recombinant congenic mouse strains (IRCSs) bearing 1–2% SEG/Pas (Mus spretus) genomic fragments disseminated in a C57Bl/6J (Mus domesticus) background were used to systematically analyze male fertility parameters. One of the most prominent advantages of this model is the possibility of analyzing stable phenotypes in living animals. Here, we demonstrate the possibility in one-step fine mapping for several fertility traits. Focusing on strains harboring a unique spretus fragment, we could unambiguously localize two testis and one prostate weight-regulating QTL (Ltw1, Ltw2, and Lpw1), four QTL controlling the sperm nucleus shape (Sh1, Sh2, Sh3, and Sh4), and one QTL influencing sperm survival (Dss1). In several cases, the spretus DNA fragment was small enough to propose sound candidates. For instance, Spata1, Capza, and Tuba7 are very strong candidates for influencing the shape of the sperm head. Identifying new genes implied in mammalian fertility pathways is a necessary prerequisite for clarifying their molecular grounds and for proposing diagnostic tools for masculine infertilities.
IN humans, infertility is defined as an inability to deliver a child within 2 years of unprotected intercourse. It is estimated that from 10 to 15% of couples consult at least once for infertility problems (Feng 2003). Fertility appears as a quantitative feature of complex genetic determinism, since numerous genes act at different levels to define the fertility potential of an individual, as shown by gene invalidation (knockout) studies in mice. For the male side of the picture, according to human clinical data, sperm count, viability, motility, and morphology are highly variable even between fertile individuals, and most infertile males have one or several of these parameters below the normal thresholds. The biological explanation of these variations is not completely elucidated. Indeed, the molecular basis of male infertility is determined in only ∼10% of the cases; these patients are essentially azoospermic or severely oligospermic, owing to microdeletions in the AZF region (the major cluster of genes influencing sperm count; for review, see Affara and Mitchell 2000) at Yq11 (Foresta et al. 2001). Genes governing sperm shape (Mendoza-Lujambio et al. 2002; Nakamura et al. 2004), acrosome formation (Yao et al. 2002), or sperm movement (Pilder et al. 1997; Neesen et al. 2001; Carlson et al. 2003) have also been shown to cause infertility in mouse-gene-targeting models.
Identifying the genes underlying infertility represents a difficult challenge in humans, since the genetic analysis stumbles over the obvious problem of building informative families. Therefore, the use of animal models has a great potential for improving our understanding of mammalian infertility. In particular, mouse models made it possible to elucidate portions of the underlying genetics and have been of great help in identifying genes putatively involved in human infertility (Matzuk and Lamb 2002). In mice, numerous laboratory strains have been created, originating mainly from a small pool of ancestors belonging to the Mus musculus species. For this reason, the different strains are genetically highly similar, an issue that sometimes constitutes a serious problem for addressing the genetic bases of complex phenotypes, due to the lack of informative markers. This lack of genetic polymorphisms can be overcome by crossing laboratory mice (M. musculus) with mice belonging to other species or subspecies of wild mice, such as Mus spretus (Avner 1998; Guenet and Bonhomme. 2003). Classical QTL analysis on a backcross population established from an original interspecific cross made it possible to locate loci involved in hybrid sterility, in meiosis control, and/or in spermatogenesis (Elliott et al. 2004).
In this study, we used an original panel of 53 mouse interspecific recombinant congenic strains (IRCSs) established at the Pasteur Institute. These lines were established by introgressing chromosomal segments of M. spretus origin into the C57BL/6J background (Churchill et al. 2004). A medium-/high-density map of the genome of the whole IRCS set has been recently established by genotyping 673 microsatellites and SNPs. This mouse genetic tool is particularly useful for mapping multi-genetic traits since deviations in phenotype between the recombinant strain and the C57BL/6 parental strain should be due to the spretus fragment(s), accounting on average for <2% of the genomic content of each strain. This tool was used to map and characterize phenotypic traits correlated with male fertility. Our IRCS mice, albeit hypo-fertile compared to the parental strains, are stabilized and constitute a permanent phenotypic and genotypic resource, clearly complementary to other QTL mapping designs. Here, we could identify significant phenotypic differences for most parameters measured and map some of them to chromosomal regions of several megabases only, making it possible to propose strong candidate genes in only one mapping step.
This study complements the analysis carried out by Oka and co-workers that made it possible to map QTL affecting sperm morphology and testis weight on the X chromosome, which is not covered in our set of strains (Oka et al. 2004), and on chromosomes 1 and 11 in interaction with the X chromosome (Oka et al. 2007). spretus segments from the X chromosome have been eliminated in our IRCS, presumably due to the presence of hybrid sterility genes on this chromosome. We believe that such genomic analyses will be the prelude to identifying relevant genes and will participate in clarifying the complex pathways leading to normal fertility in mammalian species.
MATERIALS AND METHODS
Animals:
The parental strains were M. spretus (SEG/Pas strain: SEG) and Mus musculus domesticus (C57BL/6J strain: B6). To construct the IRCSs, F1 females (B6 × SEG) were crossed with B6 males. Fertile backcross males were mated with B6 females and their progeny were brother–sister mated for over 20 generations to produce inbred strains. At the time of the study, 43 of the 53 strains investigated had >40 generations of inbreeding. The animals were bred in Pasteur's animal facility until weaning and then housed in a controlled environment (light/dark cycle, temperature, free access to mouse food and water) in the animal facility of the Cochin Institute. All the strains were sampled on several litters in homogenized environmental conditions; the observed phenotypes were highly stable through litters and generations.
All the experimental procedures were conducted in accordance with the policies of the University and the Guidelines for Biomedical Research Involving Animals.
Genotyping and improvement of the map resolution of the different strains:
In a preliminary map version, 180 microsatellites were genotyped on the 53 mouse strains, showing that, on the whole, the spretus fragments disseminated in the set of IRCSs represented ∼40% of the mouse genome. Genotyping of 1536 additional SNPs was carried out at the Centre National de Génotypage (Evry, France) using an Illumina platform. A total of 673 (∼44%) were found informative between SEG and B6. The map can be found at http://www.pasteur.fr/. Details about the construction of the new map version will be given elsewhere (G. Burgio, M. Szatanik, J. L. Guenet, M. R. Arnau, J. J. Panthier and X. Montagutelli, unpublished results). With such a map density, the existence of undetected spretus fragments is improbable. This has been checked through simulation by counting the increase of newly discovered fragment numbers, when the coverage is extended from 180 to 700 markers. For convenience, strains presenting a unique spretus fragment will later be referred to as “congenic” and strains presenting several spretus fragments will later be referred to as “pluricongenic.”
Morphological and histological analysis of the reproductive organs:
Male mice (8–9 weeks of age) never exposed to females were used for phenotyping reproductive traits. The male mice were killed by cervical dislocation, and reproductive organs (testes, epididymis, seminal vesicles, and prostates) were dissected. The organs were weighted and divided into lots that were either immersed in DF2 fixative (35% absolute alcohol, 10% acetic acid, 2% formaldehyde, in distilled water) for 24 hr at room temperature for histological analysis or deep frozen at −80° for further protein and RNA analysis.
Fixed organs were dehydrated and embedded in paraffin. Sections (4 μm) were cut, cleared by incubation in alcohol, rehydrated, and stained by hematoxylin/eosin/safran. Histology was carried out by microscopic examination of organs from a minimum of two animals for each strain. For strains presenting a testis weight significantly different from B6's, we performed a quantitative analysis of testis histology by measuring diameters of transversal sections of seminiferous tubules at stage VII–VIII under a ×20 objective using an Eclipse 600 Nikon microscope.
Epididymal sperm count:
We counted the number of spermatozoa present in the total length of the epididymis. To achieve this, the epididymis previously stored at −80° was thawed at room temperature and homogenized in 5 ml of physiological serum (9 g/liter NaCl) supplemented with 0.1% Triton X-100, with an ultra-turax for three rounds of 3 min. The sperm number in the suspension was determined by counting sperm heads under a microscope, using a Thoma cell chamber.
Sperm processing and examination:
The cauda epididymis and vas deferens were placed in 1 ml M2 medium (M7167, Sigma-Aldrich) in a petri dish and immediately punctured to allow the spermatozoa to swim out. After 15 min of incubation time at 37°, the dispersed sperm cells were recovered, diluted twice in modified M16 medium (M7292, Sigma-Aldrich) containing 50 mm sodium bicarbonate and 6% BSA (A2153, Sigma-Aldrich), and incubated for capacitation at 37°. After 1 hr incubation, sperm samples were processed for acrosome reaction (AR) induction experiments.
Sperm viability:
This parameter was assessed on sperm cells recovered 15 min after puncturing the epididymis (early viability) and later, during the evaluation of the mouse sperm acrosomal status (late viability: 2 hr after the sperm was released), using ethidium homodimer (EH; Sigma-Aldrich) as vital staining (Emiliozzi et al. 1996). A stock solution of EH was prepared at a concentration of 2 mm in DMSO and aliquots were stored at −20° in the dark. Before use, the stock solution was diluted 50 times in DMSO and added to the sperm suspension at a final concentration of 0.4 μm. After 1-min incubation with EH at room temperature, the sperm samples were centrifuged at 600 g and the pellet resuspended in PBS plus 0.1% formaldehyde until analysis with a flow cytometer (Cytomics FC500).
Acrosome reaction induction and acrosomal status evaluation:
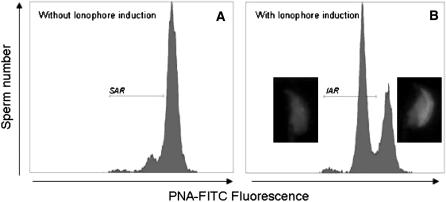
To investigate the ability of spermatozoa to undergo an acrosome reaction, capacitated spermatozoa (200 μl) were incubated for 1 hr in M16 medium in the presence of calcium ionophore (A 23187, Sigma-Aldrich) dissolved in DMSO at a final concentration of 20 μm or in the presence of DMSO only. The acrosomal status (presence or absence of acrosomal matrix corresponding to intact or acrosome-reacted spermatozoa) was determined on viable sperm cells according to the procedure of Tao et al. (1993) using the fluorescein isothiocyanate-conjugated peanut agglutinin (FITC-PNA L7381, Sigma-Aldrich). EH was used in parallel as a vital staining. Briefly, at the end of the incubation with the calcium ionophore or DMSO control, 0.4 μm EH was added to the sperm suspension before centrifugation at 600 × g and then the sperm pellet was washed with PBS supplemented with 25 μg/ml salmon sperm DNA (SSDNA; Sigma-Aldrich). Sperm membranes were permeabilized with a solution of 40 mm digitonin (D1407, Sigma-Aldrich) in PBS–SSDNA for 5 min at 37° and centrifuged. The pellet was suspended in PSB–SSDNA and stained by 10 μg/ml FITC–PNA 30 min before fluorescence measurement with a flow cytometer (Cytomics FC500). A total of 10,000 living sperm were monitored in each sample. The frequency of acrosome-reacted spermatozoa (FITC–PNA negative) in the control incubation corresponds to the spontaneous acrosome reaction (SAR in Figure 7A), and the frequency of acrosome-reacted spermatozoa in the ionophore incubation corresponds to the induced acrosome reaction (IAR in Figure 7B). IAR minus SAR corresponds to the specifically ionophore induced-acrosome reaction (IIAR).
Figure 7.—
Flow cytometer analysis of acrosomal sperm status. (A) Flow cytometer acquisition in FITC spectra of non-ionophore-treated sperm: a population of spontaneous acrosome-reacted sperm exists under the SAR marker. (B) Flow cytometer acquisition in FITC spectra of ionophore-treated sperm: a population of induced acrosome-reacted sperm appears under the IAR marker.
Sperm morphology:
A drop of 20 μl of sperm suspension was air dried on a slide and then the nuclei and the acrosomal matrix were labeled with EH (1 μm in PBS) and FITC–PNA (20 μg/ml in PBS), respectively. The sperm morphology was examined in a minimum of 300 cells/mouse with a ×100 objective, using the epifluorescence microscope.
Statistical analysis:
Identification of statistically significant differences between B6 and the IRCS:
Statistical analyses were performed with SPSS (version 8.0.1 for Windows, SPSS). Normality of data was verified using the SPSS explore function. Comparisons between IRCS and parental B6 were conducted by analysis of variance with the post-hoc Dunnett's test (α = 5%). The post-hoc Dunnett's test was performed to take into account the biases induced by multiple testing.
QTL mapping:
Phenotypic effects detected in one given IRCS can in general be assigned to the different spretus fragments present in the genome of this strain. Therefore, combining phenotype information from various IRCSs can theoretically be used to evaluate the effect of a given fragment and to eventually map the gene responsible for this effect (fragment-per-fragment analysis). However, since this analysis could be biased by epistatic interactions, we decided to focus our analysis mainly on phenotypes occurring in IRCSs possessing a unique spretus fragment. Whenever possible, strains with similar phenotype characteristics and sharing overlapping chromosome segments were compared to refine QTL location.
Potential candidate genes encoded in the region of the mapped QTL were identified using the Mouse Genome Database at http://www.informatics.jax.org. Patterns of expression of these genes were obtained using the GNF SymAtlas v1.2.4 database of the Genomics Institute of the Novartis Research Foundation at http://symatlas.gnf.org/SymAtlas (Su et al. 2002, 2004).
RESULTS
Various features of the male genital tract as well as functional and morphological characteristics of the sperm were measured in 428 individuals belonging to the 53 strains. The total body weight of the 9-week-old male mice analyzed was 20.3 ± 2.7 g for SEG and 23.7 ± 2.5 g for B6 and ranged from 20.1 ± 0.7 g (strain 144E) to 28.4 ± 1.8 (strain 137B) in the IRCS. To take into account these weight differences, the organ weights were calculated relative to body weights for statistical analysis.
Testis parameters:
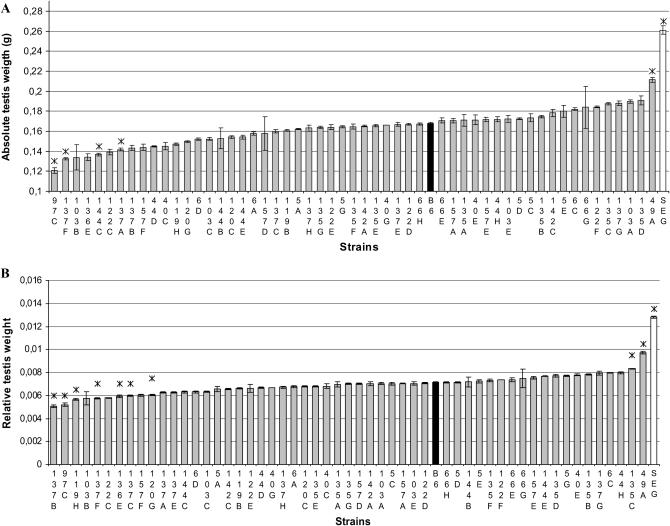
Both testes appeared comparable upon dissection, so the mass of the two testes was considered as the whole testis weight. Figure 1 shows a histogram of testis weights for each IRCS compared to B6. IRCS testes weights ranged from 0.121 ± 0.013 g (97C strain) to 0.211 ± 0.027 g (49A strain), representing differences ranging from −28 to +26% compared to the testis weight of B6 (0.168 ± 0.021 g). The largest testis weight of IRCS remained inferior to the parental SEG testis weight (0.261 ± 0.043 g). For the lines with a testis weight significantly different from B6, we conducted a histological analysis of this organ. No difference from the B6 parent was found at the interstitial testicular tissue level.
Figure 1.—
Testis weight of parental and IRCS mice. The mean values (±SEM) of absolute (A) and relative (B) testis weight are presented in order of increasing values, with mean B6 and SEG testis weights shown in solid and open bars, respectively. Asterisks (*) denote strains that showed a significant difference in mean values compared to the B6 strain (P < 0.05).
QTL positively affecting testicular weight:
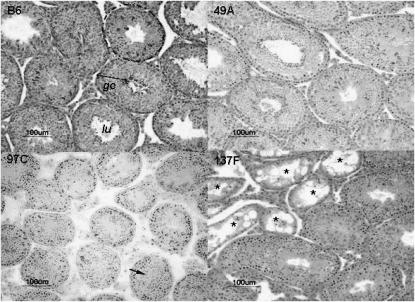
Seminiferous tubule diameter was significantly higher in 49A than in B6 (Figure 2 and Table 1), and the germinative epithelium appeared denser. These quantitative observations were consistent with an epididymal sperm count significantly higher in 49A compared to B6, corresponding to a 42% increase of testicular production. It is clear that at least one QTL of increased sperm production segregates between 49A and B6, this or these QTL accounting for ∼50% of the B6–SEG difference for absolute testis weight. This or these QTL map in the spretus fragments of the 49A genome on MMU4, MMU15, or MMU16.
Figure 2.—
Testis histology of three IRCS and B6 strains. (Top left) Seminiferous tubule sections (dotted lines) with normal spermatogenesis of B6 parental strain. These seminiferous tubules are used as a reference in the histological analysis of the IRCS (lu, lumen; ge, germinative epithelium). (Top right) Testis sections of the 49A strain, the IRCS with the heavier testes compared to B6; note the normal spermatogenesis and high cellular density in the seminiferous tubules, which have a diameter significantly superior to B6's diameter. (Bottom left) Testis section of the 97C strain. Most of the seminiferous tubules display an absence of lumen, but mature spermatozoa are detected (arrow); tubule diameters were significantly inferior to B6 ones. (Bottom right) Testis section of the 137F strain. Some tubules presented a SCO syndrome (asterisks); the remaining tubules display a normal spermatogenesis. Bar, 100 μm.
TABLE 1.
Phenotype–genotype relation for testis-related traits in IRC strains
| Quantitative traits
|
spretus segments
|
||||||
|---|---|---|---|---|---|---|---|
| Strains | n | % of B6's absolute testis weight | Qualitative traits: Testis histology | Tubule diameter (μm) ±SEM and P-value | Sperm count × 106 ±SEM (% of B6's value) and P-value | Chromosome | Localization (bp) |
| B6 | 19 | Control | 202 ± 13 | 14.4 ± 0.4 | |||
| 49A | 10 | 26 | Dense and well-organized germinative epithelium | 216 ± 25, P = 0.0429 | 20.4 ± 0.5 (42), P = 0.0005 | 4 | 18,946,683–44,005,346 |
| 15 | 30,205,799–60,861,596 | ||||||
| 16 | 57,560,000–98,252,459 | ||||||
| 135C | 13 | 16 | Normal | 186 ± 15, P = 0.001 | 19.5 ± 0.2 (35), P = 0.0066 | 6 | 38,991,008–55,936,298 |
| 18 | 0–31,055,551 | ||||||
| 120G | 12 | −11 | Partial SCO | 196 ± 16 (NS) | 14.6 ± 0.2 (1.5) (NS) | 1 | 21,009,497–54,610,000 |
| 54,610,000–65,340,594 | |||||||
| 5 | 26,457,520–31,750,835 | ||||||
| 6 | 0–15,366,869 | ||||||
| 14 | 78,826,456–107,526,001 | ||||||
| 119H | 12 | −12 | Normal | 110 ± 9, P < 0.0001 | 22.8 ± 1.8 (58) P = 0.049 | 2 | 134,728,264–164,190,990 |
| 13 | 0–13,423,656 | ||||||
| 137B | 11 | −14 | Normal | 157 ± 17, P < 0.0001 | 16.1 ± 0.5 (12) (NS) | 6 | 133,269,077–149,525,685 (Ltw2) |
| 137A | 11 | −15.5 | Normal | 182 ± 12, P < 0.0001 | 15.2 ± 0.3 (6) (NS) | 6 | 133,269,077–149,525,685 |
| 7 | 25,872,301–35,582,876 | ||||||
| 35,582,876–60,165,513 | |||||||
| 18 | 46,781,237–69,139,024 | ||||||
| 19 | 55,219,249–61,321,190 | ||||||
| 144C | 16 | −18.5 | Normal | 188 ± 24, P < 0.0001 | 11.2 ± 0.3 (−22) (NS) | 15 | 9,280,213–38,655,639 |
| 19 | 55,219,249–61,321,190 | ||||||
| 137F | 17 | −21 | Partial SCO | 167 ± 15, P < 0.0001 | 13.4 ± 0.3 (−14) (NS) | 1 | 41,416,802–73,177,650 |
| 6 | 133,269,077–146,678,290 | ||||||
| 19 | 16,994,293–20,262,739 | ||||||
| 97C | 6 | −28 | Undetected seminiferous tubule lumen | 157 ± 14, P < 0.0001 | 16.3 ± 0.7 (13) (NS) | 11 | 3,525,585–26,517,138 (Ltw1) |
n, number of analyzed mice.
Mice of the 135C strain exhibited a significant increase of relative testis weight (+16%, P = 6.7 × 10−4) associated with an increase of 35% of epididymal sperm count (Table 1). The histology of 135C testes appeared normal, with tubule diameters being slightly reduced compared to B6 and with an efficient spermatogenesis. The 135C genome carries two spretus fragments on MMU6 and MMU18, which should therefore contain QTL influencing sperm production. These QTL explain ∼20% of the B6–SEG difference for testis weight.
QTL negatively affecting testicular weight:
We identified seven IRCSs with testis weight significantly reduced compared to B6, suggesting that SEG carries alleles that negatively influence testis weight in a B6 background. This type of phenotypic deviation outside of the B6–SEG range of testis weights involves a negative epitasis between spretus fragments and domesticus genetic background for these seven IRCSs (Figure 1). The 97C line exhibited a significant reduction of absolute testis weight compared to B6 (−28%, P = 8.4 × 10−5, Table 1). 97C mice presented reduced seminiferous tubule diameters without visible lumen in most of these tubules (Figure 2). However, a normal spermatogenesis took place in this strain, as indicated by the presence of mature spermatozoa in the tubules (arrow in Figure 2). This observation was consistent with a normal epididymal sperm count. The 97C genome carries only one spretus fragment, making it possible to localize directly a QTL for low testis weight (Ltw1) on MMU11 in a 23-Mb fragment containing 141 genes. Mice of the 137B strain also present small testes (−14%, compared to B6, P = 5 × 10−5). Their genome contains a unique 16-Mb spretus fragment located on MMU6, making it possible to map another QTL of low testis weight (Ltw2, Table 1). In this strain, the low testis weight was associated with a reduced seminiferous tubule size without other discernible alterations in the testis. It is worth noting that the 137A strain, which shares the same MMU6 fragment as 137B, exhibits a similar testis phenotype. In the same way, the 137F strain, which also presented a low testis weight, carries the same but slightly shorter (∼13 Mb) MMU6 fragment, strongly supporting this region as containing one low-testis-weight QTL. This region encompasses 115 genes. The 137F strain also presents a typical “Sertoli cell only” (SCO) phenotype in several seminiferous tubule sections (asterisks in Figure 2) and a reduction in the diameter of the other tubules where spermatogenesis is visible (Table 1). The SCO phenotype is shared with the 120G strain. It is worth noting that these two strains share a common 24-Mb spretus fragment located on MMU1 (41,400–65,300 kb). Two other small testis strains are described in Table 1.
Male accessory glands:
The epididymis, seminal vesicles, and prostate are important glands for male fertility since they provide most of the seminal fluid in which sperm is ejaculated. Therefore, we dissected and weighted these glands in every strain.
Epididymis:
We found a positive correlation between epididymis weight and sperm count (r = 0.499, P = 5.6 × 10−21). Two strains, 135C and 6C, presented absolute and relative epididymis weight significantly higher than B6 [+21.6% (P = 9.4 × 10−4) and +23.2% (P = 1.1 × 10−4), respectively] (Table 2). The 135C strain also had a significant increase in the sperm count (Table 1). No IRCSs displayed any notable abnormality of epididymis histology, except the 119H strain for which 3/12 animals presented an unilateral epididymis hypertrophy and an engorgement of the cauda epididymis due to an accumulation of spermatozoa, which showed reduced motility after recovery (Table 1).
TABLE 2.
Phenotype–genotype relation for accessory-gland-related traits in IRC strains
|
spretus segments
|
|||||
|---|---|---|---|---|---|
| Strains | n | % of B6's absolute gland weight | Glandular histology | Chromosome | Localization (bp) |
| 119H | 12 | Epididymis: +17% | Normal when nonengorged | 2 | 134,728,264–164,190,990 |
| 13 | 0–13,423,656 | ||||
| 6C | 12 | Epididymis: +18% | Normal | 2 | 0–16,231,985 |
| 20,958,126–25,353,595 | |||||
| 5 | 7,469,757–9,108,473 | ||||
| 131,219,492–134,200,000 | |||||
| 7 | 61,576,601–86,973,716 | ||||
| 130,015,787–145,134,094 | |||||
| 12 | 0–16,821,081 | ||||
| 17 | 46,495,245–66,722,220 | ||||
| 135C | 13 | Epididymis: +15%; seminal vesicles: +36% | Normal | 6 | 38,991,008–55,936,298 |
| 18 | 0–31,055,551 | ||||
| 120G | 12 | Seminal vesicles: +28% | Normal | 1 | 21,009,497–54,610,000 |
| 54,610,000–65,340,594 | |||||
| 5 | 26,457,520–31,750,835 | ||||
| 6 | 0–15,366,869 | ||||
| 14 | 78,826,456–107,526,001 | ||||
| 137C | 13 | Seminal vesicles: +50% | Normal | 6 | 133,269,077–149,525,685 |
| 14 | 23,634,750–32,466,048 | ||||
| 19 | 16,994,293–20,262,739 | ||||
| 136E | 11 | Seminal vesicles: −39%; prostate: −38% | Small prostatic and seminal vesicle glands | 3 | 36,936,374–57,602,475 |
| 10 | 29,893,191–40,449,730 | ||||
| 16 | 77,136,873–98,252,459 | ||||
| 135E | 16 | Prostate: +28% | Normal | 19 | 43,916,288–61,321,190 (Hpw1) |
| 157A | 9 | Prostate: −29% | Small prostatic glands | 15 | 0–30,205,799 |
| 16 | 11,645,398–28,465,840 | ||||
n, number of analyzed mice.
Seminal vesicles:
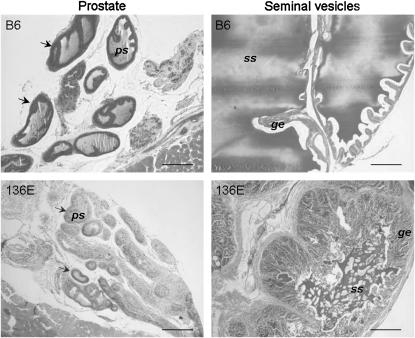
Absolute seminal vesicle weight ranged from 0.096 ± 0.051 g for 136E line to 0.238 ± 0.053 g for 137C line (Table 2). 137C is the only strain whose absolute seminal vesicle weight is significantly superior to B6 (0.159 ± 0.011 g), exhibiting a 50% increase (P = 1.3 × 10−4). Considering relative seminal vesicle weights, 135C and 120G had a significant increase [+36% (P = 5.9 × 10−5) and +28% (P = 1.7 × 10−3), respectively]. Seminal vesicle histology appeared normal under microscopic observation with a typical glandular epithelium and the presence of a seminal secretion in the lumens. Interestingly, 136E is the only IRCS with a significantly decreased seminal vesicle weight (−39%, P = 1.7 × 10−3). Seminal vesicles of most of the 136E animals (8/12) were atrophied albeit able to produce a small amount of seminal secretion (Figure 3). The 136E genome carries three spretus fragments on MMU3, MMU10, and MMU16.
Figure 3.—
Histology of seminal vesicle and prostate of the 136E and B6 strains. When compared to B6 histology, 136E seminal vesicles presented reduced glandular lumens. Seminal secretions (ss), however, are visible, but in lesser quantity than in B6 glands. Glandular epithelium (ge) displays numerous convolutions, indicating a quiescent state of the gland. Prostate glands (arrows) of 136E present the same quiescent aspect as that of the seminal vesicles: no or very few prostate secretions (ps) can be observed in the gland lumens, compared to B6. Bar, 200 μm.
Notably, we observed an intriguing external aspect of the seminal vesicles of the 97C mice: They exhibited a translucent aspect rather than the normal off-white color. However, they appeared well developed and able to produce seminal liquid, according to histological analysis. As a similar aspect can be observed in 5-week-old not fully pubescent mice, it would be a sign of a delay in the development of 97C male genital apparatus.
Prostate:
Prostate weights ranged from 0.0152 ± 0.0032 g for the 157A strain to 0.0272 ± 0.0069 g for the 135E strain. Only the 135E strain has significantly increased absolute and relative prostate weights compared to B6 (0.0213 ± 0.0042 g) (P = 5 × 10−3; Table 2). Histological analysis of 135E prostates showed a well-developed glandular tissue with a large lumen and secretion, similar to B6. As the genome of 135E carries a unique spretus fragment on MMU19, our results show that a high-prostate-weight QTL (Hpw1) maps between 43 Mb and the telomeric region of MMU19 encompassing 187 genes on 18 Mb. The 136E and 157A strains had a relative prostate weight significantly decreased compared to B6 [−38% (P = 3.7 × 10−5) and −29% (P = 7.4 × 10−4), respectively]. Histological analysis of 136E prostates showed glands smaller than B6, with a visible secretion in only a few glands (Figure 3).
Sperm functional traits:
Morphology:
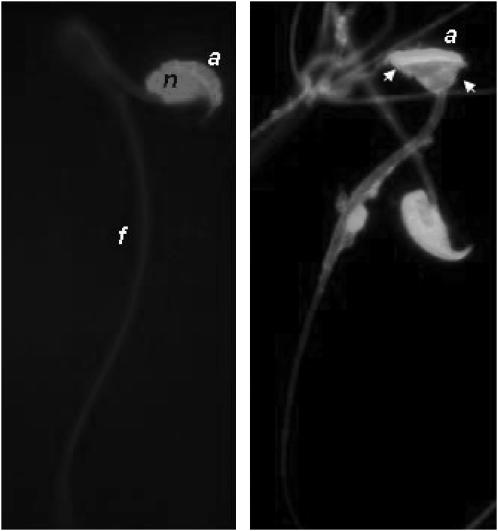
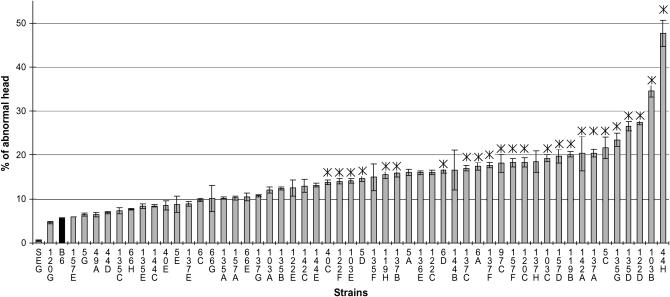
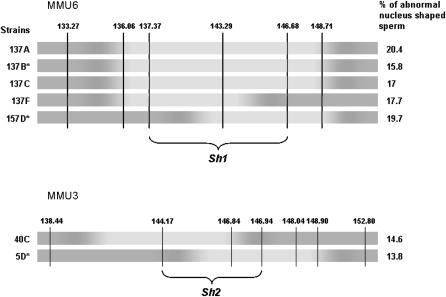
To assess the quality of the male gametes produced by the mouse strains, we analyzed the morphology of mature spermatozoa recovered from the cauda epididymis. The spermatozoa of the parental strains displayed a normal rodent morphology with a hook shape of the head and no apparent flagella abnormality. Only a very small proportion of B6 and SEG spermatozoa (5.5 ± 3.8% and 0.5 ± 0.4%, respectively) showed an abnormal head, as illustrated in Figure 4. The observed anomalies essentially affected the nucleus shape, giving it a symmetrical “hammerhead”-like appearance, with the flagellum originating from the middle of this structure. Despite this highly atypical nuclear form, these spermatozoa did present an acrosome normally capping the nucleus (Figure 4). Compared to the parental strains, 24 strains showed a significantly higher number of abnormal sperm than B6 (Figure 5), reaching up to 47.7 ± 11.7% in the 44H strain. Of these 24 strains, 5 were congenic lines, thus making it possible to map QTL influencing nuclear shape (Table 3). It is worth noting that 137B and 157D mice, which showed the same frequency of abnormal sperm heads (∼17%), share a common 12-Mb MMU6 fragment (137–149 Mb). Furthermore, this region is at least partially shared with 3 other strains, making it possible to reduce the location of the QTL involved to a 9-Mb fragment (sperm hammerhead 1, Sh1, 137–146 Mb, Figure 6). This fragment encompasses 56 genes. Two genes (Capzα3 and Tuba7) encode proteins involved in the cytoskeleton structure.
Figure 4.—
Morphology of mouse sperm from the cauda epididymis. Two fluorescent light microscope images of normal and abnormal (arrows) mouse spermatozoa stained with EH for the nucleus (n) and with FITC–PNA for the acrosome (a). (Right) Arrows point out the misshapenness of the head, which exhibited a normal-shaped acrosome (a); the flagellum (f) appears normal.
Figure 5.—
Frequency of sperm with an abnormal head in the IRCS. Mean values (±SEM) are presented in order of croissant abnormality frequency. B6 and SEG values are shown by solid bars. Asterisk (*) denotes 24 strains that showed a significant increase in mean values compared to the B6 strain.
TABLE 3.
Phenotype–genotype relation for sperm-head-shape-related traits in IRC strains
|
spretus segments
|
||||
|---|---|---|---|---|
| Strains | n | % of abnormal sperm ±SEM | Chromosome | Localization (bp) |
| B6 | 16 | 5.5 ± 0.2 | ||
| 5D | 7 | 14.6 ± 0.7 | 3 | 144,167,685–152,797,124 (Sh2) |
| 137B | 11 | 15.8 ± 0.7 | 6 | 133,269,077–149,525,685 (Sh1) |
| 97C | 5 | 18.2 ± 2 | 11 | 3,525,585–26,517,138 (Sh4) |
| 157D | 4 | 19.7 ± 1.6 | 6 | 137,373,153–149,525,685 (Sh1) |
| 135G | 4 | 23.5 ± 1.4 | 12 | 106,319,918–120,463,159 (Sh3) |
| 40C | 8 | 13.8 ± 0.5 | 2 | 20,958,126–25,353,595 |
| 3 | 138,437,876–146,939,883 | |||
| 5 | 131,219,492–134,200,000 | |||
| 10 | 104,072,802–129,959,148 | |||
| 15 | 0–30,205,799 | |||
| 122F | 12 | 14 ± 0.6 | 6 | 137,373,153–149,525,685 |
| 8 | 86,030,732–95,648,339 | |||
| 103E | 7 | 14.1 ± 0.5 | 9 | 54,777,202–65,644,180 |
| 14 | 105,008,380–123,978,870 | |||
| 19 | 3,110,000–32,998,316 | |||
| 55,219,249–61,321,190 | ||||
| 119H | 9 | 15.5 ± 0.8 | 2 | 134,728,264–164,190,990 |
| 13 | 0–13,423,656 | |||
| 6D | 16.5 ± 0.7 | 6 | 120,395,329–122,822,036 | |
| 18 | 31,494,974–41,505,256 | |||
| 19 | 16,994,293–61,321,190 | |||
| 137C | 13 | 17 ± 0.6 | 6 | 133,269,077–149,525,685 |
| 14 | 23,634,750–32,466,048 | |||
| 19 | 16,994,293–20,262,739 | |||
| 6A | 9 | 17.5 ± 0.9 | 5 | 7,469,757–9,108,473 |
| 131,219,492–134,200,000 | ||||
| 6 | 137,373,153–149,525,685 | |||
| 13 | 103,445,606–120,614,378 | |||
| 19 | 3,110,000–32,998,316 | |||
| 137F | 13 | 17.7 ± 0.6 | 1 | 41,416,802–73,177,650 |
| 6 | 133,269,077–146,678,290 | |||
| 19 | 16,994,293–20,262,739 | |||
| 157F | 6 | 18.3 ± 1 | 6 | 133,269,077–146,678,290 |
| 16 | 0–28,465,840 | |||
| 120C | 6 | 18.3 ± 1 | 2 | 153,405,225–162,891,332 |
| 4 | 0–32,266,906 | |||
| 76,112,398–83,862,809 | ||||
| 12 | 16,821,081–28,791,620 | |||
| 28,791,620–49,618,164 | ||||
| 58,263,512–74,115,351 | ||||
| 16 | 0–28,465,840 | |||
| 103C | 9 | 19.3 ± 0.8 | 7 | 55,914,049–83,429,087 |
| 9 | 54,777,202–65,644,180 | |||
| 13 | 0–38,113,893 | |||
| 19 | 3,110,000–32,998,316 | |||
| 32,998,316–55,219,249 | ||||
| 55,219,249–61,321,190 | ||||
| 119B | 11 | 20.1 ± 0.6 | 2 | 134,728,264–158,315,158 |
| 15 | 90,836,253–103,492,577 | |||
| 142A | 2 | 20.3 ± 4 | 6 | 16,464,329–28,638,293 |
| 107,874,585–118,504,993 | ||||
| 137A | 9 | 20.4 ± 0.9 | 6 | 133,269,077–149,525,685 |
| 7 | 25,872,301–35,582,876 | |||
| 35,582,876–60,165,513 | ||||
| 18 | 46,781,237–69,139,024 | |||
| 19 | 55,219,249–61,321,190 | |||
| 5C | 3 | 21.6 ± 2.5 | 2 | 16,231,985–23,819,867 |
| 5 | 128,896,990–134,200,000 | |||
| 18 | 31,494,974–41,505,256 | |||
| 122D | 6 | 27.5 ± 0.6 | 1 | 17,340,697–41,416,802 |
| 11 | 46,917,720–58,920,013 | |||
| 114,028,355–121,798,632 | ||||
| 16 | 15,456,409–48,218,872 | |||
| 44H | 5 | 47.6 ± 2.9 | 3 | 0–36,936,374 |
| 4 | 143,368,235–149,581,818 | |||
| 12 | 106,319,918–120,463,159 | |||
| 13 | 65,525,612–81,623,009 | |||
| 102,577,536–120,614,378 | ||||
| 15 | 42,535,894–46,300,641 | |||
| 19 | 13,446,378–32,998,316 | |||
n, number of analyzed mice.
Figure 6.—
Fine mapping of sperm hammerhead 1 and sperm hammerhead 2 QTL by an interstrain analysis. Marker positions are given in mega base pairs. Dark regions correspond to the B6 background and light regions to the SEG fragments in chromosomes 6 (MMU6) and 3 (MMU3) of different strains having comparable abnormal sperm-head frequency. Congenic strains are identified by an asterisk. Overlapping SEG fragments of the different strains makes it possible to map Shh1 QTL on a maximal 9-Mb fragment (137–146 Mb) on MMU6 and Shh2 QTL on a maximal 3 Mb of MMU3.
Another unique spretus fragment, on MMU3, was found in the 5D strain, which presents 14.6% of abnormal sperm. Furthermore, the 40C strain (13.8% abnormal sperm) shares a portion of this MMU3 fragment (Figure 6). The most probable location of the QTL is therefore reduced to an ∼3-Mb fragment (sperm hammerhead 2, Sh2, 144.17–146.94 Mb, Figure 6). This region encompasses 22 genes; one of them, Spata1, is specifically associated with spermatogenesis and constitutes an interesting candidate for the observed phenotype.
A unique 14-Mb MMU12 fragment was found to increase the frequency of abnormal sperm to 23.5% in the 135G strain (sperm hammerhead 3, Sh3, Table 3). This fragment is shared by the 44H mice and contains 71 genes. Interestingly, the 44H sperm harbors the highest proportion of anomalies (47.6%). It is therefore tempting to conclude that half of this effect is due to the QTL located on MMU12. Finally, a fourth QTL of abnormal sperm head was carried by the MMU11 spretus fragment of the 97C strain (sperm hammerhead 4, Sh4, Table 3).
Viability:
The viability of the spermatozoa collected early after puncturing the epididymis ranged from 58.0 ± 2% to 76.3 ± 3.2% of live spermatozoa. The 137G strain (59.0 ± 0.5%) displayed an early viability significantly reduced compared to B6 (70.9 ± 2%, P = 1 × 10−3), indicating a moderate necrospermia. After 2 hr of incubation time in the capacitating medium, the sperm viability decreased to a range of 39.5 ± 9.3% to 67.1 ± 2.0%. The 157D and 120C strains displayed a sperm late viability significantly decreased when compared to B6 (P = 7.8 × 10−3), corresponding to the loss of 24% of live spermatozoa. Since 157D is congenic for a 12-Mb fragment on MMU6 containing 86 genes, a QTL influencing sperm survival is present at this locus (decreased sperm survival 1, Dss1, Table 4).
TABLE 4.
Phenotype–genotype relation for viability and acrosome-status-related traits in IRC strains
| Viability
|
AR
|
spretus segments
|
|||||
|---|---|---|---|---|---|---|---|
| Strains | n | Early viability (%) ±SEM and P-value | Viability decrease (%) ±SEM and P-value | SAR ±SEM and P-value | IIAR ±SEM and P-value | Chromosome | Localization (bp) |
| B6 | 16 | 70.9 ± 2 | 10 ± 4 | 22.8 ± 0.5 | 19.6 ± 1.3 | ||
| 137G | 4 | 59 ± 0.5 (P = 0.003) | 12.3 ± 0.2 (NS) | 1 | 65,340,594–80,570,000 | ||
| 6 | 137,373,153–149,525,685 | ||||||
| 7 | 25,872,301–35,582,876 | ||||||
| 120C | 6 | 64 ± 0.2 (NS) | 24.2 ± 0.8 (P = 0.01) | 2 | 153,405,225–162,891,332 | ||
| 4 | 0–32,266,906 | ||||||
| 76,112,398–83,862,809 | |||||||
| 12 | 16,821,081–28,791,620 | ||||||
| 28,791,620–49,618,164 | |||||||
| 58,263,512–74,115,351 | |||||||
| 16 | 0–28,465,840 | ||||||
| 157D | 4 | 68 ± 0.5 (NS) | 24.2 ± 2 (P = 0.009) | 6 | 137,373,153–149,525,685 (Dss1) | ||
| 44D | 12 | 38.9 ± 0.9 (P = 0.0002) | 16.5 ± 1 (NS) | 6 | 0–28,638,293 | ||
| 13 | 99,827,842–111,310,000 | ||||||
| 18 | 69,139,024–90,736,837 | ||||||
| 66H | 9 | 22.1 ± 0.5 (NS) | 4.8 ± 0.8 (P = 0.0002) | 1 | 80,570,000–113,454,683 | ||
| 13 | 84,982,049–102,577,536 | ||||||
| 18 | 36,912,104–57,454,744 | ||||||
Viability decrease is early viability minus late viability after 2 hr of incubation in capaciting medium. n, number of analyzed mice.
Acrosome reaction:
We also explored the capacity of the sperm to undergo a calcium ionophore-induced AR, using a fluorescent probe specifically labeling the acrosomal matrix (Figure 7B). This labeling was analyzed by a flow cytometer, enabling an objective assessment of the acrosome status (Figure 7). Upon AR measurement, only two strains (44D and 66H) presented significant phenotypic deviations from B6 (Table 4). The average level of SAR in IRCSs ranged from 15.4 ± 3% to 38.9 ± 0.9% compared to 22.8 ± 0.5% for B6. 44D mice, with the highest SAR level, significantly differed from B6 (P = 9 × 10−5). This strain bears three fragments on MMU8, MMU13, and MMU18.
The proportion of IRCS spermatozoa that underwent an acrosome reaction in response to the ionophore (rate of IIAR) ranged from <5 to 40% (19.6 ± 1.3% for B6). One strain (66H) of 49 studied for this trait showed a strongly decreased response to ionophore treatment (IIAR: 4.8 ± 0.8%, P = 1.8 × 10−4). Phenotypic features and QTL mapping are summarized in Table 4.
DISCUSSION
Mammalian fertility appears as a quantitative feature. For instance, it is well known from human clinical data that subnormal sperm count (<5–10 × 106 sperm cells/ml) affects male fertility very strongly (Slama et al. 2002). However, studying such parameters to try to address their molecular grounds in humans is a very difficult challenge, while the laboratory mouse appears as an optimal mammalian model, due to its high prolificacy and short generation time. Also, the comprehensive knowledge of the genome sequence of humans and mice suggests that, in many cases, the interspecific transposition will be possible using comparative mapping approaches. Several QTL studies performed by genome scanning in recent years pointed to specific mouse genome regions involved in fertility. Such regions, generally very large (in the half-chromosome range), were identified for female (Krewson et al. 2004; Peripato et al. 2004; Rocha et al. 2004; Liljander et al. 2006) and male fertility parameters (Zidek et al. 1998; Le Roy et al. 2001; Oka et al. 2004; Bolor et al. 2006). Also, the genetic basis of F1 hybrid sterility has been addressed by this type of QTL approach (Elliott et al. 2004). Clearly, QTL studies are only the prelude of positional cloning projects, since eventually identifying a causal mutation requires fine-mapping efforts in a second step. An alternative to classical QTL family design is to use a stock of genetically heterogeneous mice (Valdar et al. 2006), whose genomes are composed of a mosaic originating from several classical laboratory strains, and to multiply the measured phenotypes on a very large scale. Indeed, using such a model, Valdar et al. (2006) recently demonstrated that this kind of approach is very efficient for mapping traits in the megabase range. However, genetically heterogeneous mice are unique individuals, meaning that generally, due to the short life span of mice, they are phenotyped only once. Therefore, it is not possible to come back to these animals to further refine the phenotype analysis. For instance, since Valdar and co-workers did not choose fertility parameters in their measured phenotypes, no information can be extracted from their impressive data set for such features.
In this study, we investigated male fertility parameters using the original IRCS genetic tool, developed for 15 years at the Pasteur Institute (Paris). The link between male fertility and the male parameters analyzed is clear since most of the strains where we found QTL altering strongly qualitative or quantitative sperm or testes parameters (such as 97C, 66H, and 44H) also display a long delay (often several months) of male/female promiscuity to obtain pregnancies. The IRCS model presents several advantages over classical QTL family designs: (i) a high variability between the two parental strains (species), ensuring a high level of marker informativity compared to classical laboratory mice [one SNP in every 80–100 bp between spretus and domesticus species (Guenet and Bonhomme 2003)]; (ii) a nonambiguous association between a phenotypic effect and precisely delimited spretus fragments; and (iii) the theoretical possibility of reducing relevant intervals to small genomic regions (down to 2 Mb, or even less) by combining phenotypic and mapping data from different strains, this procedure being achieved in one step.
The 53 IRCS panel used in this study do not encompass any spretus fragments from the X chromosome (∼5% of the mouse genome). Since our IRCSs cover ∼43% of the mouse genome with spretus fragments, together with the results from Oka et al. (2004, 2007) it is now close to 50% of the genome that has been analyzed for male fertility parameters in interspecific crosses.
One possible drawback of the use of stable congenic strains resides in the fact that, in some very exceptional cases, sporadic undetectable mutations that may have occurred early during the construction of the strain could affect the observed phenotype independently of the spretus fragment. In this study, two kinds of arguments may contribute to disqualifying this theoretical possibility:
In many cases, the same phenotype segregates with a given spretus fragment observed in independent congenic strains (an interesting example, among many others, is the partial “Sertoli cell only” phenotype observed in the 120G and 137F strains, which shares a 13-Mb common fragment on MMU1 from 41.4 to 54.6 Mb; see Table 1). Two independent mutations in two independent strains affecting the same phenotype in two congenic strains would be much less probable than an effect of a large spretus fragment highly divergent from the B6 orthologous region.
In one case, highly relevant for human medicine (delayed puberty, represented by a small testis weight, in the 97C strain), we produced a preliminary F2 population (n = 40). Polymorphic microsatellites discriminating the B6 (B) and Spretus (S) allele on the 97C chromosome 11 fragment were genotyped, and we could demonstrate a conservation of the small-testis-weight phenotype with the segregation of the MMU11 fragment [average testis weight: homozygous S/S, 0.116 ± 0.003 g; heterozygous B/S, 0.122 ± 0.001 g; homozygous B/B, 0.168 ± 0.021 g; p (S/S vs. B/S) = 0.52; p(S/S + B/S vs. B/B) < 1.10−8].
In this study, we chose to focus primarily on “congenic” strains to locate QTL. Then, in several cases, other strains were used to complement this primary mapping information. We have therefore deliberately chosen not to use “pluricongenic” strains to obtain primary QTL locations, so that we could get rid of complex epistatic effects susceptible to appearing in strains with two or more genomic fragments of spretus origin. This fine-mapping potential of the IRCSs is illustrated in our study by three examples: in the 137B, a MMU6 QTL is associated with a low testis weight (Ltw2) and the fragment is reduced from 16 to 13 Mb using the 137F strain information. Two QTL responsible for the sperm hammerhead phenotype, Sh1 and Sh2, had a refined localization. Sh1, first identified on the 137B strain on a 16-Mb fragment, is finally located in a 9-Mb fragment using phenotypes from the 157D and 137F strains. Similarly, Sh2, primarily located on a MMU3 9-Mb fragment, is finally located on an ∼3-Mb fragment using data from the 40C strain (Figure 6).
For most traits measured, we found lines differing significantly from the control B6 and, thanks to “congenics” strains, we could unambiguously localize three QTL regulating the testis weight (Ltw1, Ltw2) and the prostate weight (Lpw1), four QTL controlling the shape of the sperm nucleus (Sh1, Sh2, Sh3, and Sh4), and a QTL influencing sperm survival (Dss1). When the QTL location was sufficiently precise (in a 3- to 20-Mb fragment encompassing <200 genes), candidate genes were searched and proposed.
Considering the testis weight, we localized two new QTL on MMU6 and MMU11 in addition to those reported on chromosome 4 (Zidek et al. 1998; Le Roy et al. 2001; Bolor et al. 2006) and on chromosome 10 (Le Roy et al. 2001). Also, in this study, some “pluricongenic” strains with low testis weight associated with spretus MMU13 and MMU18 fragments could confirm and refine two previously suggested QTL on chromosome 13 (Zidek et al. 1998; Le Roy et al. 2001) and on chromosome 18 (Le Roy et al. 2001). The MMU4 locus described by Bolor et al. (2006) is associated with a small testis weight and giant multi-nucleated cells in degenerating germinative epithelium, whereas our “pluricongenic” 49A strain, which encompasses the homologous spretus fragment, presents an elevated testis weight. However, in preliminary experimental crosses to segregate the spretus fragments, we obtained an F2 population from the 49A strain and, among the offspring, ∼10% presented with an abnormally small testis weight and giant multi-nucleated cells. This strongly suggests the existence of epistasis between the different spretus fragments of 49A.
Two phenotypic characteristics, which could be of interest in human clinics, have been found associated with a low testis weight: the partial SCO phenotype of 137F and 120G strains and the absence of lumen in the seminiferous tubules of 97C testis. Whereas the two former strains are “pluricongenics,” making it impossible to map the SCO QTL unambiguously, we mapped the Ltw1 QTL on the 97C MMU11 26-Mb fragment. This region encompasses 147 genes among which is Otx1, a strong potential candidate gene for the puberty-delay phenotype since Otx1 knockout mice and 97C share a similar testicular phenotype (Acampora et al. 1998).
For male reproductive accessory gland weight regulation, a unique QTL of high prostate weight could be unambiguously mapped. This QTL localized on MMU19 on a 18-Mb fragment encompassing 134 genes. Among them, Mxi1 is a good candidate gene since its knockout causes a cellular proliferation of the prostatic glandular epithelium (Eagle et al. 1995).
Interestingly, we could finely map two QTL affecting the head of the spermatozoa on MMU3 and MMU6. The MMU6 region is a 9-Mb spretus fragment encompassing 68 genes. Expression profiles of these genes were obtained from the SymAtlas database. Only four genes appeared as almost strictly testis specific: Igbp1b, Plcz1, Capza3, and Tuba7, the latter two being the most promising candidates. Capza3 (actin capping protein α3) is principally localized in the neck region of ejaculated sperm in humans (Miyagawa et al. 2002). This localization matches the actin distribution in human sperm. This factor is supposed to have an important role in sperm architecture and fertility (Miyagawa et al. 2002). It is therefore an excellent candidate for the sperm anomaly that we have detected. Tuba7 (Tubulin a7) has a strict testis-specific expression controlled by the E2F6 transcription factor, since the knockout of this last gene triggers the ubiquitous expression of the tubulin gene (Pohlers et al. 2005). Tubulin is the major component of the sperm manchette, a cytoskeleton structure that plays a central role in shaping the spermatid nucleus, suggesting that Tuba7 is an interesting potential candidate for the observed phenotype. Notably, this primary QTL localization also encompasses two histone genes, Hist4H4 and H2ajf. While the expression profile of these genes is not strictly testis specific, they may be implicated in the observed phenotype since an histone H1 variant gene knockout induced similar sperm-nucleus misshapenness (Martianov et al. 2005). On MMU3, we could reduce the relevant region to a fragment of ∼2.5 Mb. In this region, only 41 genes were found, of which only one, Spata1, displayed a strictly testis-specific expression profile. While little is known about this gene, the sequence analysis reveals the presence of an ABC-SMC4 domain in the second half of the protein. SMC (structural maintenance of chromosomes) proteins are factors involved in chromosome condensation, sister-chromatid cohesion, DNA repair and recombination, and gene dosage compensation, and they function in somatic and meiotic cells (Hirano 2002; Jessberger 2002). Therefore, Spata1 is an excellent positional and functional candidate for the observed phenotype. Interestingly, the E2F6 transcription factor, responsible for Tuba7 tissue specificity, is also known as a repressor of SMC proteins (Storre et al. 2005). Therefore Tuba7 and Spata1 could belong to the same functional pathway.
The nucleus morphology anomaly that we observed in the IRCS is a major sperm-head malformation, probably strongly impairing male fertility, especially when the number of abnormal spermatozoa reaches half of the epididymal sperm population as in the 44H strain. This head deformity was different from the phenotype associated with the X-linked QTL sha1, -2 and -3, recently described by Oka et al. (2004, 2007). Indeed, the sperm-head defect that we described concerned only nucleus shape, without visible abnormality of the acrosome, whereas the Oka group's evaluation of sperm-head morphology included both the nucleus and the acrosome.
The rapid identification of strong candidates for sperm morphology, testis, and prostate weight emphasizes the exceptional power of the IRCS model for positional cloning approaches. For QTL that we could not map directly as those implicated in SCO or in the absence of AR, we will segregate the spretus fragments to precisely localize the QTL effects and potentially epistasis. The final demonstration of the involvement of candidate genes may necessitate various approaches. Sequencing the gene in B6 and SEG will be a necessary prerequisite. It is worth noting that the observed phenotypes in the IRCS study result systematically from inadequacies between two divergent genomes. These inadequacies certainly reflect molecular malfunctions between the spretus and musculus genomes at the level of either protein–protein interactions or protein–DNA interactions. Consequently, differences in coding sequences may pinpoint directly not only the protein responsible for the phenotype, but also the most relevant amino-acid position for enabling the normal interaction. This could be an interesting lead for setting up biochemical experimentations and could allow the release of more subtle biological information than the knocking out of a given gene.
Acknowledgments
We thank Isabelle Lanctin (Pasteur Institute) for her precious help in providing us with animals difficult to house and Corinne Lesaffre for her technical assistance. We also thank the department of “anatomie pathologique” of the Cochin's Hospital. This study was supported by grant no. 1752 from the Direction de la Recherche et des Etudes Doctorales and no. 06-2005-MAMMIFERT-02 from the French Agence National pour la Recherche. David L'Hôte is funded by grants from the Institut National de la Santé et de la Recherche Médicale and Limoges University.
References
- Acampora, D., S. Mazan, F. Tuorto, V. Avantaggiato, J. J. Tremblay et al., 1998. Transient dwarfism and hypogonadism in mice lacking Otx1 reveal prepubescent stage-specific control of pituitary levels of GH, FSH and LH. Development 125: 1229–1239. [DOI] [PubMed] [Google Scholar]
- Affara, N. A., and M. J. Mitchell, 2000. The role of human and mouse Y chromosome genes in male infertility. J. Endocrinol. Invest. 23: 630–645. [DOI] [PubMed] [Google Scholar]
- Avner, P., 1998. Complex traits and polygenic inheritance in the mouse. Methods 14: 191–198. [DOI] [PubMed] [Google Scholar]
- Bolor, H., N. Wakasugi, W. D. Zhao and A. Ishikawa, 2006. Detection of quantitative trait loci causing abnormal spermatogenesis and reduced testis weight in the small testis (Smt) mutant mouse. Exp. Anim. 55: 97–108. [DOI] [PubMed] [Google Scholar]
- Carlson, A. E., R. E. Westenbroek, T. Quill, D. Ren, D. E. Clapham et al., 2003. CatSper1 required for evoked Ca2+ entry and control of flagellar function in sperm. Proc. Natl. Acad. Sci. USA 100: 14864–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill, G. A., D. C. Airey, H. Allayee, J. M. Angel, A. D. Attie et al., 2004. The Collaborative Cross, a community resource for the genetic analysis of complex traits. Nat. Genet. 36: 1133–1137. [DOI] [PubMed] [Google Scholar]
- Eagle, L. R., X. Yin, A. R. Brothman, B. J. Williams, N. B. Atkin et al., 1995. Mutation of the MXI1 gene in prostate cancer. Nat. Genet. 9: 249–255. [DOI] [PubMed] [Google Scholar]
- Elliott, R. W., D. Poslinski, D. Tabaczynski, C. Hohman and J. Pazik, 2004. Loci affecting male fertility in hybrids between Mus macedonicus and C57BL/6. Mamm. Genome 15: 704–710. [DOI] [PubMed] [Google Scholar]
- Emiliozzi, C., H. Cordonier, J. F. Guerin, B. Ciapa, M. Benchaib et al., 1996. Effects of progesterone on human spermatozoa prepared for in-vitro fertilization. Int. J. Androl. 19: 39–47. [DOI] [PubMed] [Google Scholar]
- Feng, H. L., 2003. Molecular biology of male infertility. Arch. Androl. 49: 19–27. [DOI] [PubMed] [Google Scholar]
- Foresta, C., E. Moro and A. Ferlin, 2001. Y chromosome microdeletions and alterations of spermatogenesis. Endocr. Rev. 22: 226–239. [DOI] [PubMed] [Google Scholar]
- Guenet, J. L., and F. Bonhomme, 2003. Wild mice: an ever-increasing contribution to a popular mammalian model. Trends Genet. 19: 24–31. [DOI] [PubMed] [Google Scholar]
- Hirano, T., 2002. The ABCs of SMC proteins: two-armed ATPases for chromosome condensation, cohesion, and repair. Genes Dev. 16: 399–414. [DOI] [PubMed] [Google Scholar]
- Jessberger, R., 2002. The many functions of SMC proteins in chromosome dynamics. Nat. Rev. Mol. Cell Biol. 3: 767–778. [DOI] [PubMed] [Google Scholar]
- Krewson, T. D., P. J. Supelak, A. E. Hill, J. B. Singer, E. S. Lander et al., 2004. Chromosomes 6 and 13 harbor genes that regulate pubertal timing in mouse chromosome substitution strains. Endocrinology 145: 4447–4451. [DOI] [PubMed] [Google Scholar]
- Le Roy, I., S. Tordjman, D. Migliore-Samour, H. Degrelle and P. L. Roubertoux, 2001. Genetic architecture of testis and seminal vesicle weights in mice. Genetics 158: 333–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liljander, M., M. A. Sallstrom, S. Andersson, P. Wernhoff, A. Andersson et al., 2006. Identification of genetic regions of importance for reproductive performance in female mice. Genetics 173: 901–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martianov, I., S. Brancorsini, R. Catena, A. Gansmuller, N. Kotaja et al., 2005. Polar nuclear localization of H1T2, a histone H1 variant, required for spermatid elongation and DNA condensation during spermiogenesis. Proc. Natl. Acad. Sci. USA 102: 2808–2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzuk, M. M., and D. J. Lamb, 2002. Genetic dissection of mammalian fertility pathways. Nat. Cell Biol. 4(Suppl.): s41–s49. [DOI] [PubMed] [Google Scholar]
- Mendoza-Lujambio, I., P. Burfeind, C. Dixkens, A. Meinhardt, S. Hoyer-Fender et al., 2002. The Hook1 gene is non-functional in the abnormal spermatozoon head shape (azh) mutant mouse. Hum. Mol. Genet. 11: 1647–1658. [DOI] [PubMed] [Google Scholar]
- Miyagawa, Y., H. Tanaka, N. Iguchi, K. Kitamura, Y. Nakamura et al., 2002. Molecular cloning and characterization of the human orthologue of male germ cell-specific actin capping protein alpha3 (cpalpha3). Mol. Hum. Reprod. 8: 531–539. [DOI] [PubMed] [Google Scholar]
- Nakamura, T., R. Yao, T. Ogawa, T. Suzuki, C. Ito et al., 2004. Oligo-astheno-teratozoospermia in mice lacking Cnot7, a regulator of retinoid X receptor beta. Nat. Genet. 36: 528–533. [DOI] [PubMed] [Google Scholar]
- Neesen, J., R. Kirschner, M. Ochs, A. Schmiedl, B. Habermann et al., 2001. Disruption of an inner arm dynein heavy chain gene results in asthenozoospermia and reduced ciliary beat frequency. Hum. Mol. Genet. 10: 1117–1128. [DOI] [PubMed] [Google Scholar]
- Oka, A., A. Mita, N. Sakurai-Yamatani, H. Yamamoto, N. Takagi et al., 2004. Hybrid breakdown caused by substitution of the X chromosome between two mouse subspecies. Genetics 166: 913–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka, A., T. Aoto, Y. Totsuka, R. Takahashi, M. Ueda et al., 2007. Disruption of genetic interaction between two autosomal regions and the X chromosome causes reproductive isolation between mouse strains derived from different subspecies. Genetics 175: 185–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peripato, A. C., R. A. De Brito, S. R. Matioli, L. S. Pletscher, T. T. Vaughn et al., 2004. Epistasis affecting litter size in mice. J. Evol. Biol. 17: 593–602. [DOI] [PubMed] [Google Scholar]
- Pilder, S. H., P. Olds-Clarke, J. M. Orth, W. F. Jester and L. Dugan, 1997. Hst7: a male sterility mutation perturbing sperm motility, flagellar assembly, and mitochondrial sheath differentiation. J. Androl. 18: 663–671. [PubMed] [Google Scholar]
- Pohlers, M., M. Truss, U. Frede, A. Scholz, M. Strehle et al., 2005. A role for E2F6 in the restriction of male-germ-cell-specific gene expression. Curr. Biol. 15: 1051–1057. [DOI] [PubMed] [Google Scholar]
- Rocha, J. L., E. J. Eisen, F. Siewerdt, L. D. Van Vleck and D. Pomp, 2004. A large-sample QTL study in mice: III. Reproduction. Mamm. Genome 15: 878–886. [DOI] [PubMed] [Google Scholar]
- Slama, R., F. Eustache, B. Ducot, T. K. Jensen, N. Jorgensen et al., 2002. Time to pregnancy and semen parameters: a cross-sectional study among fertile couples from four European cities. Hum. Reprod. 17: 503–515. [DOI] [PubMed] [Google Scholar]
- Storre, J., A. Schafer, N. Reichert, J. L. Barbero, S. Hauser et al., 2005. Silencing of the meiotic genes SMC1beta and STAG3 in somatic cells by E2F6. J. Biol. Chem. 280: 41380–41386. [DOI] [PubMed] [Google Scholar]
- Su, A. I., M. P. Cooke, K. A. Ching, Y. Hakak, J. R. Walker et al., 2002. Large-scale analysis of the human and mouse transcriptomes. Proc. Natl. Acad. Sci. USA 99: 4465–4470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su, A. I., T. Wiltshire, S. Batalov, H. Lapp, K. A. Ching et al., 2004. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc. Natl. Acad. Sci. USA 101: 6062–6067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao, J., E. S. Critser and J. K. Critser, 1993. Evaluation of mouse sperm acrosomal status and viability by flow cytometry. Mol. Reprod. Dev. 36: 183–194. [DOI] [PubMed] [Google Scholar]
- Valdar, W., L. C. Solberg, D. Gauguier, S. Burnett, P. Klenerman et al., 2006. Genome-wide genetic association of complex traits in heterogeneous stock mice. Nat. Genet. 38: 879–887. [DOI] [PubMed] [Google Scholar]
- Yao, R., C. Ito, Y. Natsume, Y. Sugitani, H. Yamanaka et al., 2002. Lack of acrosome formation in mice lacking a Golgi protein, GOPC. Proc. Natl. Acad. Sci. USA 99: 11211–11216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zidek, V., A. Musilova, J. Pintir, M. Simakova and M. Pravenec, 1998. Genetic dissection of testicular weight in the mouse with the BXD recombinant inbred strains. Mamm. Genome 9: 503–505. [DOI] [PubMed] [Google Scholar]