Abstract
When individuals' fitnesses depend on the genetic composition of the population in which they are found, selection is then frequency dependent. Frequency-dependent selection (FDS) is often invoked as a heuristic explanation for the maintenance of large numbers of alleles at a locus. The pairwise interaction model is a general model of FDS via intraspecific competition at the genotypic level. Here we use a parameter-space approach to investigate the full potential for the maintenance of multiallelic equilibria under the pairwise interaction model. We find that FDS maintains full polymorphism more often than classic constant-selection models and produces more skewed equilibrium allele frequencies. Fitness sets with some degree of rare advantage maintained full polymorphism most often, but a wide variety of nonobvious fitness patterns were also found to have positive potential for polymorphism. An example is put forth suggesting possible explanations for multiallelic polymorphisms maintained despite positive FDS on individual alleles.
THE majority of existing models of natural selection assume that selection remains constant over time. This use of constant selection coefficients is largely for mathematical convenience (Kojima 1971), since it is much more likely that selection pressures in real populations will vary, for example, in space and time. One way to model changing selection is to make fitness frequency dependent: When individuals' fitnesses depend on the genetic composition of the population in which they are found, selection is then frequency dependent. Intuitively, negative frequency dependence (selection against common alleles) should be good at maintaining many alleles at a locus. Conversely, one would expect positive frequency dependence (selection for common alleles) to result in monomorphism. Positive and negative frequency-dependent selection (FDS) are but two extremes along a continuum; there is substantial evidence to suggest that many kinds of FDS are widespread in naturally polymorphic populations. In experiments with cultivated plants, genotypic fitnesses are affected in a variety of ways by the presence, number, and genotype of their neighbors (Antonovics and Ellstrand 1984). Studies of predator–prey choice (Allen et al. 1998; Bond and Kamil 1998, 2002; Allen and Weale 2005; for reviews see Allen 1988; Punzalan et al. 2005) have found evidence for a variety of kinds of FDS. Positive FDS specifically has been implicated in studies of Mullerian mimicry (Langham 2004). Negative FDS is found in a wide variety of systems such as host–pathogen coevolution (May and Anderson 1983; Dybdahl and Lively 1998; Carius et al. 2001; Trachtenberg et al. 2003), mate choice (Hughes et al. 1999), and Batesian mimicry (Pfennig et al. 2001; Cheney and Cote 2005; Anderson and Johnson 2006) and is generally used as a heuristic explanation for the selective maintenance of genetic variation (e.g., Wilson et al. 1994; Raymond et al. 1996; Yuste et al. 2002; Billiard et al. 2005; Olendorf et al. 2006; Piertney and Oliver 2006).
The abundance of field evidence for FDS (and not only negative FDS) in polymorphic natural systems suggests that we require a more mathematically rigorous explanation for this polymorphism than “it must be some sort of negative frequency dependence.” What kinds of FDS can maintain variation? What kinds of equilibrium allele-frequency distributions does FDS produce? How do systems with positive FDS (predator choice, mimicry, etc.) retain polymorphism? Does FDS produce a detectable signature of selection in population allele frequencies? To address these and other questions about FDS and polymorphism, we first require a general model of FDS. Here we use the pairwise interaction model (PIM) of selection via intraspecific competition as our general model of FDS.
Ever since Darwin, the concepts of intraspecific competition and natural selection have been closely linked. As natural selection is a consequence of the struggle for existence, so can its mirror image, fitness, be considered a consequence of intraspecific competitive interactions. Cockerham et al. (1972) argued that parameterizing fitness as a product of intraspecific competition at the genotype level provides a more biologically reasonable, but still mathematically tractable, framework for modeling natural selection. The PIM is such a model: A genotype's fitness is a function of its frequency in the population, its relative fitness in interactions with the other genotypes, and the frequencies of the other genotypes. Each genotype is assumed to have some constant interaction-fitness value in association with each other genotype in the population, and, assuming random mixing of individuals, the frequencies of interactions correspond to the frequencies of the interacting genotypes. The biological motivation for modeling fitness as a function of intergenotypic interactions is perhaps most obvious in plant systems, where the genotypes of neighboring plants can have a variety of immediate impacts on fitness (Antonovics and Ellstrand 1984). Its general formulation allows the PIM to include all forms of FDS (positive, negative, balancing, disruptive) as well as constant selection as a special case. One could parameterize the model by constructing structured fitness sets corresponding to commonly observed forms of FDS in nature or by presuming the frequency dependence to be due to some specific biological mechanisms (say, predation) but we are interested in the more general question of the maintenance of polymorphism under all possible forms of FDS.
Most existing models of genotype-level FDS have focused on a single diallelic locus (Asmussen and Basnayake 1990; Altenberg 1991; Gavrilets and Hastings 1995; Yi et al. 1999; Asmussen et al. 2004) and/or examine only very special cases of frequency dependence (Roff 1998; Burger and Gimelfarb 2004; Burger 2005; Schneider 2006). Asmussen and Basnayake (1990) made a full analysis of the potential for maintaining genetic variation in the diallelic PIM and our numerical results for the multiallelic PIM with two alleles agree with those findings. Investigations of the potential for chaos and cycling in the diallelic PIM have also been undertaken (Altenberg 1991; Gavrilets and Hastings 1995). Cycling behavior was observed in our model, and discussion of this phenomenon will be undertaken elsewhere.
By extending the PIM to the multiallelic case we want to clarify the potential for maintaining many alleles at a locus under FDS. Our analysis is motivated by three central questions: (1) How effective is FDS in general at maintaining genetic variation as compared to other models?, (2) What kinds of FDS are best at maintaining genetic variation?, and (3) What kinds of allele-frequency distributions does FDS produce?
THE MODEL
We use an approach similar to that of Lewontin et al. (1978) to measure the proportion of PIM parameter space, for a given number of alleles (n), that has potential to maintain all alleles. We measure the potential of a large number of randomly generated fitness sets to maintain genetic variation as a means to assess the overall potential for genetic variation under the model. Note that the measured potential for variation does not correspond to any sort of “probability” of maintaining variation. The random generation of fitnesses and initial allele frequencies simply allows us to measure the potential for variation in all regions of the parameter space. We make no assumptions about the distributions of these fitness values and genotype frequencies in nature. The measurement of overall potential, together with assumptions about the nature of fitness determination and mutational generation of allele frequencies, can give us a clearer idea of the role of FDS in maintaining polymorphisms in nature. This study is concerned with the measurement of potential under the PIM; further investigations of fitness determination and mutation in the PIM are currently underway.
Until recently, such an investigation of the potential for permanent genetic variation in PIM with multiple alleles would have been impractical due to limitations with available computational power. With the exception of special cases (Altenberg 1991; Gavrilets and Hastings 1995; Asmussen et al. 2004), the systems of recursion equations governing fitness are analytically intractable, and so numerical simulations are required to fully elucidate the behavior of the model.
The general formulation of the PIM (Cockerham et al. 1972) concerns a single diploid locus under viability selection with n alleles, in an infinite, isolated population with random mating, discrete generations, and no mutation. The central assumption of the PIM is that each genotype AiAj has distinct fitnesses in its interactions with the other genotypes AkAl in the population. These values are referred to individually as interaction fitnesses (wij,kl) and collectively as fitness sets. Note that AiAj is assumed to be equivalent to AjAi, and thus 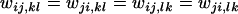 . Assuming that individuals in the population mix at random, intergenotypic interactions will occur in proportion to the genotype frequencies.
. Assuming that individuals in the population mix at random, intergenotypic interactions will occur in proportion to the genotype frequencies.
Allele frequencies are governed by the transformation 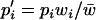 , where
, where  denotes the value of p in the following generation,
denotes the value of p in the following generation, 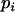 is the frequency of allele
is the frequency of allele 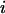 ,
, 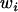 is the marginal fitness of allele
is the marginal fitness of allele 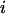 , and
, and 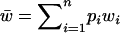 is the mean fitness of the population.
is the mean fitness of the population.
The total fitness of each genotype (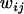 ) is a linear function of its relative fitnesses in interactions with the other genotypes in the population, weighted by the frequencies of those genotypes:
) is a linear function of its relative fitnesses in interactions with the other genotypes in the population, weighted by the frequencies of those genotypes:
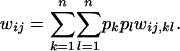 |
The marginal fitness of allele i is a sum of fitnesses for all genotypes involving i, weighted by their frequencies:
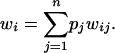 |
The properties of the model are invariant to the scaling of 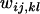 -values because their relative rather than absolute values determine the dynamics and equilibrium outcome of a fitness set. We can assume, therefore, without lack of generality that all
-values because their relative rather than absolute values determine the dynamics and equilibrium outcome of a fitness set. We can assume, therefore, without lack of generality that all 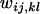 have values between 0 and 1. Due to the complexity of the recursions, numerical simulations are required to determine the equilibrium state for any given initial conditions of the system, except in special cases (Asmussen et al. 2004).
have values between 0 and 1. Due to the complexity of the recursions, numerical simulations are required to determine the equilibrium state for any given initial conditions of the system, except in special cases (Asmussen et al. 2004).
Numerical simulations were undertaken for systems of two, three, four, and five alleles. All programs were written and run in both C++ and Matlab to confirm results. For each n, we generated 100,000 random fitness sets, where each  was a uniform random number between 0 and 1, using the lagged-Fibonacci pseudo-random-number generator of Marsaglia et al. (1990). Assuming a uniform distribution of
was a uniform random number between 0 and 1, using the lagged-Fibonacci pseudo-random-number generator of Marsaglia et al. (1990). Assuming a uniform distribution of 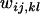 's allows us to visualize the total fitness space as an n(n + 1)/2-dimensional hypercube of unit dimensions. The random generation of fitnesses does not imply any assumption that fitness values in natural systems will be uniformly distributed; it is merely a method for measuring the proportion of the total parameter space that can maintain full polymorphism.
's allows us to visualize the total fitness space as an n(n + 1)/2-dimensional hypercube of unit dimensions. The random generation of fitnesses does not imply any assumption that fitness values in natural systems will be uniformly distributed; it is merely a method for measuring the proportion of the total parameter space that can maintain full polymorphism.
In the PIM, the equilibrium state of the system may differ depending on the initial allele-frequency vector used (Cockerham et al. 1972; Asmussen and Basnayake 1990), for example, if a fixation state and a polymorphic equilibrium are simultaneously stable. For each fitness set allele frequencies were iterated to equilibrium from at least 10n random initial allele-frequency vectors generated using a broken-stick method (as described in Marks and Spencer 1991). The population was considered to be at allele-frequency equilibrium if the maximum change in allele frequency (Δpi) fell below 10−9. The proportion of allele-frequency vectors that maintained full polymorphism for any given fitness set is henceforth referred to as the potential for genetic variation (after Asmussen and Basnayake 1990). Alleles whose frequencies fell below 10−5 were considered extinct. Iteration of allele frequencies was stopped when an extinction (pi < 10−5 for some i) occurred or the population reached a fully polymorphic equilibrium (pi > 10−5 for all i). If neither condition had occurred after 104 generations, the run was stopped and that fitness set stored for further investigation. If at least one equilibrium was reached, the final allele frequencies were recorded and counters updated for the number of, and potential for, fully polymorphic equilibria given that fitness set.
Fitness sets that maintained full polymorphism from at least one initial allele-frequency vector are referred to as “successful.” A fitness set is thus successful if and only if its potential is greater than zero. At the end of each successful run, equilibrium allele frequencies, mean fitness, and the successful fitness set were stored. The simulations kept statistics on the proportion of successful sets and, for each successful set, the potential for full polymorphism. Statistics were also kept of the overall potential for full polymorphism across all fitness sets.
To facilitate comparisons across different values of n, we partitioned the interaction-fitness values within each fitness set into nine different fitness classes. Class divisions are based on the heterozygosity of, and amount of allele sharing between, interacting genotypes. The class of homozygote-by-like-homozygote interactions, for example, is characterized by Cii,ii, and the class of homozygote-by-unlike-homozygote interactions by Cii,jj. For each fitness set, the value of each fitness class is the mean of all the interaction fitnesses belonging to that class. Since the dynamics of the model depend not only on the fitness set but on allele frequencies as well, we also measured fitness classes after weighting each fitness set by its allele frequencies at equilibrium. It is worth noting that not all classes exist for n < 4, as a system with two or three alleles cannot have heterozygote-by-unlike-heterozygote (Cij,kl) interactions, etc.
To provide a basis for comparisons, for all n we ran analogous simulations using constant genotypic fitness sets (after Lewontin et al. 1978). Since any fully polymorphic equilibrium is globally stable for this (Lewontin et al. 1978) form of selection, the proportion of fitness sets that maintain variation and the overall potential for variation are equivalent.
RESULTS
Potential for maintaining genetic variation:
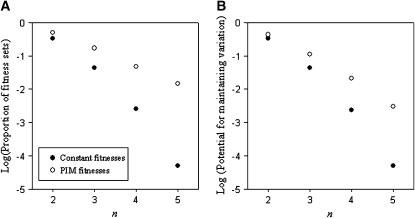
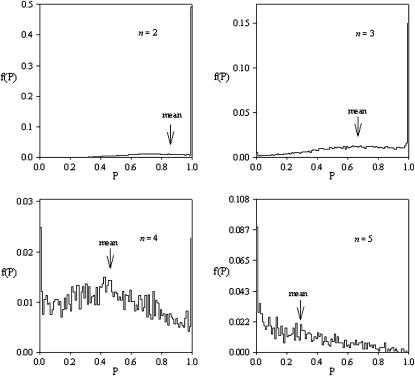
We measured the potential for maintaining genetic variation under the PIM for a given value of n using two different, nested, methods. First we recorded the proportion of the total number of random fitness sets that maintained all alleles from at least one starting allele-frequency vector (i.e., the successful fitness sets). For each fitness set, we also recorded the proportion of allele-frequency starting vectors that maintained all alleles (potential for variation) and then averaged these values across fitness sets to give the overall potential for maintaining variation. Both measures are illustrated in Figure 1, while the distributions of the potential for variation for each successful fitness set can be found in Figure 2.
Figure 1.—
Proportion of PIM parameter space that maintains variation. (A) Proportion of successful fitness sets (i.e., those with nonzero potential for full polymorphism) for n = 2,…, 5. (B) Overall potential for full polymorphism for systems with n = 2,…, 5 alleles. Log-transformed data are shown.
Figure 2.—
Distributions of P, the potential for maintaining genetic variation, for successful fitness sets with n = 2,…, 5 alleles.
As we know from Lewontin et al. (1978), and reconfirmed by our own simulations (Figure 1), the proportion of fitness sets that maintain genetic variation in constant selection models drops off drastically as n increases and is vanishingly small for n > 5. In the PIM simulations, the proportion of fitness sets maintaining genetic variation decreases more slowly as n increases. It would seem then that there are weaker constraints on what kind of PIM fitness set can maintain variation for any given n. Due to the sensitivity of equilibrium conditions to initial allele frequencies, the potential for maintaining variation in the PIM drops off more rapidly than does the proportion of successful fitness sets.
Fitness sets that led to more than one distinct polymorphic equilibrium were relatively rare (1198 cases for n = 2, 305 when n = 3, 33 when n = 4, and 4 when n = 5), and almost all of those consisted of cases with two equilibria. There were two fitness sets, both involving three alleles, that led to three distinct polymorphic equilibria. While the vast majority of successful fitness sets maintained their alleles in an equilibrium state, a small subset of fitness sets produced permanent allele-frequency cycling (0.3367% of cases when n = 3, 0.370% when n = 4, 0.197% when n = 5). The fitness sets that led to cycling are included in the measures of total potential for variation (i.e., Figure 1), but they are not included in the other analyses in this article.
As the number of alleles in the system increased the mean within-fitness-set potential for variation dropped off (Figure 2). More exactly, as the number of alleles in the system increased the distribution of the proportion of initial allele-frequency vectors that led to a fully polymorphic equilibrium changed from being strongly skewed toward 1.0 for n = 2 to being strongly skewed toward 0.0 for n = 5. This result makes sense if one considers that, as the number of alleles increases, the number of interaction fitnesses increases drastically and so too does the number of possible evolutionary outcomes, particularly the number of fixation equilibria. Consequently, as numbers of alleles increase, we expect a randomly generated allele frequency vector to be less likely to fall within the domain of attraction of a fully polymorphic equilibrium. The overall mean proportion of points converging to a fully polymorphic equilibrium decreases linearly with the number of alleles while the total allele-frequency space increases exponentially. This apparent limitation on systems with large numbers of alleles makes it nearly impossible to find successful fitness sets for n > 5 by randomly searching fitness space.
One might reasonably expect the proportion of vectors that converge to a fully polymorphic equilibrium to be related to the values of the interaction fitnesses. Fitness sets with negative FDS might have larger domains of attraction for their equilibria, for example. In the context of the PIM, we consider negative FDS to mean that genotypes have their highest fitness when they are rare, which intuitively should promote polymorphism by making extinction of alleles difficult. This kind of negative FDS is most likely in fitness sets where Cii,ii- and Cij,ij-values are low relative to the other Cij,kl's, where low-fitness self-interactions drive down the fitness of genotypes at high frequencies.
We examined correlations between within-fitness set potential for variation (P) and the various C class values using Spearman's nonparametric ρ (rs). C–P correlations are summarized in Table 1. Each of the nine fitness classes was significantly correlated with P for at least one n. Due to large sample sizes most fitness classes had significant correlations with P, but all were weak (rs < 0.5). The only relationship consistent across all n is a negative correlation between P and Cii,ii. For all n > 2, P is also negatively correlated with Cii,ij, which also implies negative FDS since Cii,ij-values will also drag down the fitness of allele i when it is common. These correlative analyses suggest that fitness sets with some degree of negative FDS (due to low Cii,ii- and/or Cii,ij-values) do tend to have larger domains of attraction around their polymorphic equilibria.
TABLE 1.
Correlations (Spearman's ρ) between within-set potential (P) and interaction fitness-class means (Cij,kl) for different numbers of alleles (n)
|
rs correlations between C class values and P, the proportion of initial vectors that converged to a polymorphic equilibrium for that fitness set
|
||||||||
|---|---|---|---|---|---|---|---|---|
|
n = 2
|
n = 3
|
n = 4
|
n = 5
|
|||||
| Fitness class | rs | P | rs | P | rs | P | rs | P |
| Cii,ii | −0.4951 | 0.0000 | −0.4282 | 0.0000 | −0.2820 | 0.0000 | −0.2022 | 0.0000 |
| Cii,jj | 0.0447 | 0.0000 | 0.0037 | 0.6338 | −0.0273 | 0.0746 | −0.0325 | 0.3237 |
| Cii,ij | −0.0825 | 0.0000 | −0.2644 | 0.0000 | −0.2737 | 0.0000 | −0.2013 | 0.0000 |
| Cii,jk | NA | NA | −0.0101 | 0.1954 | −0.0420 | 0.0060 | −0.0912 | 0.0056 |
| Cij,jj | 0.4471 | 0.0000 | 0.1362 | 0.0000 | 0.0083 | 0.5886 | −0.0139 | 0.6729 |
| Cij,kk | NA | NA | 0.1888 | 0.0000 | 0.1634 | 0.0000 | 0.1383 | 0.0000 |
| Cij,jk | NA | NA | 0.2873 | 0.0000 | 0.1896 | 0.0000 | 0.0754 | 0.0220 |
| Cij,kl | NA | NA | NA | NA | 0.1843 | 0.0000 | 0.1948 | 0.0000 |
| Cij,ij | 0.1203 | 0.0000 | −0.0071 | 0.3605 | −0.0916 | 0.0000 | −0.1861 | 0.0000 |
Distribution of polymorphic equilibria:
Centrality of allele-frequency distributions:
For all n, we compared the equilibrium allele-frequency distributions generated by PIM selection, constant selection, and random chance (i.e., no selection), using the variance of the allele frequencies 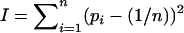 as a measure of their centrality. The “random chance” allele frequencies were generated using the same “broken-stick method” (as described in Marks and Spencer 1991) we used to produce initial allele frequencies scattered uniformly throughout allele-frequency space.
as a measure of their centrality. The “random chance” allele frequencies were generated using the same “broken-stick method” (as described in Marks and Spencer 1991) we used to produce initial allele frequencies scattered uniformly throughout allele-frequency space.
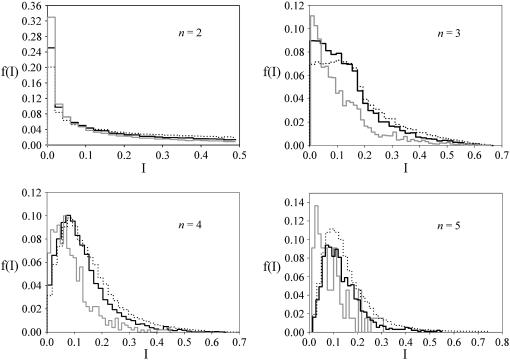
If all alleles are present at equal frequency, I = 0. If one allele is common and the others are vanishingly rare, 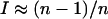 , as n increases. Distributions of I-values from allele-frequency equilibria generated by the different models are summarized in Figure 3. For all n, PIM equilibria tend to be less centralized than those generated by constant selection, but more centralized than randomly generated allele frequencies. At the equilibria maintained by constant selection, heterozygote fitnesses are in general greater than those of homozygotes (Lewontin et al. 1978). Because heterozygotes each affect two allele frequencies equally, the frequencies at equilibrium are likely to be more similar than randomly chosen values. Under FDS, some homozygote fitnesses may also be large, so equilibrium allele frequencies tend to be less even than under constant selection (but still more so than randomly selected values). In nature, truly centered allele-frequency distributions are rare (Keith 1983; Keith et al. 1985), and thus the ability of the PIM to generate skewed distributions is an encouraging argument for its plausibility as a model.
, as n increases. Distributions of I-values from allele-frequency equilibria generated by the different models are summarized in Figure 3. For all n, PIM equilibria tend to be less centralized than those generated by constant selection, but more centralized than randomly generated allele frequencies. At the equilibria maintained by constant selection, heterozygote fitnesses are in general greater than those of homozygotes (Lewontin et al. 1978). Because heterozygotes each affect two allele frequencies equally, the frequencies at equilibrium are likely to be more similar than randomly chosen values. Under FDS, some homozygote fitnesses may also be large, so equilibrium allele frequencies tend to be less even than under constant selection (but still more so than randomly selected values). In nature, truly centered allele-frequency distributions are rare (Keith 1983; Keith et al. 1985), and thus the ability of the PIM to generate skewed distributions is an encouraging argument for its plausibility as a model.
Figure 3.—
The proportion of equilibria falling within successive shells around the centroid of the allele-frequency state space. Solid line, results for PIM fitnesses; shaded line, results for constant genotypic fitnesses; dotted line, proportion of allele-frequency vectors scattered randomly in the space using the broken-stick method.
Homozygosity test of neutrality:
We wished to know if the kinds of FDS produced by PIM fitnesses leave a detectable signature of selection in the equilibrium allele frequencies they produce. The equilibrium allele frequencies generated both by the PIM and by the classic constant selection model, for n = 2,…, 5, were subjected to the Ewens–Watterson homozygosity test of neutrality (Ewens 1979), using the method of Marks and Spencer (1991). For each set of equilibrium allele frequencies, we generated 200 multinomial samples of 50, 100, and 200 genes and noted the number of these samples that were rejected by the Ewens–Watterson (EW) test at the 5% level of significance. For a set of truly neutral allele frequencies, we would expect ∼5% of multinomial samples (10 of 200) to be rejected as nonneutral. Each sample, then, represents a single binomial experiment with a probability of success, π, of 0.05. The null hypothesis (that π = 0.05) will be rejected at the 5% significance level, given 200 trials, if the number of successes is ≥18. The Ewens–Watterson test is a two-tailed test, where smaller values of the homozygosity statistic, F, indicate an excess of genetic diversity usually explained by invoking balancing selection, and larger values of F indicate a dearth of genetic diversity, explained by directional or purifying selection. Therefore Table 2 records the proportion of PIM equilibrium allele frequencies that had ≥18 neutrality rejections from either rejection region. All observed allele-frequency equilibria had significant numbers of rejections from only one rejection region, but never from both.
TABLE 2.
For different numbers of alleles (n), proportions of the equilibria generated by the constant-fitness model and the PIM that produced significant numbers of rejections of neutrality under the homozygosity test of neutrality
| Sample size
|
|||||||
|---|---|---|---|---|---|---|---|
| 50
|
100
|
200
|
|||||
| n | No. of equilibria tested | Low F | High F | Low F | High F | Low F | High F |
| PIM | |||||||
| 2 | 51,565 | 0.11252 | 0.067875 | 0.22358 | 0.033046 | 0.20169 | 0.02717 |
| 3 | 16,836 | 0.27940 | 0.016393 | 0.29455 | 0.003326 | 0.30768 | 0.000713 |
| 4 | 4,304 | 0.3223 | 0.007203 | 0.3771 | 0.001394 | 0.4315 | 0.000232 |
| 5 | 927 | 0.303 | 0.00108 | 0.393 | 0 | 0.561 | 0 |
| Constant fitness | |||||||
| 2 | 33,091 | 0.1570 | 0.04790 | 0.29981 | 0.022151 | 0.2737 | 0.01837 |
| 3 | 4,292 | 0.4089 | 0.0119 | 0.4303 | 0.002796 | 0.4466 | 0.00117 |
| 4 | 232 | 0.496 | 0 | 0.547 | 0 | 0.608 | 0 |
| 5 | 5 | 0.4 | 0 | 0.4 | 0 | 0.4 | 0 |
Although selection was acting in all cases, the homozygosity test detected selection in at most 56% of cases (n = 5, 2k = 200). For all n, when neutrality was rejected, it was most often rejected due to an excess of genetic diversity (low F). This result aligns with the standard expectation that FDS is most likely to maintain variation when it is in the form of negative FDS (or balancing selection). Interestingly, all n also had at least one equilibrium for which the EW test rejected neutrality in the high-F rejection region. High F-values result from highly skewed allele-frequency distributions (i.e., one very common allele and many rare alleles) and imply directional or purifying selection. High-F rejections are far more numerous in the equilibria produced by PIM than in those produced by constant selection, further evidence that FDS is more apt to produce skewed allele frequencies at equilibrium.
Analysis of fitness sets:
Strength of frequency dependence:
For each successful fitness set, and for 50,000 random fitness sets per n, we calculated within- and among-row and -column (ANOVA-style) variances. Within-row/column variances are the means of the variances within rows/columns, and among-row/column variances are the variances of the within-row/column means. In random fitness sets, as n increases, the among-row and among-column variances converge to 0 (as within-row/column means all approach 0.5) while the within-row and within-column variances converge on 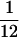 ∼ 0.083 (the variance of the uniform distribution). The mean values of the four fitness set variance measures across all successful and random sets are in Table 3. Of the four metrics, within- and among-row variances differed between successful and random sets while within- and among-column variances were indistinguishable.
∼ 0.083 (the variance of the uniform distribution). The mean values of the four fitness set variance measures across all successful and random sets are in Table 3. Of the four metrics, within- and among-row variances differed between successful and random sets while within- and among-column variances were indistinguishable.
TABLE 3.
Overall means of within- and among-row and within- and among-column variances for successful and random PIM fitness sets
| Successful sets
|
Random sets
|
|||||||
|---|---|---|---|---|---|---|---|---|
| n | 2 | 3 | 4 | 5 | 2 | 3 | 4 | 5 |
| ARV | 0.0238 | 0.0117 | 0.0074 | 0.0049 | 0.0279 | 0.0139 | 0.0083 | 0.0055 |
| WRV | 0.0874 | 0.0854 | 0.0844 | 0.0839 | 0.0832 | 0.0833 | 0.0833 | 0.0833 |
| ACV | 0.0279 | 0.014 | 0.0083 | 0.0053 | 0.0275 | 0.0139 | 0.0083 | 0.0055 |
| WCV | 0.0832 | 0.0832 | 0.0834 | 0.0834 | 0.0835 | 0.0834 | 0.0833 | 0.0834 |
ARV, among-row variance; WRV, within-row variance; ACV, among-column variance; WCV, within-column variance.
In the context of the PIM, within-row variance of fitness sets corresponds to the strength of frequency dependence in the set. If all values in a row are equal, the genotype corresponding to that row will have constant (frequency-independent) fitness. Conversely, if the values within a row are highly variable, the fitness of that row's genotype will depend more strongly on the frequencies of interacting genotypes, being thus under selection that is strongly frequency dependent.
Among-row variances (variance of within-row means) can be interpreted as measuring the strength of frequency-independent fitness. If the among-row variance is zero, all genotypes will have identical frequency-independent fitnesses. If the among-row variance is large, the genotypes will have widely differing fitnesses independent of frequency, being thus under strong frequency-independent selection. We compared both within- and among-row variances of successful sets with their values under random expectation. Within- and among-row variances for successful sets are both significantly different (P < 0.001 for all n) from their expected values of 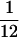 and 0 and from the values generated by simulation of random matrices, respectively. Within-row variances were consistently larger in successful sets, whereas among-row variances were higher in random sets. Successful sets are therefore more strongly frequency dependent and more weakly selected than random chance predicts.
and 0 and from the values generated by simulation of random matrices, respectively. Within-row variances were consistently larger in successful sets, whereas among-row variances were higher in random sets. Successful sets are therefore more strongly frequency dependent and more weakly selected than random chance predicts.
Mean fitness:
For all successful fitness sets we recorded the mean fitness (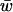 ) of the population at all equilibria. For all n, mean fitnesses were normally distributed. The range of mean fitnesses at equilibrium became more narrow as n increased, and the average
) of the population at all equilibria. For all n, mean fitnesses were normally distributed. The range of mean fitnesses at equilibrium became more narrow as n increased, and the average 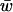 decreased from 0.54 at n = 2 to 0.51 at n = 5 (Table 4).
decreased from 0.54 at n = 2 to 0.51 at n = 5 (Table 4).
TABLE 4.
Summary statistics for mean fitnesses (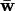 ) at all N equilibria produced by the PIM with different (n) numbers of alleles
) at all N equilibria produced by the PIM with different (n) numbers of alleles
| n | N | Minimum | Maximum | Mean | Variance |
|---|---|---|---|---|---|
| 2 | 51,565 | 0.00 | 1.00 | 0.5448 | 0.021 |
| 3 | 16,836 | 0.10 | 0.93 | 0.5332 | 0.007 |
| 4 | 4,304 | 0.21 | 0.86 | 0.5216 | 0.003 |
| 5 | 927 | 0.38 | 0.73 | 0.5156 | 0.001 |
Fitness classes:
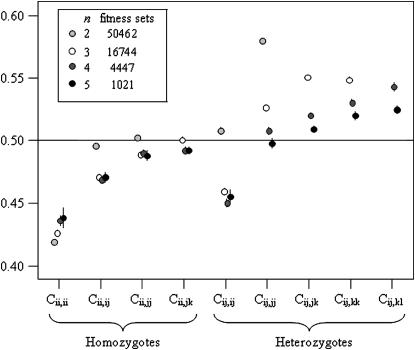
The general pattern of fitness across all successful fitness sets is summarized in Figure 4. All fitness class values are normally distributed and all but two are mutually uncorrelated, across all n, the exception being a weak significant correlation (r = 0.213, P < 0.001) between Cii,ii and Cij,jj when n = 2. Interaction fitnesses were originally drawn from the uniform distribution [0, 1] and each fitness class value was an average of all fitnesses in that class; thus the expectation across all fitness sets and classes is 0.5. There are two obvious patterns in the fitness set data (Figure 4): heterozygote advantage and negative FDS.
Figure 4.—
Fitness class means with 95% confidence intervals for successful fitness sets with n = 2,…, 5.
While they are often cited separately as mechanisms for maintaining large amounts of genetic variation, heterozygote advantage and negative FDS are not mutually exclusive. The results of the PIM simulations suggest that the two mechanisms may often act together to maintain polymorphism. For all n, among successful PIM fitness sets, the overall mean of heterozygote fitnesses, Cij, is on average significantly higher than the mean homozygote fitness within the same set (P < 0.001 for all n).
This pattern is also visible in Figure 4, where the majority of heterozygote fitness classes have means above 0.5, while homozygote fitness class means tend to fall below 0.5. The exception to the rule, Cij,ij, being low when n > 2, suggests that negative FDS is also often a factor in PIM polymorphism. Cii,ii (all n) and Cij,ij (n > 2) are on average significantly <0.5 (P < 0.001 for all n > 2) and, more importantly, tend to have lower values than the other C within sets (Figure 4). Low fitnesses in interactions with like genotypes are an indicator of negative FDS, as these will drag down a genotype's total fitness when it is at high frequencies, due to more frequent self-interactions.
Flavors of frequency dependence:
The summary data from Figure 4 confirm that negative FDS is a good way to maintain polymorphism, but further analyses suggest it is not the only way. Intuitively, one would expect fitness sets with negative FDS or heterozygote advantage to be most likely to maintain variation, while positive FDS and homozygote advantage should eliminate variation and so be absent from our collection of “successful” fitness sets. We searched the successful sets for various different kinds of selection, using schemes and criteria as listed in Table 5. If a genotype has low fitness in interactions with like genotypes, its fitness will tend to decrease as its own frequency increases. A fitness set is thus considered to be under negative FDS if the fitness class values for self–self-interactions (Cii,ii and Cij,ij) are the lowest fitness class values. For strict negative FDS, we extend the above definition to include low fitness in interactions between genotypes that share similar alleles (Cii,ii < Cii,ij < Cii,jj, Cij,jj < Cij,ik < Cij,kk, etc.). We considered a fitness set to contain heterozygote advantage if all heterozygote fitness class values were greater than all homozygote fitness class values. Positive FDS schemes are the mirror images of the negative FDS schemes, and homozygote advantage is the opposite of heterozygote advantage.
TABLE 5.
Frequencies of selection schemes in successful and random fitness sets
| Selection scheme | Definition | Proportion PIM sets (n = 3) | Proportion random sets (n = 3) |
|---|---|---|---|
| Negative FDS | Cii,ii and Cij,ij less than all others | 0.17263 | 0.0803 |
| Positive FDS | Cii,ii and Cij,ij greater than all others | 0.027413 | 0.0803 |
| Strict negative FDS | More shared alleles, less fitness | 0.014160 | 0.0058 |
| Strict positive FDS | More shared alleles, more fitness | 0.0007867 | 0.0058 |
| Heterozygote advantage | Cij,__ greater than all Cii,__ | 0.034493 | 0.0140 |
| Homozygote advantage | Cij,__ less than all Cii,__ | 0.0036309 | 0.0140 |
| Totals | All special cases above | 0.253114 | 0.2002 |
For simplicity, Table 5 lists results only for the n = 3 case. Results were nearly identical for all other numbers of alleles. The only notable difference among n was that for larger n, the proportion of sets meeting the negative FDS criteria increased from 17% for n = 3 to 20% for n = 4 and to almost 25% for n = 5. The fitness schemes we used, while informative, accounted for at most (when n = 5) 32% of the total fitness sets that maintained polymorphism. The majority, then, of successful fitness sets had nonobviously quantifiable patterns of fitness. Of the above selection schemes, negative FDS was by far the most common pattern found in successful sets. Negative FDS is commonly cited as a form of balancing selection and therefore might be expected to produce even allele-frequency distributions at equilibrium. Neither the distributions of I-values for equilibria from sets with negative FDS nor any of the other selective scenarios differ qualitatively from the overall distribution pictured in Figure 3.
Contrary to intuitive expectation, positive FDS occurred in 2.7% of successful fitness sets. The maintenance of variation by positive FDS ceases to be surprising when one recalls the multidimensional nature of the PIM. Our conventional definitions of positive (and negative) FDS assume that the fitness of a given allele depends primarily on its own frequency in the population, which is necessarily so in systems with two alleles. While the assumption of self-FDS may be biologically plausible, there is no mathematical reason for it to be valid in the context of the PIM. Alleles may be under positive FDS with respect to their own frequencies, which should eliminate polymorphism, but this effect can be swamped by the influence of their interactions with other alleles in the system.
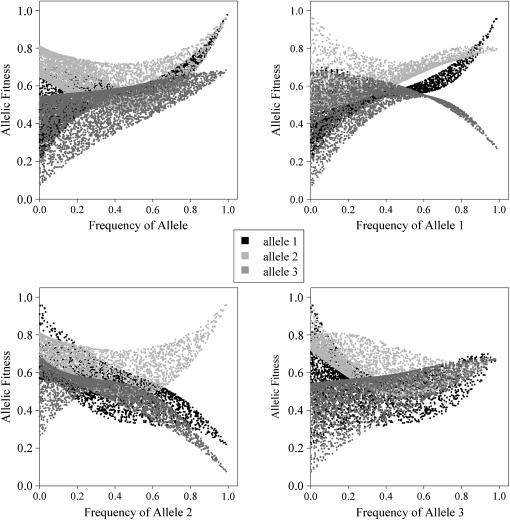
In the following example (Figure 5) data come from a successful PIM fitness set with n =3, where the criteria were met for positive FDS, and a fully polymorphic equilibrium is maintained from ∼10% of starting points. The allele interactions responsible for the maintenance of the fully polymorphic equilibrium are apparent if one examines plots of allele fitnesses against the frequency for each allele. While all alleles are under weak positive FDS with respect to their own frequency (Figure 5, top left plot), there is strong negative FDS on alleles 1 and 3 with respect to allele 2 (Figure 5, bottom left plot), which favors an equilibrium point with allele 2 at very low frequency.
Figure 5.—
Allele fitnesses as functions of allele frequency for an example fitness set with positive FDS, where n = 3.
DISCUSSION
Our investigation of the PIM confirmed that, in general, FDS is better at maintaining genetic variation than constant selection. Furthermore, our findings uphold the longstanding intuitive hypothesis that negative FDS is the best way to maintain genetic variation. More interesting, however, is the result that a wide range of FDS regimes, including but not limited to the “obvious” negative FDS and heterozygote advantage, can maintain genetic variation. The vast majority of successful fitness sets contain patterns of interaction fitnesses that defy classification into standard selective regimes (Table 4). Our definitions of obvious selective regimes, like negative FDS, all assume that an allele's fitness depends primarily on its own frequency. While self-FDS is often biologically plausible, in the purely mathematical context of the PIM there is no particular reason why an allele's fitness should depend more on its own frequency than on the frequencies of other alleles in the system. The tendency to focus on self-FDS is understandable, since—in contrast to more complex scenarios of non-self-FDS like those often produced by the PIM—it is both more amenable to mathematical analysis and easier to identify and test in natural populations. There are indeed very few known examples of this kind of non-self-FDS in nature, with the exception of the rock–paper–scissors dynamic found in some species with alternative male mating strategies (Sinervo and Lively 1996). Whether this sort of nonintuitive FDS is important biologically in multiallelic systems, or merely a mathematical oddity of the PIM, remains to be seen.
To date, the majority of models of natural selection have focused on the diallelic case of polymorphism. It is intuitively tempting to presume that results from such diallelic models will apply in a general way to multiple-allele cases. Unfortunately generalizing from n = 2 to all n is often unrealistic, as often the two-allele case is a special case mathematically. In our analysis of the PIM the results for the n = 2 case were in many ways qualitatively and quantitatively different from those for n = 3, 4, and 5. Most notably, for n > 2, heterozygote advantage is neither necessary nor sufficient for the maintenance of fully polymorphic equilibria under constant selection. To move beyond n = 2, we investigated the PIM parameter space by generating many random matrices of PIM fitnesses, with different numbers of alleles, and numerically assessing their potential to maintain full polymorphism. For all n, a much larger proportion of PIM fitness space had the potential for full polymorphism than in equivalent constant fitness models. The difference in potential becomes more pronounced as the number of alleles increases. For systems with two alleles, PIM selection maintained polymorphism 1.67 times more often than constant selection, whereas with five alleles PIM maintained polymorphism more than 60 times as often. This marked difference suggests there are far fewer constraints on the kinds of PIM fitnesses that can keep polymorphism.
Among PIM fitness sets allowing fully polymorphic equilibria there was wide variation in the proportion of initial allele-frequency vectors that converged to equilibrium. We expected to find some correlation between the contents of a fitness set and its potential to maintain full polymorphism. The probability of convergence to a fully polymorphic equilibrium is significantly but weakly correlated (rs < 0.3) to a slightly different subset of fitness classes for each n (see Table 1). This inconsistency further underlines one of the complications created by partitioning fitness sets into classes: Not all interaction classes can exist for n < 4, and so patterns of fitness class correlations can be hard to discern. This complication is also highlighted in Figure 4, where in several places data for n = 2 and 3 do not align with the strong patterns in the n ≥ 4 cases. In light of these observations, it makes more sense to consider n = 2 and n = 3 as distinct special cases of PIM and all n ≥ 4 as the “general” case.
The location of polymorphisms in allele-frequency space is at least as interesting as their number. Allele-frequency distributions for naturally occurring polymorphisms tend to be skewed, with a few common alleles and many rare alleles (Keith et al. 1985). The tendency for constant-selection models to produce balanced allele-frequency distributions (e.g., Lewontin et al. 1978; Clark and Feldman 1986) has been used to argue that selection cannot maintain many unevenly distributed alleles (e.g., Keith et al. 1985). Equilibria produced by PIM have a tendency to be more skewed than those produced by analogous constant-selection models (Figure 3). The ability of PIM to produce biologically reasonable equilibria is a reassuring testament to its biologically motivated underpinnings as an intraspecific competition model. Some of the most skewed allele-frequency vectors were produced by fitness sets with strong negative FDS, suggesting that balancing selection does not necessarily produce balanced allele frequencies.
The partitioning of fitness sets into fitness classes was intended to allow us to make statements about the kinds of fitness that maintain polymorphism regardless of n. Negative FDS was by far the most common pattern observed in successful fitness sets (Table 5), which is hardly surprising since it is also the most “intuitively obvious” explanation for polymorphisms in nature. Also intuitively, one would expect selection against rare alleles to accelerate the loss of alleles and eliminate polymorphism. Regardless of our intuition, positive FDS is known to occur in a variety of polymorphic natural systems such as groups of Mullerian mimics (Langham 2004), antiapostatic predator choice (Lindstrom et al. 2001), and mate choice (van Gossum et al. 2001) including positive assortative mating (Nevo et al. 2000). The potential for full polymorphism in PIM fitness sets with both weak and strong positive FDS suggests that positive selection and genetic polymorphism are not always mutually exclusive. The formulation of the PIM also allows for allelic fitnesses to be more strongly dependent on the frequencies of other alleles in the system than on their own frequency (Figure 5). Strong non-self-FDS is one possible explanation for the ability of fitness sets with positive self-FDS to maintain variation.
While the fitness classes are a useful tool for finding qualitative patterns in the fitnesses that lead to full polymorphism, they do leave much to be desired. The sheer number of intergenotypic interactions involved in the multiallelic PIM results in most fitness sets having fitness relationships too intricate to be described using such simplistic definitions as “rare advantage” or even heterozygote advantage. This relative weakness of constraint, while supporting the idea of FDS as a strong mechanism for maintaining polymorphism, also makes it impossible to propose any general conditions for polymorphism.
Another obvious shortcoming of this formulation of the PIM is its static equilibrium focus. Natural populations experience drift, mutation, migration, and other forces of dynamic change. Further investigations are underway to incorporate mutation into a more dynamic model of PIM polymorphism.
Acknowledgments
The authors thank Rick Stoffels, Kristan Schneider, and an anonymous reviewer for helpful discussion and comments on the manuscript. This work was supported by the Marsden Fund of the Royal Society of New Zealand (contract U00315) and by the Allan Wilson Centre for Molecular Evolution and Ecology. M.V.T was the recipient of a scholarship from the Division of Sciences of the University of Otago.
References
- Allen, J. A., 1988. Frequency-dependent selection by predators. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 319: 485–503. [DOI] [PubMed] [Google Scholar]
- Allen, J. A., and M. E. Weale, 2005. Anti-apostatic selection by wild birds on quasi-natural morphs of the land snail Cepaea hortensis: a generalised linear mixed models approach. Oikos 108: 335–343. [Google Scholar]
- Allen, J. A., H. E. Raison and M. E. Weale, 1998. The influence of density on frequency-dependent selection by wild birds feeding on artificial prey. Proc. R. Soc. Lond. Ser. B Biol. Sci. 265: 1031–1035. [Google Scholar]
- Altenberg, L., 1991. Chaos from linear frequency-dependent selection. Am. Nat. 138: 51–68. [Google Scholar]
- Anderson, B., and S. D. Johnson, 2006. The effects of floral mimics and models on each others' fitness. Proc. R. Soc. Lond. Ser. B Biol. Sci. 273: 969–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonovics, J., and N. C. Ellstrand, 1984. Experimental studies of the evolutionary significance of sexual reproduction. 1. A test of the frequency-dependent selection hypothesis. Evolution 38: 103–115. [DOI] [PubMed] [Google Scholar]
- Asmussen, M. A., and E. Basnayake, 1990. Frequency-dependent selection - the high-potential for permanent genetic-variation in the diallelic, pairwise interaction-model. Genetics 125: 215–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asmussen, M. A., R. A. Cartwright and H. G. Spencer, 2004. Frequency-dependent selection with dominance: a window onto the behavior of the mean fitness. Genetics 167: 499–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billiard, S., C. Faurie and M. Raymond, 2005. Maintenance of handedness polymorphism in humans: a frequency-dependent selection model. J. Theor. Biol. 235: 85–93. [DOI] [PubMed] [Google Scholar]
- Bond, A. B., and A. C. Kamil, 1998. Apostatic selection by blue jays produces balanced polymorphism in virtual prey. Nature 395: 594–596. [Google Scholar]
- Bond, A. B., and A. C. Kamil, 2002. Visual predators select for crypticity and polymorphism in virtual prey. Nature 415: 609–613. [DOI] [PubMed] [Google Scholar]
- Burger, R., 2005. A multilocus analysis of intraspecific competition and stabilizing selection on a quantitative trait. J. Math. Biol. 50: 355–396. [DOI] [PubMed] [Google Scholar]
- Burger, R., and A. Gimelfarb, 2004. The effects of intraspecific competition and stabilizing selection on a polygenic trait. Genetics 167: 1425–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carius, H. J., T. J. Little and D. Ebert, 2001. Genetic variation in a host-parasite association: potential for coevolution and frequency-dependent selection. Evolution 55: 1136–1145. [DOI] [PubMed] [Google Scholar]
- Cheney, K. L., and I. M. Cote, 2005. Frequency-dependent success of aggressive mimics in a cleaning symbiosis. Proc. R. Soc. Lond. Ser. B Biol. Sci. 272: 2635–2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark, A. G., and M. W. Feldman, 1986. A numerical-simulation of the one-locus, multiple-allele fertility model. Genetics 113: 161–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockerham, C. C., T. Prout, S. S. Young and P. M. Burrows, 1972. Frequency-dependent selection in randomly mating populations. Am. Nat. 106: 493–515. [Google Scholar]
- Dybdahl, M. F., and C. M. Lively, 1998. Host-parasite coevolution: evidence for rare advantage and time-lagged selection in a natural population. Evolution 52: 1057–1066. [DOI] [PubMed] [Google Scholar]
- Ewens, W. J., 1979. Mathematical Population Genetics. Springer, Berlin.
- Gavrilets, S., and A. Hastings, 1995. Intermittency and transient chaos from simple frequency-dependent selection. Proc. R. Soc. Lond. Ser. B Biol. Sci. 261: 233–238. [DOI] [PubMed] [Google Scholar]
- Hughes, K. A., L. Du, F. H. Rodd and D. N. Reznick, 1999. Familiarity leads to female mate preference for novel males in the guppy, Poecilia reticulata. Anim. Behav. 58: 907–916. [DOI] [PubMed] [Google Scholar]
- Keith, T. P., 1983. Frequency-distribution of esterase-5 alleles in 2 populations of Drosophila-pseudoobscura. Genetics 105: 135–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keith, T. P., L. D. Brooks, R. C. Lewontin, J. C. Martinezcruzado and D. L. Rigby, 1985. Nearly identical allelic distributions of xanthine dehydrogenase in 2 populations of Drosophila-pseudoobscura. Mol. Biol. Evol. 2: 206–216. [DOI] [PubMed] [Google Scholar]
- Kojima, K. I., 1971. Is there a constant fitness value for a given genotype - No. Evolution 25: 281–285. [DOI] [PubMed] [Google Scholar]
- Langham, G. M., 2004. Specialized avian predators repeatedly attack novel color morphs of Heliconius butterflies. Evolution 58: 2783–2787. [DOI] [PubMed] [Google Scholar]
- Lewontin, R. C., L. R. Ginzburg and S. D. Tuljapurkar, 1978. Heterosis as an explanation for large amounts of genic polymorphism. Genetics 88: 149–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindstrom, L., R. V. Alatalo, A. Lyytinen and J. Mappes, 2001. Strong antiapostatic selection against novel rare aposematic prey. Proc. Natl. Acad. Sci. USA 98: 9181–9184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks, R. W., and H. G. Spencer, 1991. The maintenance of single-locus polymorphism. 2. The evolution of fitnesses and allele frequencies. Am. Nat. 138: 1354–1371. [Google Scholar]
- Marsaglia, G., A. Zaman and W. W. Tsang, 1990. Toward a universal random number generator. Stat. Probab. Lett. 9: 35–39. [Google Scholar]
- May, R. M., and R. M. Anderson, 1983. Epidemiology and genetics in the coevolution of parasites and hosts. Proc. R. Soc. Lond. Ser. B Biol. Sci. 219: 281–313. [DOI] [PubMed] [Google Scholar]
- Nevo, E., A. Beiles, A. B. Korol, Y. I. Ronin, T. Pavlicek et al., 2000. Extraordinary multilocus genetic organization in mole crickets, Gryllotalpidae. Evolution 54: 586–605. [DOI] [PubMed] [Google Scholar]
- Olendorf, R., F. H. Rodd, D. Punzalan, A. E. Houde, C. Hurt et al., 2006. Frequency-dependent survival in natural guppy populations. Nature 441: 633. [DOI] [PubMed] [Google Scholar]
- Pfennig, D. W., W. R. Harcombe and K. S. Pfennig, 2001. Frequency-dependent Batesian mimicry - predators avoid look-alikes of venomous snakes only when the real thing is around. Nature 410: 323. [DOI] [PubMed] [Google Scholar]
- Piertney, S. B., and M. K. Oliver, 2006. The evolutionary ecology of the major histocompatibility complex. Heredity 96: 7–21. [DOI] [PubMed] [Google Scholar]
- Punzalan, D., F. H. Rodd and K. A. Hughes, 2005. Perceptual processes and the maintenance of polymorphism through frequency-dependent predation. Evol. Ecol. 19: 303–320. [Google Scholar]
- Raymond, M., D. Pontier, A. B. Dufour and A. P. Moller, 1996. Frequency-dependent maintenance of left handedness in humans. Proc. R. Soc. Lond. Ser. B Biol. Sci. 263: 1627–1633. [DOI] [PubMed] [Google Scholar]
- Roff, D. A., 1998. The maintenance of phenotypic and genetic variation in threshold traits by frequency-dependent selection. J. Evol. Biol. 11: 513–529. [Google Scholar]
- Schneider, K. A., 2006. A multilocus-multiallele analysis of frequency-dependent selection induced by intraspecific competition. J. Math. Biol. 52: 483–523. [DOI] [PubMed] [Google Scholar]
- Sinervo, B., and C. M. Lively, 1996. The rock-paper-scissors game and the evolution of alternative male strategies. Nature 380: 240–243. [Google Scholar]
- Trachtenberg, E., B. Korber, C. Sollars, T. B. Kepler, P. T. Hraber et al., 2003. Advantage of rare HLA supertype in HIV disease progression. Nat. Med. 9: 928–935. [DOI] [PubMed] [Google Scholar]
- van Gossum, H., R. Stoks and L. De Bruyn, 2001. Reversible frequency-dependent switches in male mate choice. Proc. R. Soc. Lond. Ser. B Biol. Sci. 268: 83–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, D. S., A. B. Clark, K. Coleman and T. Dearstyne, 1994. Shyness and boldness in humans and other animals. Trends Ecol. Evol. 9: 442–446. [DOI] [PubMed] [Google Scholar]
- Yi, T., R. Cressman and B. Brooks, 1999. Nonlinear frequency-dependent selection at a single locus with two alleles and two phenotypes. J. Math. Biol. 39: 283–308. [DOI] [PubMed] [Google Scholar]
- Yuste, E., A. Moya and C. Lopez-Galindez, 2002. Frequency-dependent selection in human immunodeficiency virus type 1. J. Gen. Virol. 83: 103–106. [DOI] [PubMed] [Google Scholar]